Abstract
Background
Observational studies linking vitamin D deficiency with increased prostate cancer mortality and the pleiotropic anticancer effects of vitamin D in malignant prostate cell lines have initiated trials examining potential therapeutic benefits of vitamin D metabolites. There have been some successes but efforts have been hindered by risk of inducing hypercalcemia. A limited number of studies have investigated associations between variants in vitamin D pathway genes with aggressive forms of prostate cancer. Increased understanding of relevant germline genetic variation with disease outcome could aid in development of vitamin D-based therapies.
Methods
We undertook a comprehensive analysis of 48 tagging single nucleotide polymorphisms (tagSNPs) in genes encoding for vitamin D receptor (VDR), vitamin D activating enzyme 1-α-hydroxylase (CYP27B1), and deactivating enzyme 24-hydroxylase (CYP24A1) in a cohort of 1,294 Caucasian cases with an average of 8 years of follow-up. Disease recurrence/ progression and prostate cancer-specific mortality risks were estimated using adjusted Cox proportional hazards regression.
Results
There were 139 cases with recurrence/progression events and 57 cases who died of prostate cancer. Significantly altered risks of recurrence/progression were observed in relation to genotype for two VDR tagSNPs (rs6823 and rs2071358) and two CYP24A1 tagSNPs (rs927650 and rs2762939). Three VDR tagSNPs (rs3782905, rs7299460 and rs11168314), one CYP27B1 tagSNP (rs3782130) and five CYP24A1 tagSNPs (rs3787557, rs4809960, rs2296241, rs2585428, and rs6022999) significantly altered risks of prostate cancer death.
Conclusions
Genetic variations in vitamin D pathway genes were found to alter both risk of recurrence/progression and prostate cancer-specific mortality.
Keywords: Vitamin D Receptor, 1-alpha-Hydroxylase, 24-Hydroxylase, Prostatic Neoplasms, Outcomes
Introduction
Vitamin D has been shown to reduce cellular proliferation, increase apoptosis and inhibit invasion, migration, metastasis and angiogenesis [1–3]. The potential role of vitamin D in prostate tumor growth and aggressiveness is supported by ecological and case-control studies demonstrating an inverse relationship between prostate cancer (PCa) mortality and advanced disease with ultraviolet (UV) exposure, which is the primary source of vitamin D [4–8]. There is also an increased PCa mortality in African American and older men where skin has a reduced capacity for absorbing UV [9,10]. In early clinical trials vitamin D analogs such as calcitriol have shown some success as therapeutic agents for patients with androgen-independent PCa lesions through reduction in prostate-specific antigen (PSA) levels and increasing patient survival, however the problem of circumventing hypercalcemia has not been overcome [11,12].
Whether ingested or produced by the skin via UV exposure, vitamin D is first hydroxylated in the liver to form the metabolically inactive form of the prohormone, 25-hydroxyvitamin D [25(OH)D]. The rate limiting step is secondary conversion into the active form of the hormone, 1α,25-dihydroxy-vitamin D [1α,25(OH)2D], by 1-α-hydroxylase (1α-OHase), which is encoded by the gene CYP27B1. There is autocrine or paracrine synthesis of 1α,25(OH)2D by 1α-OHase within normal prostate cells, however expression of 1α-OHase is greatly reduced early in the neoplastic process of PCa cells [13,14]. Binding to the nuclear vitamin D receptor (VDR), which is widely expressed in prostate cells, mediates all functions of 1α,25(OH)2D. The enzyme 24-hydroxylase, encoded by the gene CYP24A1, metabolizes 1α,25(OH)2D into its excretion product calcitroic acid. Some prostate cancer cell lines have shown increased expression of the catabolic CYP24A1 in vitro [15].
Few genetic association studies have been conducted that specifically examined prostate cancer progression or mortality in association with vitamin D metabolism pathway genes. Some studies have attempted to address the potential association of vitamin D pathway genetic polymorphisms with prostate tumor growth and aggressiveness by examining prostate cancer risk within clinically defined subsets. For the more commonly studied polymorphisms, poly(A) microsatellite, FokI (rs10735810), BsmI (rs1544410), and TaqI (rs731236), no clear consensus has emerged from studies reporting genotype associations for men with advanced stage disease or higher Gleason scores [16–19]. Only one study has reported associations between CYP27B1 genotypes and prostate cancer risk, and no association with disease aggressiveness was noted [20]. To better answer the question of whether germline variants within vitamin D metabolism pathway genes predict disease outcome, a study design that follows prostate cancer cases over time is a more accurate predictor of disease recurrence and survival. Three studies have looked at associations between VDR genotypes in cases who underwent radical prostatectomy and went on to exhibit recurrence, as defined by PSA failure [21–23]. For the FokI polymorphism, one study observed an increased risk of recurrence for homozygote carriers of the wildtype (G) allele, or “FF” individuals, while the other study did not corroborate the finding of an increased risk for recurrence, but did report an association with more aggressive tumors [21,22]. The third study, which looked at recurrence risk by BsmI and TaqI genotypes in Caucasians and African Americans, did not observe any association with disease outcome overall; however, they did report a significant decrease in recurrence risk for carriers of the BsmI variant (A) allele, or “B” allele, among Caucasian men with locally advanced disease [23].
To more carefully dissect these issues, we conducted a genetic association study of prostate cancer recurrence/progression and prostate cancer-specific mortality using a population-based cohort of men with long-term follow-up. Our study focused on three vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, and used tagSNPs to better capture genetic variability within each gene. The long-term follow-up was used to access association with both recurrence/progression and mortality.
Materials and Methods
Study Population
Study subjects were enrolled in one of two population-based prostate cancer case-control studies that have been described previously [24,25]. Cases were newly diagnosed with histologically confirmed prostate cancer in two study periods, either January 1, 1993 to December 31, 1996 (Study I, age range 40–64 years) or January 1, 2002 to December 31, 2005 (Study II, age range 35–74 years). Prostate cancer cases were identified from the metropolitan Seattle-Puget Sound population-based tumor registry that part of the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program. Of the 1,754 eligible, interviewed cases we obtained peripheral blood leukocyte samples for genotyping from 1,458 men. From this group we excluded 16 cases because they did not have sufficient DNA for this specific study and further limited this dataset to Caucasians for a total of 1,294 cases (Study I: n=585; Study II: n=709).
The recurrence/progression analysis was limited to a subset of 458 cases in Study I; subjects were excluded if they were alive but did not fill out a follow-up questionnaire or give access to medical record review (n=109) or had an initial diagnosis of metastatic disease (n=18). In January 2004 a self-administered follow-up questionnaire collecting information on use of secondary therapies, follow-up PSA results, and evidence for prostate cancer recurrence/ progression was sent to Study I cases. The overall response level was 82% and showed no association between clinical parameters and non-response [26]. Data from this survey was used to determine recurrence/progression status for 426 cases. An additional 32 cases who were diagnosed with local/regional disease and who were deceased at the time of the follow-up survey had recurrence/progression data available; 21 of these cases had next-of-kin provided consent for medical record review, which was used to determine recurrence/progression status, and 11 of these cases died of metastatic prostate cancer and were coded as having recurred. The survival analysis included all 1,294 cases from both studies. The SEER registry provided information on tumor characteristics, primary therapy, vital status, and underlying cause of death. Death certificates were obtained to confirm fatal prostate cancer. The agreement between the SEER registry and death certificate has been reported to be excellent [27]. The most recent registry linkage update for mortality was June 15, 2009. This study was approved by Fred Hutchinson Cancer Research Center’s Institutional Review Board, all subjects had informed consent, and genotyping was approved by the Internal Review Board of the National Human Genome Research Institute.
SNP Selection and Genotyping
SNPs that captured the genetic variability in the VDR [28] and CY27B1 [29] genes were selected using resequencing data, while SNP selection for CYP24A1 [30] used publicly available from HapMap consortium data (www.hapmap.org). Using parameters of r2 ≥ 0.8 and minor allele frequency ≥ 5% [31], a total of 25 tagSNPs for VDR (chromosome 12q13, length 63.4 kb, 9 exons), 3 tagSNPs for CYP27B1 (chromosome 12q13, length 4.8 kb, 9 exons), and 20 tagSNPs for CYP24A1 (chromosome 20q13; length 20.5 kb, 12 exons) were chosen.
The Applied Biosystems (ABI) SNPlex™ Genotyping System was used for genotyping, and proprietary GeneMapper® software (www.appliedbiosystems.com) was used for calling alleles. Discrimination of specific SNP alleles was determined by an ABI 3730xl DNA Analyzer and is based on presence of a unique sequence assigned to the original allele-specific oligonucleotide. The SNPlex assay could not be designed for 7 tagSNPs and 3 tagSNPs failed genotyping after the design stage; accordingly, we present results for 22 tagSNPs for VDR, 2 tagSNPs for CYP27B1, and 14 tagSNPs for CYP24A1. Quality control included genotyping of 76 blind duplicate samples, which revealed 99% agreement on genotyping calls across all SNPs assayed. In addition, each batch of DNA aliquots genotyped incorporated similar numbers of case and control samples, and laboratory personnel were blinded to case-control status of samples. The call rate was ≥97% for all but two SNPs (VDR rs2238139, 96%; CYP24A1 rs6127118, 95%). Further details of genotyping methods are described elsewhere [32]. All SNPs included in this study were consistent (p > 0.05) with Hardy-Weinberg equilibrium (HWE) with the exception of CYP24A1 rs13038432 (p < 0.001).
Statistical Analysis
For each SNP we classified homozygote carriers of the common allele as the referent group and carriers of the less common variant allele as the exposure group. We used both dominant and co-dominant models, except when no individuals were homozygous for the variant genotype. Trend tests, which used a single indicator variable coded as the number of variant alleles for each SNP, were used to assess gene dosage. To examine associations between individual SNPs with prostate cancer recurrence/progression and prostate cancer-specific mortality we used Cox proportional hazards (PH) regression models adjusting for age, Gleason score [2–6, 7(3+4), or 7(4+3)-10], stage at diagnosis (local, regional, or distant), diagnostic PSA level (0–9.9 or ≥10.0 ng/mL), and primary treatment [radical prostatectomy, radiation with or without androgen deprivation therapy (ADT), ADT only, other treatment, or active surveillance]. Analysis was also done limiting dataset to men treated with radical prostatectomy.
Time to prostate cancer recurrence/progression was defined as time from diagnosis to the first reported evidence of recurrence as described previously [26]. For those with follow-up but without an event, the censoring date was the date that the follow-up questionnaire was returned. For the 11 cases who were diagnosed with localized/regional disease but died of prostate cancer prior to follow-up survey administration, time to recurrence was imputed [26]. Time to prostate cancer-specific mortality was defined as the time from diagnosis to death. Living cases were censored on date of most recent linkage with the cancer registry. Cases that died from other or unknown causes were censored at the time of death.
Gene-environment interactions were assessed for first-degree family history of prostate cancer, body mass index (BMI), and vitamin D supplement use/dietary intake for both disease recurrence/progression and survival in Cox PH regression models with and without interaction terms. Models were compared using the likelihood ratio test. Vitamin D intake was calculated using food frequency and supplement use questionnaires administered separately from the original interview for 1,305 of the 1,442 cases. For the interaction analyses, PH models were limited to risk estimates assuming a dominant genetic model, combining heterozygotes and homozygous variants as the exposed group.
To account for the effect of multiple testing, sets of outcomes (PCa mortality or recurrence/progression) and clinical covariates were permuted in order to approximate distribution of covariate adjusted p-values under the null hypothesis. For each permutation, dominant, codominant and trend models were fit for all SNPs and the minimum p-values kept for each SNP. P-values were ordered to approximate null distribution of the order statistics, i.e., minimum p-value, second smallest p-value, etc. The original p-values were also ordered and permutation p-values were calculated by comparing the ordered p-values to the null distribution for the appropriate order statistic. Permutation p-values can be interpreted as the probability of observing a p-value less than or equal to what was observed for the given order statistic under the null hypothesis of no association with disease outcomes for any of the 38 SNPs. A SNP was considered to be significantly associated with prostate cancer mortality or recurrence/progression if the nominal p-value and the permuted p-value were both less than 0.05.
A stepwise AIC (Akaike’s Information Criterion) regression procedure was used to develop predictive models for prostate cancer mortality and recurrence/progression. The procedure was restricted to include all clinical predictors in the final model (age, stage, Gleason score, diagnostic PSA and primary treatment). Empirical ROC (receiver operating characteristic) curves for 5- and 10-year prostate cancer mortality and recurrence/progression were used to compare the prognostic accuracy of models containing SNPs and clinical covariates versus baseline models with only clinical covariates. Improvement of prediction accuracy due to addition of optimal SNPs selected by the AIC stepwise regression approach was summarized by comparing ROC (.2), sensitivity at a specificity of 80%, and area under the ROC curve (AUC). Bootstrap confidence intervals were presented for differences in ROC (.2) and AUC. All analyses were done using R version 2.8.1 and the STATA statistical package (version 10.1, STATA Corp., College Station, TX).
Results
There were 139 events of recurrence/progression, with an average 8.2 years of follow-up (range 0.1–12.8 years) after diagnosis. For cases having recurrence/progression events, a greater proportion were diagnosed at younger ages, with regional stage of disease (cases with distant stage were excluded), higher Gleason scores, and diagnostic PSA values greater than 10 ng/mL (Table 1). There were 57 cases who died of prostate cancer in the average 8.5 years of follow-up (range 0.8–15.9 years). For cases who died of prostate cancer, a greater proportion were at diagnosed at younger ages, with regional or distant stages of disease, higher Gleason scores, diagnostic PSA values greater than 10 ng/mL, and a BMI greater than or equal to 30 kg/m2 (Table 1).
Table 1.
Selected Demographic and Clinical Characteristics of Prostate Cancer Patients by Disease Recurrence/Progression and Mortality.
| PC Recurrence/Progression |
Vital Status2 |
|||
|---|---|---|---|---|
| No | Yes1 | Alive | Prostate Cancer- Specific Deaths |
|
| (n=319) | (n=139) | (n=1136) | (n=57) | |
| Time to event (years), mean (SD) | 9.4 (1.3) | 5.4 (3.3) | 8.8 (4.4) | 5.8 (3.6) |
| Age group | ||||
| 35–49 | - | - | 88 (7.7) | 9 (15.8) |
| 50–54 | 9 (2.8) | 13 (9.4) | 174 (15.3) | 7 (12.3) |
| 55–59 | 74 (23.2) | 21 (15.1) | 278 (24.5) | 16 (28.1) |
| 60–64 | 109 (34.2) | 50 (36) | 330 (29) | 18 (31.6) |
| 65–69 | 127 (39.8) | 55 (39.6) | 139 (12.2) | 3 (5.3) |
| 70–74 | - | - | 127 (11.2) | 4 (7) |
| Family History of PC | ||||
| No | 247 (77.4) | 118 (84.9) | 878 (77.3) | 49 (86) |
| Yes | 72 (22.6) | 21 (15.1) | 258 (22.7) | 8 (14) |
| Total vitamin D (ug/d) 3 | ||||
| 0–3.6 | 90 (28.2) | 39 (28.1) | 241 (21.2) | 8 (14) |
| 3.6–5.6 | 83 (26) | 29 (20.9) | 258 (22.7) | 13 (22.8) |
| 5.6–8.3 | 70 (21.9) | 35 (25.2) | 276 (24.3) | 15 (26.3) |
| >8.3 | 50 (15.7) | 27 (19.4) | 277 (24.4) | 15 (26.3) |
| Missing | 86 (8.2) | 9 (6.5) | 84 (7.4) | 6 (10.5) |
| Body Mass Index (kg/m2) | ||||
| <25.0 | 114 (35.7) | 52 (37.4) | 376 (33.1) | 23 (40.4) |
| 25.0–29.9 | 156 (48.9) | 65 (46.8) | 559 (49.2) | 21 (36.8) |
| ≥30.0 | 49 (15.4) | 22 (15.8) | 201 (17.7) | 13 (22.8) |
| Stage of PC at diagnosis | ||||
| Local | 254 (79.6) | 86 (61.9) | 922 (81.2) | 14 (24.6) |
| Regional | 65 (20.4) | 53 (38.1) | 205 (18) | 22 (38.6) |
| Distant | - | - | 9 (0.8) | 21 (36.8) |
| Gleason score at diagnosis | ||||
| 2–6 | 222 (69.6) | 66 (47.5) | 672 (59.2) | 9 (15.8) |
| 7 (3+4) | 74 (23.2) | 39 (28.1) | 317 (27.9) | 11 (19.3) |
| 7( 4+3), 8–10 | 23 (7.2) | 34 (24.5) | 145 (12.8) | 35 (61.4) |
| Unknown | - | - | 2 (0.2) | 2 (3.5) |
| Diagnostic PSA | ||||
| 0–9.9 ng/mL | 227 (71.2) | 78 (56.1) | 818 (72) | 12 (21.1) |
| 10+ ng/mL | 64 (20.1) | 50 (36) | 233 (20.5) | 41 (71.9) |
| Unknown | 28 (8.8) | 11 (7.9) | 85 (7.5) | 4 (7) |
| Composite Aggressiveness Score 4 | ||||
| Low | 233 (73) | 62 (44.6) | 798 (70.2) | 6 (10.5) |
| High | 86 (27) | 77 (55.4) | 338 (29.8) | 51 (89.5) |
| Primary Treatment | ||||
| Radical prostatectomy | 248 (77.7) | 85 (61.2) | 696 (61.3) | 15 (26.3) |
| Radiation with or without ADT 5 | 51 (16) | 34 (24.5) | 305 (26.8) | 14 (24.6) |
| ADT only | 6 (1.9) | 9 (6.5) | 28 (2.5) | 26 (45.6) |
| Other treatment | 3 (0.9) | 0 (0) | 4 (0.4) | 0 (0) |
| Active surveillance | 11 (3.4) | 11 (7.9) | 103 (9.1) | 2 (3.5) |
Category includes a self-reported physician’s diagnosis of prostate cancer recurrence/progression, a positive bone scan, biopsy, or MRI showing cancer after primary treatment, presence of secondary treatment, or biochemical failure.
Deaths from other causes (n=89) and deaths from unknown causes (n=12) were censored at the time of death and accounted for in the analyses.
Total daily intake from diet and supplement use.
Composite aggressiveness classification parameters for "low" include cases diagnosed at local stage, a Gleason score of 2–6 or 7 (3+4), and diagnostic PSA < 20 ng/mL. "High" includes cases diagnosed at regional/distant stage, Gleason score 7(4+3) or 8–10, or diagnostic PSA ≥ 20 ng/mL.
ADT, androgen deprivation therapy.
Two VDR tagSNPs (rs6823 and rs2071358) and one CYP24A1 tagSNP (rs2762939) showed increased risks of disease recurrence/progression for carriers of the less common alleles (Table 2). When cases were limited to men treated with radical prostatectomy, the increased risks were still significant for carriers of less common alleles for VDR rs6823 [HR 2.0 (95% CI 1.2–3.3)] and CYP24A1 rs2762939 [HR 1.3 (95% CI 1.0–2.4)], but not VDR rs2071358. One CYP24A1 tagSNP (rs927650) showed decreased risk of disease recurrence/progression for carriers of the less common allele, but this decreased risk estimate was not apparent when cases were limited to those treated with radical prostatectomy (Table 2). Only CYP24A1 rs2762939 retained significance in a logistic model unadjusted for clinical parameters. None of the tagSNPs retained significance after adjustment for multiple comparisons. There was no evidence of interaction with self-report of family history of prostate cancer, BMI, or vitamin D supplement use/dietary intake.
Table 2.
Hazard ratios (95% CI) for Prostate Cancer Recurrence/Progression and Prostate Cancer-Specific Deaths by VDR (Vitamin D Receptor) CYP27B1 (1-alpha-Hydroxlase) and CYP24A1 (24-Hydroxylase) Genotypes Among Caucasian Men.
| Recurrence/Progression |
Prostate Cancer- Specific Deaths |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Genotype | No. of events1 (n=139) |
HR (95% CI)2 | Ptrend3 | No. of events1 (n=57) |
HR (95% CI)2 | Ptrend3 |
| VDR | rs2544038 4 | TT | 47 | ref | 16 | ref | ||
| Block A | CT | 63 | 1.0 (0.7–1.5) | 30 | 1.6 (0.8–3.1) | |||
| (23295bp 3’ of STP) | CC | 24 | 0.8 (0.5–1.4) | 9 | 1.6 (0.7–3.6) | |||
| CT+CC | 87 | 1.0 (0.7–1.4) | 0.45 | 39 | 1.6 (0.8–3.0) | 0.22 | ||
| rs739837 | TT | 37 | ref | 17 | ref | |||
| Block B | GT | 51 | 0.9 (0.6–1.4) | 20 | 0.8 (0.4–1.6) | |||
| (Ex11+568) | GG | 42 | 1.2 (0.7–1.9) | 17 | 1.3 (0.6–2.7) | |||
| GT+GG | 93 | 1.0 (0.7–1.5) | 0.46 | 37 | 1.0 (0.5–1.8) | 0.57 | ||
| rs731236 4, 5 | TT | 60 | ref | 24 | ref | |||
| Block B | CT | 50 | 0.8 (0.5–1.1) | 22 | 0.7 (0.4–1.3) | |||
| (Ex11+32) | CC | 19 | 0.8 (0.5–1.4) | 8 | 0.5 (0.2–1.3) | |||
| CT+CC | 69 | 0.8 (0.5–1.1) | 0.23 | 30 | 0.6 (0.4–1.1) | 0.11 | ||
| rs1544410 | GG | 19 | ref | 21 | ref | |||
| Block B | AG | 57 | 0.9 (0.5–1.6) | 23 | 0.8 (0.4–1.5) | |||
| (IVS10+283) | AA | 56 | 1.1 (0.6–1.8) | 10 | 0.8 (0.4–1.9) | |||
| AA+AG | 113 | 1.0 (0.6–1.6) | 0.57 | 33 | 0.8 (0.5–1.5) | 0.58 | ||
| rs2239182 | GG | 31 | ref | 19 | ref | |||
| Block B | AG | 54 | 0.9 (0.6–1.4) | 22 | 0.6 (0.3–1.1) | |||
| (IVS5+3419) | AA | 42 | 1.3 (0.8–2.1) | 15 | 1.0 (0.5–2.1) | |||
| AG+AA | 96 | 1.0 (0.7–1.5) | 0.27 | 37 | 0.7 (0.4–1.3) | 0.91 | ||
| rs2107301 | CC | 66 | ref | 31 | ref | |||
| Block B | CT | 45 | 0.9 (0.6–1.3) | 20 | 0.7 (0.4–1.4) | |||
| (IVS5+3260) | TT | 19 | 1.1 (0.7–1.9) | 4 | 1.0 (0.3–3.0) | |||
| CT+TT | 64 | 0.9 (0.7–1.4) | 0.97 | 24 | 0.8 (0.4–1.4) | 0.53 | ||
| rs2239181 | TT | 104 | ref | 44 | ref | |||
| Block B | GT | 23 | 1.0 (0.6–1.6) | 11 | 0.9 (0.4–1.8) | |||
| (IVS5+2881) | GG | 3 | 1.6 (0.5–5.4) | 0 | ||||
| GT+GG | 26 | 1.0 (0.7–1.6) | 0.75 | 11 | 0.8 (0.4–1.7) | 0.54 | ||
| rs2238139 | TT | 76 | ref | 30 | ref | |||
| Block B | CT | 37 | 1.1 (0.7–1.7) | 22 | 1.3 (0.7–2.4) | |||
| (IVS5+2550) | CC | 6 | 0.8 (0.3–1.9) | 2 | 1.5 (0.3–7.0) | |||
| CT+CC | 43 | 1.1 (0.7–1.6) | 0.93 | 24 | 1.3 (0.7–2.4) | 0.35 | ||
| rs3782905 4, 5 | CC | 64 | ref | 23 | ref | |||
| Block B | CG | 56 | 1.2 (0.9–1.8) | 26 | 1.0 (0.5–1.7) | |||
| (IVSR+6584) | GG | 11 | 0.9 (0.5–1.8) | 7 | 3.0 (1.2–7.7) | |||
| CG+GG | 67 | 1.2 (0.8–1.7) | 0.62 | 33 | 1.1 (0.6–2.0) | 0.20 | ||
| rs7974708 | TT | 63 | ref | 22 | ref | |||
| Block B | CT | 55 | 1.1 (0.8–1.6) | 24 | 1.1 (0.6–2.1) | |||
| (IVS4+2586) | CC | 13 | 0.9 (0.5–1.6) | 9 | 1.2 (0.5–3.0) | |||
| CT+CC | 68 | 1.1 (0.8–1.5) | 0.97 | 33 | 1.2 (0.7–2.1) | 0.58 | ||
| rs11168275 | AA | 78 | ref | 31 | ref | |||
| Block B | AG | 51 | 1.0 (0.7–1.5) | 22 | 1.3 (0.7–2.4) | |||
| (IVS4+476) | GG | 5 | 0.9 (0.4–2.3) | 2 | 0.7 (0.2–2.9) | |||
| AG+GG | 56 | 1.0 (0.7–1.5) | 0.96 | 24 | 1.2 (0.7–2.1) | 0.81 | ||
| rs10735810 | GG | 44 | ref | 18 | ref | |||
| No Block | AG | 65 | 1.2 (0.8–1.8) | 28 | 1.6 (0.9–3.0) | |||
| (Ex4+4) | AA | 22 | 0.9 (0.5–1.5) | 10 | 0.8 (0.3–1.8) | |||
| AG+AA | 87 | 1.1 (0.8–1.6) | 0.80 | 38 | 1.3 (0.7–2.3) | 0.81 | ||
| rs2408876 5 | TT | 54 | ref | 22 | ref | |||
| Block C | CT | 57 | 0.8 (0.6–1.2) | 24 | 0.9 (0.5–1.7) | |||
| (IVS3−667) | CC | 19 | 0.7 (0.4–1.2) | 9 | 1.6 (0.7–3.8) | |||
| CT+CC | 76 | 0.8 (0.6–1.2) | 0.23 | 33 | 1.0 (0.6–1.9) | 0.44 | ||
| rs2238135 | GG | 74 | ref | 28 | ref | |||
| Block C | CG | 49 | 0.9 (0.6–1.2) | 22 | 1.3 (0.7–2.3) | |||
| (IVS2−1633) | CC | 9 | 0.7 (0.3–1.3) | 3 | 1.5 (0.4–5.4) | |||
| CG+CC | 58 | 0.8 (0.6–1.2) | 0.22 | 25 | 1.3 (0.7–2.3) | 0.39 | ||
| rs10875694 | AA | 86 | ref | 34 | ref | |||
| Block C | AT | 40 | 0.9 (0.6–1.4) | 21 | 1.7 (0.9–3.0) | |||
| (IVS2−5103) | TT | 6 | 1.0 (0.4–2.3) | 1 | 0.8 (0.1–6.1) | |||
| AT+TT | 46 | 1.0 (0.7–1.4) | 0.87 | 22 | 1.6 (0.9–2.8) | 0.20 | ||
| rs11168287 | TT | 30 | ref | 13 | ref | |||
| Block C | CT | 68 | 1.1 (0.7–1.7) | 24 | 0.8 (0.4–1.6) | |||
| (IVS2−8206) | CC | 33 | 0.9 (0.6–1.6) | 17 | 1.3 (0.6–2.7) | |||
| CT+CC | 101 | 1.0 (0.7–1.6) | 0.73 | 41 | 1.0 (0.5–1.9) | 0.46 | ||
| rs7299460 4, 5 | CC | 65 | ref | 30 | ref | |||
| Block C | CT | 53 | 0.9 (0.6–1.3) | 16 | 0.5 (0.3–1.0) | |||
| (IVS1+2470) | TT | 7 | 0.5 (0.2–1.3) | 8 | 1.9 (0.8–4.6) | |||
| CT+TT | 60 | 0.9 (0.6–1.2) | 0.14 | 24 | 0.7 (0.4–1.2) | 0.92 | ||
| rs11168314 | CC | 78 | ref | 35 | ref | |||
| Block C | CT | 43 | 1.0 (0.7–1.5) | 14 | 0.7 (0.4–1.3) | |||
| (−27390) | TT | 7 | 1.1 (0.4–2.5) | 6 | 2.8 (1.1–7.3) | |||
| CT+TT | 50 | 1.0 (0.7–1.5) | 0.94 | 20 | 0.9 (0.5–1.6) | 0.60 | ||
| rs4073729 | CC | 84 | ref | 38 | ref | |||
| Block C | CT | 38 | 1.0 (0.7–1.5) | 8 | 0.6 (0.3–1.3) | |||
| (−20950) | TT | 7 | 1.5 (0.6–3.7) | 5 | 2.0 (0.8–5.4) | |||
| CT+TT | 45 | 1.1 (0.7–1.5) | 0.65 | 13 | 0.8 (0.4–1.6) | 0.85 | ||
| rs4760674 | CC | 46 | ref | 22 | ref | |||
| Block C | AC | 68 | 1.4 (1.0–2.0) | 21 | 0.6 (0.3–1.2) | |||
| (−1005) | AA | 14 | 1.1 (0.6–2.0) | 9 | 0.9 (0.4–2.0) | |||
| AC+AA | 82 | 1.3 (0.9–1.9) | 0.37 | 30 | 0.7 (0.4–1.2) | 0.43 | ||
| rs6823 5 | CC | 39 | ref | 19 | ref | |||
| Block C | CG | 66 | 1.5 (1.0–2.3) | 21 | 0.7 (0.4–1.4) | |||
| (Ex7–250) | GG | 26 | 1.7 (1.0–2.9) | 14 | 0.9 (0.4–1.9) | |||
| CG+GG | 92 | 1.6 (1.1–2.3) | 0.02 | 35 | 0.8 (0.4–1.4) | 0.67 | ||
| rs2071358 | CC | 79 | ref | 35 | ref | |||
| Block C | AC | 48 | 1.5 (1.0–2.2) | 19 | 1.4 (0.8–2.5) | |||
| (740bp 3’ of STP) | AA | 2 | 0.8 (0.2–3.2) | 0 | ||||
| AC+AA | 50 | 1.5 (1.0–2.1) | 0.06 | 19 | 1.2 (0.7–2.1) | 0.94 | ||
| CYP27B1 | rs3782130 4 | CC | 57 | ref | 27 | ref | ||
| Block A | CG | 61 | 1.1 (0.7–1.5) | 24 | 0.6 (0.3–1.1) | |||
| GG | 16 | 0.7 (0.4–1.2) | 4 | 0.4 (0.1–1.2) | ||||
| CG+GG | 77 | 1.0 (0.7–1.4) | 0.36 | 28 | 0.5 (0.3–0.9) | 0.03 | ||
| rs4646537 | AA | 121 | ref | 49 | ref | |||
| No Block | AC | 8 | 1.1 (0.5–2.2) | 6 | 2.3 (1.0–5.5) | |||
| (IVS8+113) | CC | 0 | -- | 0 | -- | |||
| AC+CC | 8 | 1.1 (0.5–2.2) | 0.82 | 6 | 2.3 (1.0–5.5) | 0.06 | ||
| CYP24A1 | rs927650 5 | CC | 41 | ref | 15 | ref | ||
| Block A | CT | 60 | 0.7 (0.5–1.0) | 26 | 1.1 (0.6–2.1) | |||
| (IVS11+967) | TT | 24 | 0.7 (0.4–1.1) | 13 | 0.8 (0.4–1.8) | |||
| CT+TT | 84 | 0.7 (0.5–1.0) | 0.07 | 39 | 1.0 (0.5–1.8) | 0.61 | ||
| rs912505 | AA | 74 | ref | 30 | ref | |||
| Block A | AG | 58 | 1.2 (0.8–1.6) | 23 | 0.8 (0.5–1.5) | |||
| (IVS7−1179) | GG | 3 | 0.6 (0.2–2.1) | 2 | 1.3 (0.3–5.8) | |||
| AG+GG | 61 | 1.1 (0.8–1.6) | 0.74 | 25 | 0.8 (0.5–1.5) | 0.69 | ||
| rs6127118 | GG | 65 | ref | 25 | ref | |||
| Block B | AG | 56 | 1.1 (0.7–1.6) | 28 | 1.1 (0.6–2.0) | |||
| (IVS7+204) | AA | 0 | n/a | 0 | n/a | |||
| AG+AA | 56 | 1.1 (0.7–1.6) | 0.75 | 28 | 1.1 (0.6–2.0) | 0.68 | ||
| rs6068816 | CC | 99 | ref | 41 | ref | |||
| Block B | CT | 30 | 1.0 (0.6–1.5) | 14 | 1.6 (0.8–3.0) | |||
| (Ex6+12) | TT | 3 | 1.5 (0.5–4.7) | 0 | n/a | |||
| CT+TT | 33 | 1.0 (0.7–1.5) | 0.98 | 14 | 1.5 (0.8–2.9) | 0.29 | ||
| rs2762939 5 | GG | 62 | ref | 30 | ref | |||
| Block B | CG | 56 | 1.7 (1.1–2.4) | 21 | 1.0 (0.5–1.8) | |||
| (IVS5−149) | CC | 6 | 1.1 (0.5–2.5) | 2 | 1.8 (0.4–7.7) | |||
| CG+CC | 62 | 1.6 (1.1–2.3) | 0.06 | 23 | 1.0 (0.6–1.9) | 0.72 | ||
| rs2244719 | TT | 41 | ref | 15 | ref | |||
| Block C | CT | 61 | 0.8 (0.6–1.3) | 29 | 1.1 (0.5–2.2) | |||
| (IVS4−486) | CC | 31 | 0.8 (0.5–1.3) | 11 | 1.1 (0.5–2.5) | |||
| CT+CC | 92 | 0.8 (0.6–1.2) | 0.58 | 40 | 1.1 (0.6–2.1) | 0.76 | ||
| rs3787557 | TT | 101 | ref | 39 | ref | |||
| Block C | CT | 31 | 1.0 (0.7–1.6) | 15 | 0.5 (0.3–1.0) | |||
| (IVS4−763 ) | CC | 1 | 0.5 (0.1–3.7) | 1 | 3.1 (0.4–23.5) | |||
| CT+CC | 32 | 1.0 (0.6–1.5) | 0.88 | 16 | 0.6 (0.3–1.1) | 0.13 | ||
| rs2181874 | GG | 73 | ref | 34 | ref | |||
| No Block | AG | 46 | 1.0 (0.7–1.5) | 19 | 1.2 (0.7–2.2) | |||
| (IVS4+1653) | AA | 10 | 1.4 (0.7–2.7) | 3 | 0.8 (0.2–2.9) | |||
| AG+AA | 56 | 1.1 (0.8–1.5) | 0.59 | 22 | 1.1 (0.6–2.0) | 0.87 | ||
| rs4809960 | TT | 76 | ref | 33 | ref | |||
| Block D | CT | 50 | 0.9 (0.6–1.3) | 19 | 0.6 (0.3–1.0) | |||
| (IVS4+58) | CC | 4 | 0.4 (0.1–1.1) | 4 | 1.1 (0.4–3.3) | |||
| CT+CC | 54 | 0.8 (0.6–1.2) | 0.21 | 23 | 0.6 (0.3–1.1) | 0.24 | ||
| rs2296241 | AA | 35 | ref | 17 | ref | |||
| Block D | AG | 70 | 1.1 (0.7–1.7) | 24 | 0.4 (0.2–0.9) | |||
| (Ex4+9) | GG | 22 | 0.7 (0.4–1.3) | 14 | 0.6 (0.3–1.2) | |||
| AG+GG | 92 | 1.0 (0.7–1.5) | 0.25 | 38 | 0.5 (0.3–0.9) | 0.14 | ||
| rs2245153 | TT | 85 | ref | 36 | ref | |||
| No Block | CT | 42 | 1.0 (0.7–1.4) | 17 | 0.7 (0.4–1.2) | |||
| (IVS3−179) | CC | 3 | 0.7 (0.2–2.3) | 3 | 0.8 (0.2–2.7) | |||
| CT+CC | 45 | 0.9 (0.6–1.4) | 0.88 | 20 | 0.7 (0.4–1.2) | 0.25 | ||
| rs2585428 | GG | 33 | ref | 19 | ref | |||
| No Block | AG | 69 | 1.4 (0.9–2.2) | 25 | 1.9 (1.0–3.7) | |||
| (IVS3−670) | AA | 26 | 1.3 (0.8–2.2) | 11 | 2.2 (0.9–5.2) | |||
| AG+AA | 95 | 1.4 (0.9–2.1) | 0.19 | 36 | 2.0 (1.1–3.8) | 0.04 | ||
| rs13038432 6 | AA | 106 | ref | 44 | ref | |||
| No Block | AG | 22 | 1.2 (0.8–2.0) | 10 | 1.5 (0.7–3.1) | |||
| (IVS3+814) | GG | 1 | 0.3 (0.0–1.9) | 0 | n/a | |||
| AG+GG | 23 | 1.1 (0.7–1.7) | 0.66 | 10 | 1.4 (0.7–2.9) | 0.51 | ||
| rs6022999 4 | AA | 73 | ref | 35 | ref | |||
| No Block | AG | 54 | 1.1 (0.8–1.6) | 18 | 2.4 (1.2–4.7) | |||
| (IVS3+103) | GG | 6 | 0.8 (0.4–2.0) | 2 | 1.3 (0.3–5.8) | |||
| AG+GG | 60 | 1.1 (0.8–1.6) | 0.86 | 20 | 2.2 (1.1–4.2) | 0.07 | ||
Variable numbers of events reflect instances of failed genotyping.
Hazard Ratios (HR) adjusted for age, Gleason, stage, diagnostic PSA level, and primary treatment. Hazard Ratios significant at p ≤ 0.05 are bolded. These did not remain significant after adjustment for multiple comparisons. If there were no homozygote carriers of less common allele, then only the dominant model risk estimate is shown.
Analysis for linear trend according to the number of variant alleles. If there are no homozygote carriers of less common allele this analysis is omitted. P-values ≤ 0.05 are bolded
These polymorphisms were included in panel of SNPs included in the ROC curves for prostate cancer-specific mortality at 10 years from diagnosis.
These polymorphisms were included in panel of SNPs included in the ROC curves for recurrence/progression at 5 years from diagnosis.
This polymorphism was found to be out of Hardy-Weinberg equilibrium.
Two VDR tagSNPs (rs3782905 and rs11168314) and two CYP24A1 tagSNPs (rs2585428 and rs6022999) showed increased risks of prostate cancer-specific mortality for carriers of the less common alleles (Table 2). One VDR tagSNP (rs7299460), one CYP27B1 tagSNP (rs3782130) and three CYP24A1 tagSNPs (rs3787557, rs4809960, and rs2296241) showed decreased risks of prostate cancer-specific mortality (Table 2). When cases were limited to men treated with radical prostatectomy, risks of prostate cancer-specific mortality did not remain significant for carriers of the less common alleles for any of the SNPs. Only VDR rs11168314 retained significance in a logistic model unadjusted for clinical parameters. None of these tagSNPs remained significant after adjustment for multiple comparisons. There was no evidence of interaction with self-report of family history of prostate cancer or vitamin D supplement use/dietary intake. For BMI there was a suggestion of effect modification for three CYP24A1 tagSNPs, rs3787557 (p < 0.01), rs4809960 (p < 0.001), and rs2296241 (p = 0.05), but the sample size was too small to reliably report hazard ratios for each BMI strata.
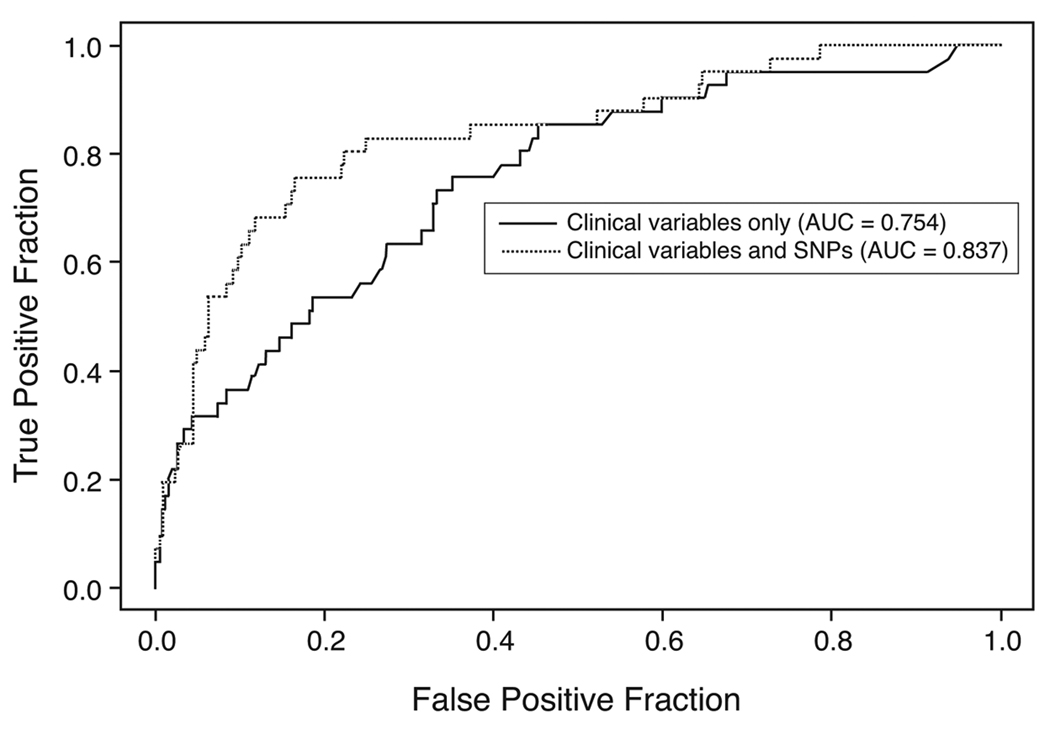
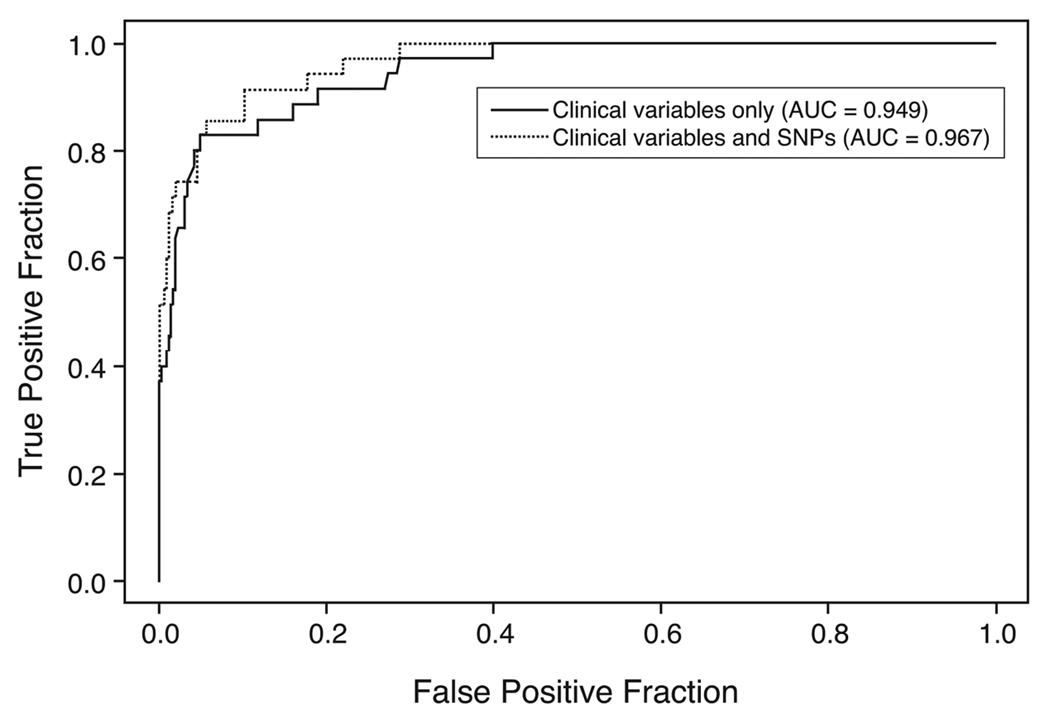
The ROC curves in figure 1 illustrate the prognostic value of clinical parameters alone versus these same parameters plus a panel of the 7 “optimal” SNPs (VDR: rs731236, rs3782905, rs2408876, rs7299460 and rs6823; CYP24A1: rs927650 and rs2762939) for predicting recurrence/progression at 5 years after diagnosis. The sensitivity for 5-year recurrence/ progression at a specificity of 80% was 53.7% for the model using only the clinical predictors, however, the sensitivity increased to 75.6% when the 7 SNP panel was added (difference: 21.9, 95% CI: 0.0%, 40.9%, p = 0.044). The difference in the AUC between the two curves was 0.082 (95% CI 0.016, 0.180, p = 0.020). The ROC curves in figure 2 illustrate the prognostic value of clinical parameters alone versus these same parameters and a panel of 6 “optimal” SNPs (VDR: rs2544038, rs731236, rs3782905, rs7299460; CYP27B1: rs3782130; CYP24A1: rs6022999) for predicting prostate cancer-specific mortality at 10 years after diagnosis. At 80% specificity, the sensitivity for 10-year prostate cancer-specific mortality was 91.4% using only the clinical predictors and increased to 94.3% when the 6 SNPs were included (difference: 2.9, 95% CI: −4.0%, 15.5%, p = 0.2). The difference in AUC between the two ROC curves was 0.018 (95% CI 0.005, 0.050, p = 0.128).
Figure 1.
Figure 2.
Discussion
We found some significant associations between risk of both tumor recurrence/progression and prostate cancer death for several SNPs in the VDR, CYP27A1 and CYP24A1 genes, however, associations at for individual SNPs did not remain significant after adjustment for multiple comparisons. Comparison of ROC curves suggests that addition of an optimal panel of SNPs to existing clinical predictors may improve predictive models for prostate cancer recurrence/ progression. Addition of the 7 SNP panel (VDR: rs731236, rs3782905, rs2408876, rs7299460 and rs6823; CYP24A1: rs927650 and rs2762939) significantly improved sensitivity at a specificity of 80% and AUC for a predictive model of 5-year prostate cancer recurrence/progression. Our previous work has not supported a consistent association between genetic variation in VDR, CYP27B1, and CYP24A1 genes and prostate cancer risk [32,33]. However, since vitamin D may play a different role in disease initiation versus disease progression, polymorphisms that predict disease outcomes are likely to be different than those that predict disease risk. Research has consistently shown a link between vitamin D and prostate cancer progression specifically; cellular studies show vitamin D can inhibit the carcinogenic progression of prostate cells, while ecologic studies provide evidence that UV exposure affects survival and prognosis [1–8].
Several VDR SNPs were identified to be associated with recurrence/progression, either by individual genotype before adjustment for multiple comparisons (rs6823 and rs2071358) or by the AIC approach for the ROC analysis (rs731236, rs3782905, rs2408876, rs7299460 and rs6823). Likewise, several VDR SNPs were shown to be associated with prostate cancer death, either at an individual level before adjustment for multiple comparisons (rs3782905, rs11168314, and rs7299460) or by the AIC approach for the ROC analysis (rs2544038, rs731236, rs3782905, and rs7299460). All of these SNPs, with the exception of rs731236, a synonymous coding SNP also known as the TaqI polymorphism (conservation score 0.175), are intronic, are not found to be evolutionarily conserved (conservation score < 0.001), and their potential biological consequences are unknown. Three of these SNPs, rs731236, rs3782905, and rs7299460 are included in both ROC models for recurrence/prediction and mortality. The TaqI polymorphism, rs731236, is not functional but is in linkage disequilibrium (LD) with a poly(A) microsatellite repeat in the 3’ untranslated region (UTR) that is thought to be important in post-transcriptional control gene expression. The SNP rs3782905 is in the DNA binding domain responsible for interaction with vitamin D response elements (VDRE) in target genes. The SNPs rs2408876, rs7299460, rs11168314, rs6828 and rs2071358 are located in either the promoter region or 5’ UTR and could be in LD with several recently reported novel SNPs that have both high conservation and apparent functional consequence affecting VDR transcription [34–37]. The SNP rs11168314 is in LD (r2 = 0.82) with Cdx-2 (rs11568820) that has been shown to alter transcription [36].
Polymorphisms in the genes involved in 1α,25(OH)2D metabolism were also found to be associated with both tumor recurrence/progression or prostate cancer-specific mortality. The one polymorphism in CYP27B1, rs3782130, associated with prostate cancer death, was not found to be evolutionarily conserved, but is located in the 3’ UTR. To date, there is limited evidence identifying functional polymorphisms in CYP27B1. It is interesting to note that CYP27B1 is down-regulated early in the neoplastic process of prostate cancer cells by epigenetic regulation, thus germline genetic variation affecting either gene expression or protein function may not have as large an effect within the malignant cell milieu [13,14,38]. The seven CYP24A1 SNPs that were associated with either tumor recurrence/progression (rs927650 and rs2762939) or prostate cancer-specific mortality (rs3787557, rs4809960, rs2296241, rs2585428, and rs6022999) were intronic and were not evolutionarily conserved with the exception of rs2296241 (conservation score 0.99), a synonymous polymorphism in exon 4. CYP24A1 was the only gene for which we used publicly available data as compared to resequencing data; consequently we are limited by the publically available genetic variants. The functional effects of indentified SNPs remain unclear but several novel SNPs have been recently identified in the promoter region 5' of exon 1 in CYP24A1 that have demonstrated a functional impact on VDRE binding and transactivation in vitro and altered expression of CYP24A1 gene expression in vivo [39]. The SNPs near this region, rs4809960, rs2296241, rs2585428, and rs6022999, could potentially be in LD with the true functional SNPs.
There were no SNPs found to be associated with both recurrence/progression and prostate cancer-specific mortality at the individual genotype level although there were three VDR SNPs, rs7311236, rs3782905, and rs7299460, that were included in both ROC models. The lack of corresponding associations with both outcomes for a given genotype does not necessarily diminish the potential association with individual outcomes because the outcome measures are not synonymous. Only 20% (n=28) of the 139 cases with recurrence/progression events also died of prostate cancer. This is mostly likely because patients who had an initial diagnosis of metastatic disease were excluded from recurrence/progression analysis. It could also be due to the fact that almost half (n=66) of the events were based on evidence of biochemical recurrence alone which is not predicative of PCa-specific mortality since the natural history of biochemical recurrence is so heterogeneous [40].
Excess body weight, measured by BMI, may be associated with increased risk of prostate cancer progression, higher risk of biochemical failure after treatment of disease, and an increased risk of dying from the disease [41,42]. Increased BMI is also associated with decreased levels of bioavailable 25(OH)D [43]. While the apparent association between adiposity and poorer prognosis is complex and multi-factorial, is has been suggested that vitamin D levels may play a role thus BMI could potentially modify the effect of these genotypes. There have been reported interactions between VDR polymorphisms and adiposity with risk of colon cancer [44]. Our findings were not appreciably different when we controlled for BMI, however we did find a suggestion of effect modification for CYP24A1 rs3787557 (p < 0.01), rs4809960 (p < 0.001), and rs2296241 (p = 0.05) with BMI for risk of recurrence/progression.
While we were able to account for dietary intake of vitamin D, we were not able to measure serum levels of 25(OH)D or UV light exposure, the primary source of vitamin D. Studies examining serum vitamin D levels with risk of PCa overall have had inconsistent results and do not seem suggest an association between levels 25(OH)D or 1α,25(OH)2D in the blood with risk of the disease [19,29,30,45,46]. This could be because measured serum levels may not correspond to vitamin D exposure during the long latency period of prostate cancer or reflect the prostate tissue-specific levels since local synthesis of 1α,25(OH)2D is independent of the tight regulation in the endocrine system [47]. Despite the lack of evidence for an association between Vitamin D levels in the blood and prostate cancer risk, there is evidence that vitamin D could be a potential effect modifier of disease risk within VDR genotypes [6,7,45,46,48]. It may be worthwhile for future studies to quantify vitamin D level in cases not only because of this potential interaction, but because serum levels of 1α,25(OH)2D may have a larger impact of cancer progression as a consequence of the down-regulation of CYP27A1 in the malignant prostate cells. A study that examined relationship between vitamin D status and survival rather than disease risk supports this hypothesis; a deceased risk of prostate cancer-specific mortality was observed in subjects with medium to high levels of 25(OH)D in serum collected upon admission to hospital for treatment [49].
This was a unique analysis with respect to vitamin D-related genes and prostate cancer prognosis in that it included long-term follow-up of all cases in a population-based study. Inclusion of tagSNPs to better capture genetic variation in each gene within this pathway allowed for a more comprehensive analysis than past studies. We were able to include relevant demographics and treatment variables to find a SNP panel that was predictive of outcome beyond clinical parameters currently used by clinicians. Although there was treatment heterogeneity among our cases, we were able to limit analyses to men receiving radical prostatectomy and replicate some of our findings. The primary weakness of our study was sample size, thus we must recognize the lack of precision in these findings and underscores the need for replication especially with respect to the ROC curves. Sample size also limited our analysis with respect to potential effect modifiers such as BMI. We did not include a stratified analysis by race because there were only 10 recurrence/progression events and nine prostate cancer deaths within African Americans in our dataset. Future studies should examine risk within African Americans since it has been postulated that the higher fraction of vitamin D deficiency observed in African American men may contribute to the increased mortality of prostate cancer observed in this population [10,50].
By studying a group of SNPs that more fully captured the genetic variability in the VDR, CYP27B1, and CYP24A1 genes, we found some evidence that genetic variation in these genes may be associated with both disease recurrence/progression and prostate cancer death and lead to an improved risk prediction. These findings should lead to future replication studies of germline genetic polymorphisms that can be used for risk prediction and to improve prostate cancer patient outcomes. These studies should include greater coverage of the promoter regions, especially for CYP24A1, in addition to examining coactivators and corepressers that modulate the response of VDR antiproliferative effects on prostate cells.
Acknowledgments
FINANCIAL SUPPORT: This work was supported by grants RO1-CA56678, RO1-CA092579, RO1-CA082554 and P50-CA097186 from the National Cancer Institute; additional support was provided by the Fred Hutchinson Cancer Research Center, the Intramural Program of the National Human Genome Research Institute, and the Prostate Cancer Foundation Young Investigator Award Grant.
ABBREVIATIONS
- PCa
Prostate cancer
- SNP
single nucleotide polymorphisms
- VDR
vitamin D receptor
- 25(OH)D
25-hydroxyvitamin D
- 1α,25(OH)2D
1α,25-dihydroxy-vitamin D
- 1α-OHase
1-α-hydroxylase
- VDRE
vitamin D response element
- SEER
Surveillance Epidemiology and End Results
- UV
ultraviolet
- PSA
prostate-specific antigen
- BMI
body mass index
- ADT
androgen deprivation therapy
- MRI
magnetic resonance imaging
- PH
proportional hazards
- HR
hazard ratio
- CI
confidence interval
- HWE
Hardy-Weinberg equilibrium
- AIC
Akaike’s information criterion
- ROC
receiver operating characteristics
- AUC
area under curve
- DNA
deoxyribonucleic acid
- LD
linkage disequilibrium
- UTR
untranslated region
Footnotes
Disclosure Statement
The authors of this manuscript do not have any affiliations that are relevant or important with any organization that to our knowledge has a direct interest, financial or otherwise, in the subject matter discussed.
References
- 1.Holick MF. Vitamin D: its role in cancer prevention and treatment. Prog Biophys Mol Biol. 2006;92(1):49–59. doi: 10.1016/j.pbiomolbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Krishnan AV, Peehl DM, Feldman D. Inhibition of prostate cancer growth by vitamin D: regulation of target gene expression. J Cell Biochem. 2003;88(2):363–371. doi: 10.1002/jcb.10334. [DOI] [PubMed] [Google Scholar]
- 3.Bao BY, Yao J, Lee YF. 1alpha 25-dihydroxyvitamin D3 suppresses interleukin-8-mediated prostate cancer cell angiogenesis. Carcinogenesis. 2006;27(9):1883–1893. doi: 10.1093/carcin/bgl041. [DOI] [PubMed] [Google Scholar]
- 4.Lim HS, Roychoudhuri R, Peto J, Schwartz G, Baade P, Moller H. Cancer survival is dependent on season of diagnosis and sunlight exposure. Int J Cancer. 2006;119(7):1530–1536. doi: 10.1002/ijc.22052. [DOI] [PubMed] [Google Scholar]
- 5.Robsahm TE, Tretli S, Dahlback A, Moan J. Vitamin D3 from sunlight may improve the prognosis of breast- colon- and prostate cancer (Norway) Cancer Causes Control. 2004;15(2):149–158. doi: 10.1023/B:CACO.0000019494.34403.09. [DOI] [PubMed] [Google Scholar]
- 6.John EM, Schwartz GG, Koo J, Van Den Berg D, Ingles SA. Sun exposure vitamin D receptor gene polymorphisms and risk of advanced prostate cancer. Cancer Res. 2005;65:5470–5479. doi: 10.1158/0008-5472.CAN-04-3134. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz GG, Hanchette CL. UV Latitude and spatial trends in prostate cancer mortality: all sunlight is not the same (United States) Cancer Causes Control. 2006;17(8):1091–1101. doi: 10.1007/s10552-006-0050-6. [DOI] [PubMed] [Google Scholar]
- 8.Lagunova Z, Porojnicu AC, Dahlback A, Berg JP, Beer TM, Moan J. Prostate cancer survival is dependent on season of diagnosis. Prostate. 2007;67(12):1362–1370. doi: 10.1002/pros.20577. [DOI] [PubMed] [Google Scholar]
- 9.Johnson MA, Davey A, Park S, Hausman DB, Poon LW. Age race and season predict vitamin D status in African American and white octogenarians and centenarians. J Nutr Health Aging. 2008;12(10):690–695. doi: 10.1007/BF03028616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Cancer Society. Cancer facts & figures 2008. American Cancer Society; 2008. pp. 1–68. [Google Scholar]
- 11.Vijayakumar S, Mehta RR, Boerner PS, Packianathan S, Mehta RG. Clinical trials involving vitamin D nalogs in prostate cancer. Cancer J. 2005;11(5):362–373. doi: 10.1097/00130404-200509000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Beer TM, Ryan CW, Venner PM, Petrylak DP, Chatta GS, Ruether JD, Redfern CH, Fehrenbacher L, Saleh MN, Waterhouse DM, Carducci MA, Vicario D, Dreicer R, Higano CS, Ahmann FR, Chi KN, Henner WD, Arroyo A, Clow FW. Double-blinded randomized study of high-dose calcitriol plus docetaxel compared with placebo plus docetaxel in androgen-independent prostate cancer: a report from the ASCENT Investigators. J Clin Oncol. 2007;25(6):669–674. doi: 10.1200/JCO.2006.06.8197. [DOI] [PubMed] [Google Scholar]
- 13.Chen TC, Wang L, Whitlatch LW, Flanagan JN, Holick MF. Prostatic 25-hydroxyvitamin D-1alpha-hydroxylase and its implication in prostate cancer. J Cell Biochem. 2003;88(2):315–322. doi: 10.1002/jcb.10342. [DOI] [PubMed] [Google Scholar]
- 14.Hsu JY, Feldman D, McNeal JE, Peehl DM. Reduced 1alpha-hydroxylase activity in human prostate cancer cells correlates with decreased susceptibility to 25-hydroxyvitamin D3-induced growth inhibition. Cancer Res. 2001;61(7):2852–2856. [PubMed] [Google Scholar]
- 15.Lou YR, Qiao S, Talonpoika R, Syvala H, Tuohimaa P. The role of vitamin D3 metabolism in prostate cancer. J Steroid Biochem Mol Biol. 2004;92(4):317–325. doi: 10.1016/j.jsbmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Luscombe CJ, French ME, Liu S, Saxby MF, Jones PW, Fryer AA, Strange RC. Outcome in prostate cancer; associations with skin type and polymorphism in pigmentation-related genes. Carcinogenesis. 2001;22(9):1343–1347. doi: 10.1093/carcin/22.9.1343. [DOI] [PubMed] [Google Scholar]
- 17.Cheteri MB, Stanford JL, Friedrichsen DM, Peters MA, Iwasaki L, Langlois MC, Feng Z, Ostrander EA. Vitamin D Receptor Gene polymorphisms and prostate cancer risk. Prostate. 2004;59(4):409–418. doi: 10.1002/pros.20001. [DOI] [PubMed] [Google Scholar]
- 18.Cicek MS, Liu X, Schumacher FR, Casey G, Witt JS. Vitamin D receptor genotypes/haplotypes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15(12):2549–2552. doi: 10.1158/1055-9965.EPI-06-0409. [DOI] [PubMed] [Google Scholar]
- 19.Mikhak B, Hunter DJ, Spiegelman D, Platz EA, Hollis BW, Giovannucci E. Vitamin D receptor (VDR) gene polymorphisms and haplotypes interactions with plasma 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D and prostate cancer risk. Prostate. 2007;67(9):911–923. doi: 10.1002/pros.20570. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins GA, Cramer SD, Zheng SL, Isaacs SD, Wiley KE, Chang BL, Bleecker ER, Walsh PC, Meyers DA, Isaacs WB, Xu J. Sequence variants in the human 25-hydroxyvitamin D3 1-alpha-hydroxylase (CYP27B1) gene are not associated with prostate cancer risk. Prostate. 2002;53(3):175–178. doi: 10.1002/pros.10144. [DOI] [PubMed] [Google Scholar]
- 21.Huang SP, Huang CY, Wu WJ, Pu YS, Chen J. Association of vitamin D receptor FokI polymorphism with prostate cancer risk clinicopathological features and recurrence of prostate specific antigen after radical prostatectomy. Int J Cancer. 2006;199(8):1902–1907. doi: 10.1002/ijc.22053. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Shibata A, McNeal JE, Stamey TA, Feldman D, Peehl DM. Vitamin D receptor start codon polymorphism (FokI) and prostate cancer progression. Cancer Epidemiol Biomarkers Prev. 2003;12(1):23–27. [PubMed] [Google Scholar]
- 23.Williams H, Powell IJ, Land SJ, Sakr WA, Hughes MR others. Vitamin D receptor gene polymorphisms and disease free survival after radical prostatectomy. Prostate. 2004;61(3):267–275. doi: 10.1002/pros.20103. [DOI] [PubMed] [Google Scholar]
- 24.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 8(10):881–886. 99. [PubMed] [Google Scholar]
- 25.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008;168(3):250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt SK, Kwon EM, Lin DW, Ostrander EA, Stanford JL. Association of Hepsin gene variants with prostate cancer risk and prognosis. Prostate. doi: 10.1002/pros.21135. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penson DF, Albertsen PC, Nelson PS, Barry M, Stanford JL. Determining cause of death in prostate cancer: are death certificates valid? J Natl Cancer Inst. 2001;93(23):1822–1823. doi: 10.1093/jnci/93.23.1822. [DOI] [PubMed] [Google Scholar]
- 28.Nejentsev S, Godfrey L, Snook H, Rance H, Nutland S, Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Tuomilehto-Wolf E, Tuomilehto J, Newport MJ, Clayton DG, Todd JA. Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet. 2004;13(15):1633–1639. doi: 10.1093/hmg/ddh169. [DOI] [PubMed] [Google Scholar]
- 29.Platz EA, Leitzmann MF, Hollis BW, Willett WC, Giovannucci E. Plasma 1,25-dihydroxy- and 25-hydroxyvitamin D and subsequent risk of prostate cancer. Cancer Causes Control. 2004;15(3):255–265. doi: 10.1023/B:CACO.0000024245.24880.8a. [DOI] [PubMed] [Google Scholar]
- 30.Tuohimaa P, Tenkanen L, Ahonen M, Lumme S, Jellum E, Hallmans G, Stattin P, Harvei S, Hakulinen T, Luostarinen T, Dillner J, Lehtinen M, Hakama M. Both high and low levels of blood vitamin D are associated with a higher prostate cancer risk: a longitudinal nested case-control study in the Nordic countries. Int J Cancer. 2004;108(1):104–108. doi: 10.1002/ijc.11375. [DOI] [PubMed] [Google Scholar]
- 31.Clayton D, Leung HT. An R package for analysis of whole-genome association studies. Hum Hered. 2007;64:45–51. doi: 10.1159/000101422. [DOI] [PubMed] [Google Scholar]
- 32.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S. Comprehensive association analysis of the vitamin D pathway genes VDR CYP27B1 and CYP24A1 in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1990–1999. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 33.Holt SK, Kwon EM, Peters U, Ostrander EA, Stanford JL. Vitamin D pathway gene variants and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2009 Jun;18(6):1929–1933. doi: 10.1158/1055-9965.EPI-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jehan F, d'Alesio A, Garabedian M. Exons and functional regions of the human vitamin D receptor gene around and within the main 1a promoter are well conserved among mammals. J Steroid Biochem Mol Biol. 2007;103(3–5):361–367. doi: 10.1016/j.jsbmb.2006.12.057. [DOI] [PubMed] [Google Scholar]
- 35.Ukaji M, Saito Y, Fukushima-Uesaka H, Maekawa K, Katori N, Kaniwa N, Yoshida T, Nokihara H, Sekine I, Kunitoh H, Ohe Y, Yamamoto N, Tamura T, Saijo N, Sawada J. Genetic variations of VDR/NR1I1 encoding vitamin D receptor in a Japanese population. Drug Metab Pharmacokinet. 2007;22(6):462–467. doi: 10.2133/dmpk.22.462. [DOI] [PubMed] [Google Scholar]
- 36.Fang Y, van Meurs JB, d'Alesio A, Jhamai M, Zhao H, Rivadeneira F, Hofman A, van Leeuwen JP, Jehan F, Pols HA, Uitterlinden AG. Promoter and 3'-untranslated-region haplotypes in the vitamin d receptor gene predispose to osteoporotic fracture: the rotterdam study. Am J Hum Genet. 2005;77(5):807–823. doi: 10.1086/497438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidd LC, Paltoo DN, Wang S, Chen W, Akereyeni F, Isaacs W, Ahaghotu C, Kittles R. Sequence variation within the 5' regulatory regions of the vitamin D binding protein and receptor genes and prostate cancer risk. Prostate. 2005;64(3):272–282. doi: 10.1002/pros.20204. [DOI] [PubMed] [Google Scholar]
- 38.Khorchide M, Lechner D, Cross HS. Epigenetic regulation of vitamin D hydroxylase expression and activity in normal and malignant human prostate cells. J Steroid Biochem Mol Biol. 2005;93(2–5):167–172. doi: 10.1016/j.jsbmb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 39.Roff A, Wilson RT. A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding transactivation and gene expression. J Steroid Biochem Mol Biol. 2008;112(1–3):47–54. doi: 10.1016/j.jsbmb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedland SJ, Humphreys EB, Mangold LA, Eisenberger M, Dorey FJ, Walsh PC, Partin AW. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294(4):433–439. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]
- 41.Freedland SJ, Giovannucci E, Platz EA. Are findings from studies of obesity and prostate cancer really in conflict? Cancer Causes Control. 2006;17:5–9. doi: 10.1007/s10552-005-0378-3. [DOI] [PubMed] [Google Scholar]
- 42.Gong Z, Agalliu I, Lin DW, Stanford JL, Kristal AR. Obesity is associated with increased risks of prostate cancer metastasis and death after initial cancer diagnosis in middle-aged men. Cancer. 2007;109(6):1192–1202. doi: 10.1002/cncr.22534. [DOI] [PubMed] [Google Scholar]
- 43.Wortsman J, Matsuoka LY, Chen TC, Lu Z, Holick MF. Decreased bioavailability of vitamin D in obesity. Am J Clin Nutr. 2000;72(3):690–693. doi: 10.1093/ajcn/72.3.690. [DOI] [PubMed] [Google Scholar]
- 44.Ochs-Balcom HM, Cicek MS, Thompson CL, Tucker TC, Elston RC, J Plummer S, Casey G, Li L. Association of vitamin D receptor gene variants adiposity and colon cancer. Carcinogenesis. 2008;29(9):1788–1793. doi: 10.1093/carcin/bgn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Stampfer MJ, Hollis JB, Mucci LA, Gaziano JM, Hunter D, Giovannucci EL, Ma J. A prospective study of plasma vitamin D metabolites, vitamin D receptor polymorphisms and prostate cancer. PLoS Medicine / Public Library of Science. 2007;4(3):e103. doi: 10.1371/journal.pmed.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn J, Albanes D, Berndt SI, Peters U, Chatterjee N, Freedman ND, Abnet CC, Huang WY, Kibel AS, Crawford ED, Weinstein SJ, Chanock SJ, Schatzkin A, Hayes RB. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis. 2009;30(5):769–776. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young MV, Schwartz GG, Wang L, Jamieson DP, Whitlatch LW, Flanagan JN, Lokeshwar BL, Holick MF, Chen TC. Prostate 25-hydroxyvitamin D-1 alpha-hydroxylase is not influenced by parathyroid hormone and calcium: implications for prostate cancer chemoprevention by vitamin D. Carcinogenesis. 2004;25(6):967–971. doi: 10.1093/carcin/bgh082. [DOI] [PubMed] [Google Scholar]
- 48.Rukin NJ, Luscombe C, Moon S, Bodiwala D, Liu S, Saxby MF, Fryer AA, Alldersea J, Hoban PR, Strange RC. Prostate cancer susceptibility is mediated by interactions between exposure to ultraviolet radiation and polymorphisms in the 5' haplotype block of the vitamin D receptor gene. Cancer Letters. 2007;247(2):328–335. doi: 10.1016/j.canlet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Tretli S, Hernes E, Berg JP, Hestvik UE, Robsahm TE. Association between serum 25(OH)D and death from prostate cancer. Br J Cancer. 2009;100(3):450–454. doi: 10.1038/sj.bjc.6604865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan KM, Signorello LB, Munro HM, Hargreaves MK, Hollis BW others. Vitamin D insufficiency among African-Americans in the southeastern united states: implications for cancer disparities (united states) Cancer Causes Control. 2008;19(5):527–535. doi: 10.1007/s10552-008-9115-z. [DOI] [PubMed] [Google Scholar]