Abstract
Maternally contributed mRNAs and proteins control the initial stages of development following fertilization. During this time, most of the zygotic genome remains transcriptionally silent. The initiation of widespread zygotic transcription is coordinated with the degradation of maternally provided mRNAs at the maternal-to-zygotic transition (MZT). While most of the genome is silenced prior to the MZT, a small subset of zygotic genes essential for the future development of the organism is transcribed. Previous work in our laboratory and others identified the TAGteam element, a set of related heptameric DNA-sequences in the promoters of many early-expressed Drosophila genes required to drive their unusually early transcription. To understand how this unique subset of genes is regulated, we identified a TAGteam-binding factor Grainyhead (Grh). We demonstrated that Grh and the previously characterized transcriptional activator Zelda (Zld) bind to different TAGteam sequences with varying affinities, and that Grh competes with Zld for TAGteam occupancy. Moreover, overexpression of Grh in the early embryo causes defects in cell division, phenocopying Zld depletion. Our findings indicate that during early embryonic development the precise timing of gene expression is regulated by both the sequence of the TAGteam elements in the promoter and the relative levels of the transcription factors Grh and Zld.
Keywords: Drosophila, blastoderm, transcription, Grainyhead, Zelda, TAGteam
Introduction
In many organisms the zygotic genome is quiescent following fertilization. During this time maternally contributed products regulate development. Only later, at the maternal-to-zygotic transition (MZT), is widespread zygotic transcription initiated, and its initiation is concurrent with the degradation of maternally provided mRNAs (Newport and Kirschner, 1982b). Prior to the MZT, the genome undergoes a series of rapid duplications without cytoplasmic growth. In frogs and flies this changing nucleo-cytoplasmic ratio in the young embryo determines the onset of general zygotic transcription (Foe and Alberts, 1983; Newport and Kirschner, 1982a, 1982b; Pritchard and Schubiger, 1996). The importance of the nucleo-cytoplasmic ratio has led to the proposal that the early zygotic genome is silenced by a maternally contributed transcriptional repressor that is titrated by the increasing DNA content. Despite years of study, however, the definitive mechanism by which the early zygotic genome is silenced remains speculative.
While general zygotic genome activation does not initiate until the MZT, in many organisms a small number of zygotic genes are transcribed prior to widespread zygotic transcription (De Renzis et al., 2007; Mathavan et al., 2005; Nakakura et al., 1987; Pilot et al., 2006). Genes in this subset are involved in processes that are essential for the future development of the organism and allow for progression through the MZT. Thus it is imperative that expression of these early genes be precisely controlled both temporally, as well as spatially. Little is currently known about how this subset of genes is selectively activated when the remainder of the genome is not expressed, and whether the mechanisms of activation are similar to those utilized to drive transcription later in development.
In Drosophila melanogaster, the fertilized egg undergoes a series of replication cycles without cytoplasmic divisions, generating a syncytial blastoderm. At cycle 14, the blastoderm nuclei cellularize, and it is at this developmental stage that general zygotic transcription initiates (Foe and Alberts, 1983; Lamb and Laird, 1976; McKnight and Miller, 1976). A subset of genes required for developmental patterning, sex determination, and cellularization are expressed prior to cellular blastoderm formation. Expression at this unusually early stage is essential for many of these genes. For example, genes that determine sex must be expressed early to allow for dosage compensation to ensure the proper ratio of × chromsome to autosome expression occurs upon zygotic genome activation. In addition, genes required for cellularization must also be expressed prior to the MZT as this process is coordinated with zygotic genome activation. The earliest of these pre-cellular blastoderm (pre-CB) expressed genes initiate transcription during the eighth nuclear cycle. However not all pre-CB genes are first expressed at this time, and some do not initiate transcription until cycles 12 and 13.
Previous work in our laboratory and others identified the TAGteam elements, characterized by CAGGTAG and related sequences, that are over-represented among the promoters of genes expressed in the pre-CB embryo (De Renzis et al., 2007; Li et al., 2008; ten Bosch et al., 2006). These sequence elements have been shown to be critical for the pre-CB expression of scute (sc), Sex lethal (Sxl), and zerknüllt (zen) and increasing the number of TAGteam elements preceding a gene causes transcription to initiate earlier in development (ten Bosch et al., 2006). Recently, the transcription factor Zelda (Zld) has been shown to be a TAGteam-binding factor required for the proper activation of a number of genes in the pre-CB embryo (Liang et al., 2008). Here we report the identification and characterization of a second TAGteam-binding factor, Grainyhead (Grh). grh mRNA is maternally produced, and Grh protein is expressed in the pre-cellular blastoderm embryo. Grh and Zld compete for TAGteam binding and overexpression of Grh in the early embryo results in defects in nuclear divisions, a phenotype similar to Zld depletion. In addition, we show that Grh and Zld differentially recognize TAGteam-sequence variants, suggesting that while TAGteam elements are highly related in sequence they may not function identically in driving pre-CB gene expression. From these data, we propose that the competition of these two transcription factors for DNA binding acts to fine tune expression of the subset of genes transcribed prior to the MZT.
Materials and methods
Grh purification
Nuclear extract was prepared from ~200 g of 0–12 hr Drosophila embryos as described in Lewis et al. (2004) and cut with 60% ammonium sulfate. The resulting precipitate was resuspended in buffer A-40 (25 mM Hepes pH 7.6, 40 mM KCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 0.4 mM PMSF, 1 mM DTT) and dialyzed into the same buffer. The dialyzed extract was loaded onto a heparin column and step eluted. Proteins from the 0.15 M KCl elution were loaded onto a Superdex 200 column. In this and all subsequent steps, 0.05% Igepal (Nonidet P-40 subsitute, Sigma) was added to buffer A. The Superdex 200 fractions containing the DNase I protection activity were pooled and loaded onto a DEAE Sepharose column. The flow through from this column was applied to a Mono S column. Pooled fractions from the Mono S column were dialyzed to 100 mM KCl and applied to a DNA-affinity column. The DNA-affinity column was prepared by binding biotin 5’ end-labeled repeats of the zen VRE to Dynal Streptavidin M280 beads (Invitrogen). The end-labeled repeats were produced by PCR using a biotinylated primer. The template for this reaction was four copies of a 100 base pair region of the zen VRE (surrounding the TAGGTAG and CAGGCAG sites) cloned into the plasmid pTL18U-PV with BamHI and BglII. The column was washed with buffer A-100 (buffer A with 0.1 M KCl) and stepwise eluted with 0.25 M, 0.5 M, 0.75 M, and 1 M KCl.
Recombinant protein expression and purification
The full-length open reading frames for grh-RH (from pBSSXNTF-1 (Attardi and Tjian, 1993)) and zld (from the DGRC clone #LD47819) were PCR amplified and cloned into pENTR/D-TOPO (Invitrogen) with an N-terminal FLAG or HA-affinity tag. These constructs were subsequently inserted into pDEST8 (Invitrogen) using LR clonase (Invitrogen). Baculovirus was prepared using the Bac-to-Bac expression system (Invitrogen). Infections and protein purifications were essentially as described in Ilves et al. (2009). However, infections were performed with 1 ml of freshly amplified virus. Elution was performed in buffer with 150 mM KCl. Eluted protein was then applied to either a Mono S PC 1.6/5 column (Grh) or a Mono Q PC 1.6/5 (Zld) connected to a SMART micropurification system (Pharmacia) equilibrated in buffer A-150 (buffer A with 150 mM KCl and 0.05% Igepal). Protein was eluted from the columns by gradient elution. Grh eluted from the Mono S column at ~250 mM KCl. Zld eluted from the MonoQ column at ~400 mM KCl. Protein concentration was measured by laser densitometry of SYPRO-Red (Invitrogen) stained SDS-PAGE gels. Known concentrations of bovine serum albumin were used as a standard or proteins were standardized to previously determined concentrations of proteins from an earlier preparation.
DNA-binding assays
DNase I protection assays were performed essentially as described in Jones et al. (1885). Probes were 5’ end-labeled DNA fragments of the sc, Sxl or zen regulatory regions containing TAGteam elements. Sequence of the individual probes was determined using fmol DNA Cycle Sequencing (Promega) and 5’ end-labeled sequencing primers equivalent to the 5’ end-labeled portion of the probe. The sequencing primers used in Supplemental Fig. S1 were the following: Sxl AGCTTCCCGCTAGGAAGTG, zen AGCTTCCCGTTATTGGAC, and sc AATTCGTTTTCCTGTGGG. These sequencing reactions were run on TBE urea gels in parallel with DNase I digested probes. EMSAs were set up essentially as previously described (Beall et al., 2002), except 25 mM ZnCl2 was added and 0.6 µg/ ml poly [d(I-C)][d(I-C] was substituted for poly [d(G-C)][d(G-C]. Reactions were incubated on ice for 20’. Samples were electrophoresed for 1.5–2.5 hours (depending on the resolution required) at 150 V at 4°C in 4% (60:1) polyacrylamide gels containing 1 × Tris glycine buffer (12.5 mM Tris, 190 mM glycine). For the competition experiments, 1 pmol of FLAG-Zld and between 0.3 and 5 pmol of Grh(603–1032) were used. The probes used in Figures 2C and 3C were CTATTTAGGTAGACACTGTA with the underlined bases substituted to change the TAGteam sequence variant. The TAGteam element is bolded. These probes contain a single TAGteam element and no additional Grh-binding sites.
Fig. 2.
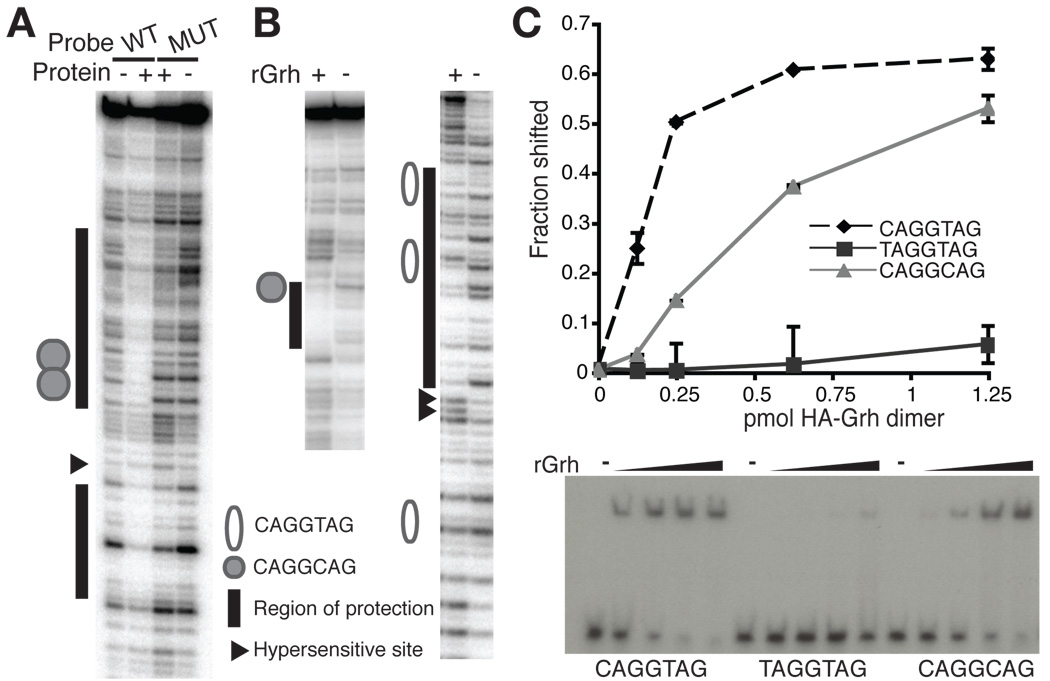
Grh binds to TAGteam elements. (A) DNase I protection by protein purified from embryonic nuclear extract of a fragment of SxlPe containing either wild-type (WT) or mutated (MUT) overlapping CAGGCAG elements. +, protein from Mono S peak. −, no protein. (B) DNase I protection by rGrh using portions of the sc promoter containing either a CAGGCAG element (left) or multiple CAGGTAG elements (right). +, rGrh. −, no protein. (C) Quantitation of EMSAs using rGrh and probes containing different TAGteam elements. Data are the mean +/− s.d. from two independent EMSAs. A representative EMSA is shown below. −, no protein.
Fig. 3.
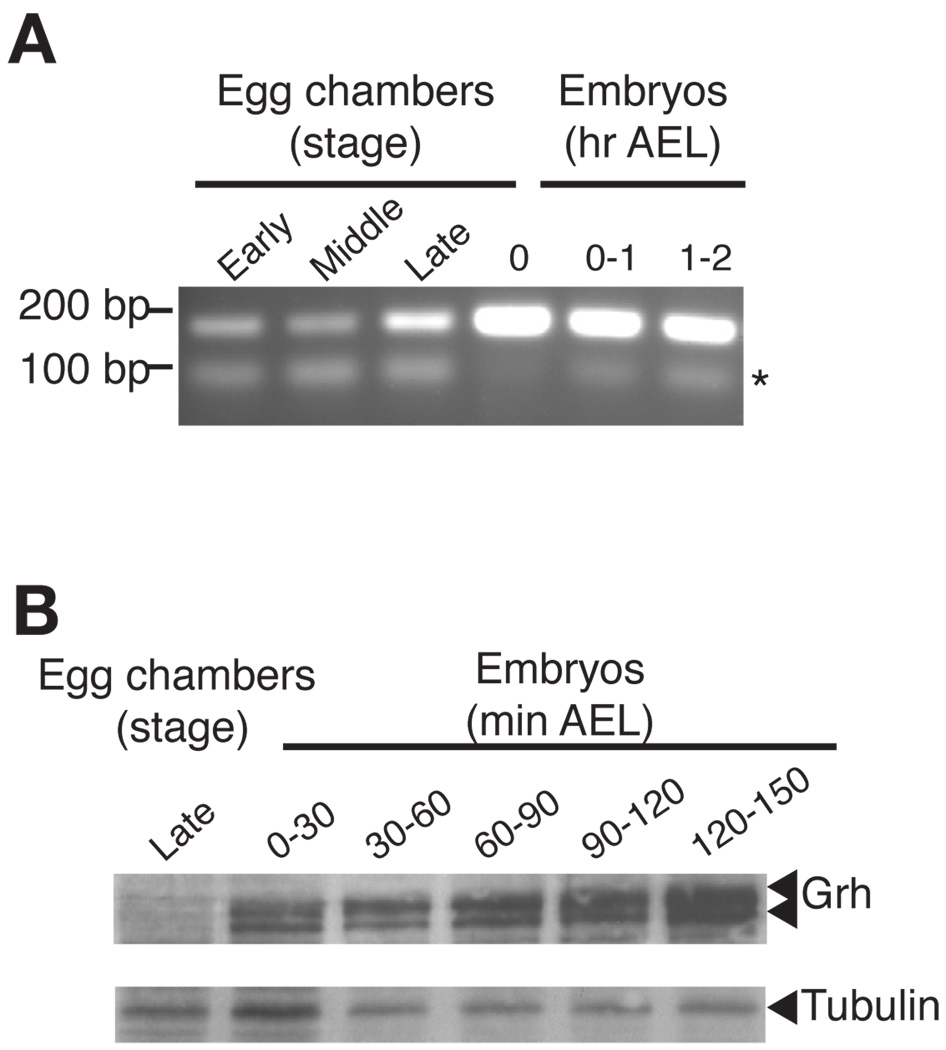
Grh is expressed in the pre-cellular blastoderm embryo. (A) RT-PCR, detecting all possible grh mRNAs, on RNA extracted from wild-type egg chambers and embryos at the stages of development indicated above each lane. The asterisk indicates a primer dimer as evidenced by a reaction containing no cDNA (not shown). AEL, after egg laying. (B) An immunoblot using αGrh 25 antibodies on protein extracted from equal volumes of egg chambers and embryos at various stages. Tubulin was included as a loading control.
RT-PCR
Equal volumes of embryos and egg chambers were physically disrupted using a Wheaton 2 ml dounce homogenizer, and RNA was extracted as directed for the RNeasy Kit (Qiagen). Absorbance 260 was used to determine the concentration of recovered RNA and equivalent amounts were used to prepare cDNA using Superscript II (Invitrogen).
Antibody production and immunoblots
Rabbits were immunized by Josman, LLC with either GST:Zld (amino acids 1117–1487) or GST:Grh (amino acids 603–1032). Antibodies were affinity purified against the MBP-tagged versions of these proteins and specificity was determined by immunoblots on extract from Drosophila S2 cells transfected with empty vector or expression plasmids for the full-length proteins. Rat anti-Grh was kindly provided by Robert Tjian (Attardi et al., 1993) and used at 1:500 dilutions in the immunoblots shown in Fig. 1C and 2D. Rabbit anti-Grh and anti-Zld were used at a 1:1000 and 1:500 dilutions, respectively, in the immunoblots in Fig. 5B.
Fig. 1.
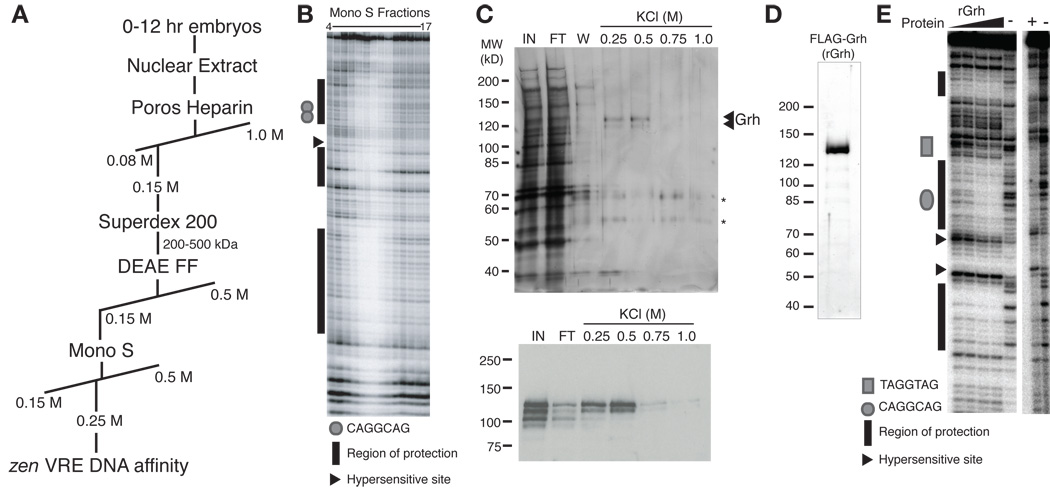
Purification and identification of the SxlPe and zen VRE-binding factor Grainyhead. (A) Purification scheme used to identify factor(s) binding to SxlPe and the zen VRE. (B) DNase I protection by Mono S fractions (indicated above each lane) using a SxlPe DNA fragment. (C) Analysis of proteins eluting from the DNA-affinity column by SDS-PAGE and silver staining (top). The two polypeptides eluting from the column at 0.25 M and 0.5 M KCl were identified as Grh by mass spectrometry. Asterisks denote keratin. 15% of each elution was loaded. IN, 1% of input. FT, 1% of flow through. W, 1.5% of wash. Shown below is an immunoblot using αGrh antibodies on protein from the DNA-affinity column. 5% of each elution was loaded. (D) SDS-PAGE and silver stain analysis of the purified recombinant Grh. Molecular weight (left) is indicated in kD. (E) DNase I protection by increasing amounts of recombinant Grh (rGrh) (left) or purified protein from nuclear extract (right) of a zen VRE DNA fragment. +, protein from embryonic nuclear extract. −, no protein.
Fig. 5.
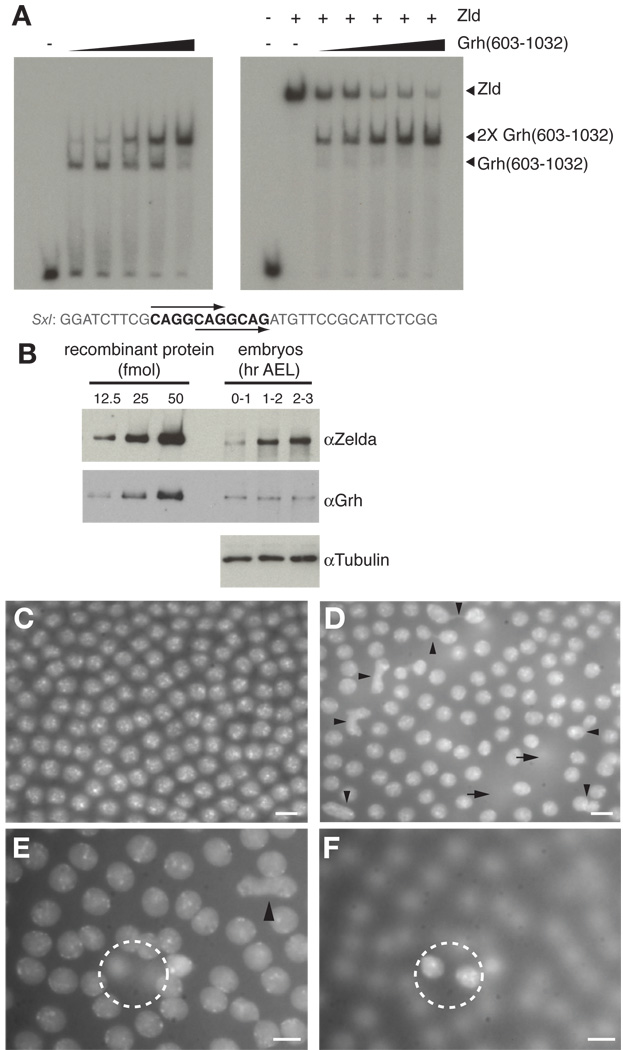
Grh and Zld compete for binding to TAGteam elements. (A) EMSAs using (Grh(603–1032), rZld, or both proteins combined (as indicated above each lane). The sequence of the probe used in the assays is shown below with the TAGteam elements bolded. Arrows indicate the two overlapping CAGGCAG elements. (B) Immunoblots using the antibodies indicated on either full-length recombinant protein (left; Zld, upper panel; Grh, middle panel) or protein extract from the equivalent of five embryos (right). Tubulin was included as a loading control. AEL, after egg laying. (C, D, E, F) DAPI-stained cycle 13 embryos. (C) A wild-type embryo. (D) An embryo with overexpressed Grh. Arrowheads indicate anaphase bridges, and arrows indicate perpendicular cell divisions that have resulted in nuclei below the plane of focus. (E, F) Two different focal planes of a single embryo overexpressing Grh. The dotted circle encompasses the identical area in each focal plane highlighting two nuclei resulting from a perpendicular cell division. Scale bars, 5 µm.
Calculation of protein levels in the early embryo
At the indicated time points following deposition, 100 embryos were collected into 1.5 microcentrifuge tubes containing PBST at 4°C. The embryos were allowed to settle, the PBST was removed, and 50 µl of 1× PAGE sample loading buffer (63 mM Tris pH6.8, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol) was added. The embryos were manually disrupted, boiled for 5 minutes, and centrifuged briefly. The embryo lysate was loaded onto an SDS-PAGE gel along with 12.5, 25, and 50 fmol of purified full-length recombinant Grh or Zld (as determined by SYPRO-Red staining, see above). Proteins were transferred to nitrocellulose and immunobloted using either anti-Grh or anti-Zld antibodies. Approximate protein concentrations for the embryonic levels of Zld and Grh were determined by comparison of signal intensities for these proteins to the signal intensities of known amounts of recombinant full-length protein. Signal intensities were within a linear range of detection. R2 for the recombinant Grh standards was 0.999 and for the recombinant Zld standards was 0.988. In an independent replicate of the experiment, similar results were observed.
Grh overexpression and maternal depletion
For overexpression of maternal grh, the full-length open reading frame for grh-RH was cloned into pENTR/D-TOPO (Invitrogen) and subsequently inserted into pPFW (Drosophila Gateway Vector Collection) using LR clonase (Invitrogen). This construct was injected into w1118 embryos by BestGene Inc and stocks were established. Transgenic flies were crossed to flies expressing GAL-VP16 under the control of the alphaTub67C promoter driving expression in the maternal germline (Matthews et al., 1989). Embryos from mother’s overexpressing FLAG-Grh were harvested, fixed in formaldehyde, and stained with DAPI to label DNA. To deplete maternal grh, we utilized the FLP-FRT system to induce mitotic recombination in the maternal germline using an FRT-containing chromosome marked by ubi-GFP that is expressed in the maternal germline and deposited in the early embryo. Flies including females of the genotype hsFlp122; FRT grh/ FRT ubi-GFP were heatshocked for 45’ at 37°C as third instar larvae. These females were collected and crossed to wild-type males. GFP negative embryos were sorted and fixed for in situ hybridization. GFP positive embryos were collected separately as controls. The grh alleles used were grhIM, grhB37, and grhB32.
Results and Discussion
To understand how a subset of genes are uniquely transcribed in the pre-CB Drosophila embryo when the remainder of the genome is not, we sought to identify proteins that bind to TAGteam elements in the regulatory regions of pre-CB-expressed genes. We fractionated nuclear extract prepared from wild-type Drosophila embryos and assayed for activity using DNase I protection of a portion of the early-expressed Sxl establishment promoter, SxlPe, containing two overlapping CAGGCAG sites (Fig. 1A) (Keyes et al., 1992). Partially purified protein(s) protected multiple regions of SxlPe from DNase I digestion, including the two TAGteam elements (Fig. 1B, and Supplemental Fig. S1A). As a final purification step, fractions were applied to a DNA-affinity column composed of oligonucleotides corresponding to four repeats of a portion of the zen ventral repression element (VRE), a TAGteam-containing sequence shown to regulate the pre-CB expression of zen (Jiang et al., 1992; Kirov et al., 1993). Using the zen VRE for the DNA-affinity column rather than SxlPe ensured that the purified protein(s) would bind to at least two sequences driving pre-CB gene expression. Two polypeptides of ~130 kD and 120 kD specifically eluted from the column (Fig. 1C). Mass spectrometry identified these two polypetides as the products of two splice isoforms generated from the single gene, grainyhead (grh). This identification was confirmed by immunoblotting (Fig. 1C bottom).
Grh is a transcription factor conserved from worms to humans that acts in mediating both transcriptional repression and activation (Dynlacht et al., 1989; Huang et al., 1995; Liaw et al., 1995; Venkatesan et al., 2003). Previous work has demonstrated that Drosophila Grh can bind to the promoters of three additional pre-CB expressed genes, fushi tarazu (ftz) (Dynlacht et al., 1989), tailless (tll) (Liaw et al., 1995), and decapentaplegic (dpp) (Huang et al., 1995), and the evidence suggests that Grh binding to these promoters results in transcriptional repression (Huang et al., 1995; Liaw et al., 1995). However, we are the first to demonstrate that Grh binds to TAGteam sites, greatly increasing the number of pre-CB genes Grh may regulate. In the early embryo, Grh may act as a repressor, in part, through its interactions with Polycomb-group proteins (Blastyak et al., 2006; Tuckfield et al., 2002). Later in both fly and mammalian embryonic development, Grh is expressed in the epidermis (Bray et al., 1989) and functions as an important transcriptional activator during the wound-healing response (Mace et al., 2005; Ting et al., 2005). Thus, whether Grh binding results in transcriptional activation or repression depends on developmental context.
While the immunoblots confirmed that Grh bound to the zen VRE DNA-affinity column, it was important to determine if Grh provided the DNA-binding activity present in embryonic nuclear extract. Anti-Grh antibodies raised against the DNA-binding domain disrupted the SxlPe DNA-binding activity present in nuclear extract, whereas non-specific IgG did not, confirming that Grh was responsible for the activity (Supplemental Fig. S2). In addition, purified full-length recombinant Grh (rGrh) provided DNase I protection of the zen VRE and SxlPe indistinguishable from that of the activity in nuclear extract (Figs. 1D, E and data not shown).
Because Grh bound to TAGteam elements in both SxlPe and the zen VRE (Figs. 1B, E and Supplemental Figs. S1A, B), we determined whether Grh specifically required TAGteam sequences for binding. We used both heparin-fractionated nuclear extract and rGrh for DNase I protection assays with a fragment of SxlPe identical to that used in the purification described above except that the overlapping CAGGCAG elements were mutated. Grh binding to the mutated CAGGCAG elements was severely inhibited demonstrating that these TAGteam sequences are essential for Grh binding (Fig. 2A and Supplemental Fig. S3).
Grh bound to additional sequences outside the TAGteam elements in SxlPe and the zen VRE as determined by DNase I protection assays (Fig. 1B, E and Supplemental Table 1). These additional binding sites do not contain sequences highly similar to the TAGteam sequences. While a consensus Grh binding site has been defined as ACYGGTT(T) (Mace et al. 2005), there is considerable variability among previously defined Grh binding sites. MEME searches on the Grh binding sites defined by both our and previous DNase I protection experiments failed to identify a strong consensus site, despite some similarity between the previously defined consensus site and TAGteam elements (Bailey and Elkan, 1994; Supplemental Table 1).
We focused our studies on the TAGteam-binding activity of Grh as these sequences have been shown to have important functions in the pre-CB embryo (ten Bosch et al., 2006). The TAGteam elements have been defined as a group of related sequences including CAGGTAG, CAGGCAG and TAGGTAG, with CAGGTAG being the most enriched in the promoters of pre-CB genes (De Renzis et al., 2007; Li et al., 2008; ten Bosch et al., 2006). To determine if Grh could bind to the prevalent CAGGTAG sequence, we used rGrh in protection assays on a region of the sc promoter containing three CAGGTAG elements and one CAGGCAG element. These experiments demonstrated protection of the CAGGCAG element as well as at least two of the three CAGGTAG elements (Fig. 2B and Supplemental Fig. S1C). We used electromobility shift assays (EMSAs) to test the affinity of Grh for different members of the TAGteam family. EMSAs with probes that only differed by the sequence of the TAGteam element showed that Grh binds strongly to CAGGTAG and CAGGCAG elements, but only weakly to TAGGTAG sequences (Fig. 2C), demonstrating the importance of the initial cytosine in Grh recognition. Previous work analyzing the ability of Grh to bind to the closely related sequence GCAGGTAA also showed the importance of the cytosine in Grh recognition. Furthermore, this cytosine was critical for the pre-CB ventral repression of a transgene reporter driven by the dpp ventral repression region (VRR) (Huang et al., 1995). Together our data show that Grh specifically binds to TAGteam elements within the promoters of three genes expressed in the early embryo, although preferentially to specific TAGteam sequences.
Given that Grh can bind to TAGteam elements, we asked if Grh was present in the early embryo. Using RT-PCR we showed that grh transcripts are present in early embryos as well as in egg chambers (ovaries) (Fig. 3A). In agreement with these data, in situ hybridizations had previously identified grh mRNA in these tissues (Huang et al., 1995). There are two well-characterized examples of alternative splicing of the grh pre-mRNA (Uv et al., 1997), and we determined that alternative splicing results in multiple mRNAs present in the early embryo (Supplemental Fig. S4). Immunoblots showed that these mRNAs are translated producing Grh proteins (Fig. 3B). Notably, Grh protein appears absent or at very low levels in late-stage egg chambers, suggesting that maternal grh mRNA, but not protein, is deposited into the embryo.
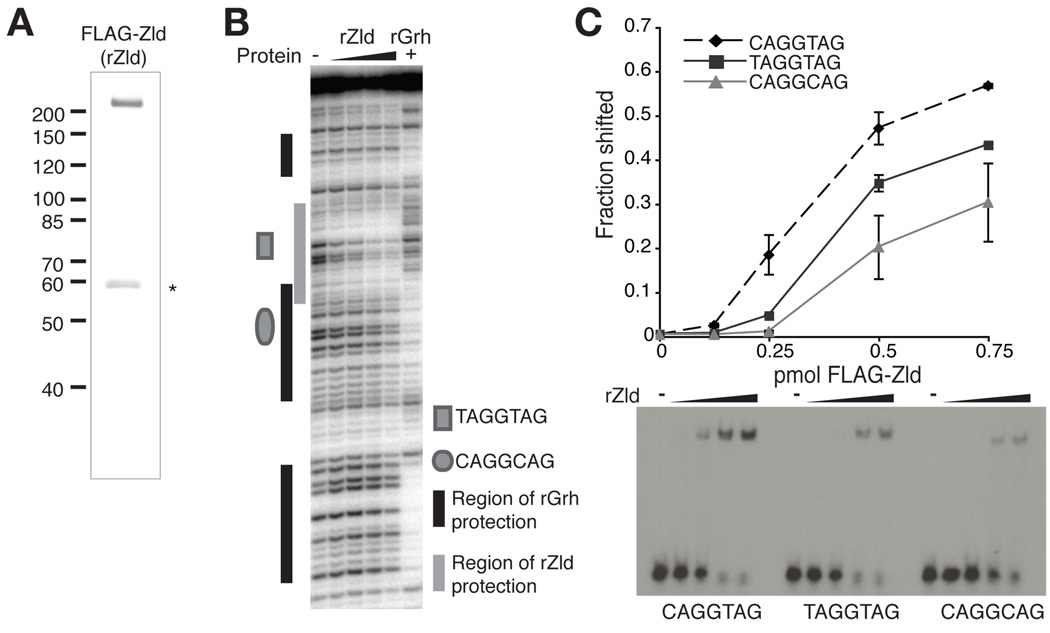
While we were characterizing the TAGteam-binding factor Grh, another TAGteam-binding protein Zelda (Zld), previously published as Vielfaltig (Staudt et al., 2006), was identified (Liang et al., 2008). Zld is a zinc-finger protein that binds to TAGteam elements in the zen VRE and is required for the proper activation of more than 100 genes in the pre-CB embryo (Liang et al., 2008). To test whether Grh and Zld have similar binding profiles for the zen VRE, we purified full-length recombinant Zld (rZld, Fig. 4A) and used it in DNase I protection assays. Interestingly, whereas Grh showed strong protection of the CAGGCAG element and little to no protection of the TAGGTAG element, Zld showed protection of the TAGGTAG and not the CAGGCAG element (Fig. 4B). To further determine if Zld and Grh had different binding affinities for TAGteam family members, we tested the affinity of Zld for distinct TAGteam elements using EMSAs. Similar to the binding profile for Grh, Zld bound most strongly to oligonucleotides containing the canonical CAGGTAG element (Fig. 4C). However, the affinity of Zld for the two additional TAGteam elements was reversed from that of Grh: Zld bound the TAGGTAG element more strongly than the CAGGCAG element. These data show that at least two TAGteam-binding factors are present in the early embryo, and that the affinities of these factors for various TAGteam elements differ. While it was previously unknown if all of the related TAGteam elements are equally effective in driving gene expression, our data demonstrating that the transcriptional activator Zld as well as the transcription factor Grh have different affinities for the related TAGteam sequences (Figs. 2 and 4) suggest that it is unlikely they are. Although all three TAGteam sequences are enriched in promoters of pre-CB expressed genes, their differential recognition by these two transcription factors may result in distinct effects on the levels or timing of gene expression.
Fig. 4.
Divergent TAGteam elements are bound by Grh and Zld with different affinities. (A) SDS-PAGE and silver stain analysis of the purified recombinant Zld. Molecular weight (left) is indicated in kD. The asterisk indicates a copurifying degradation product. (B) DNase I protection by rGrh or increasing amounts rZld on a fragment of the zen VRE. −, no protein. (C) Quantitation of EMSAs using rZld and probes containing different TAGteam elements. Data are the mean +/− s.d. from two independent EMSAs. A representative EMSA is shown below. −, no protein.
Because both Grh and Zld bind to TAGteam elements, we tested whether both proteins could bind these sequences simultaneously or whether instead they compete for binding. For these assays it was imperative that we be able to distinguish probe bound by Zld from that bound by Grh. While Grh is smaller than Zld, it binds DNA as a dimer (Attardi and Tjian, 1993; Uv et al., 1994), and binding of the Grh dimer in EMSAs resulted in shifted species that were difficult to distinguish from those shifted by Zld binding. We therefore expressed and purified a C-terminal portion of Grh containing the DNA-binding and dimerization domains (amino acids 603–1032) (Supplemental Fig. S5A) (Attardi and Tjian, 1993; Uv et al., 1994). This truncated form of Grh binds to TAGteam-containing sequences, but its binding is easily distinguishable by EMSA from that of Zld (Fig. 5A and Supplemental Fig. S5B). EMSAs performed with a probe containing two overlapping CAGGCAG elements from SxlPe, and low amounts of Grh(603–1032) resulted in a single shifted species. Increasing amounts of protein produced a slower migrating species, likely due to the binding of a second Grh dimer (Fig. 5A), suggesting that Grh binds to each of the CAGGCAG elements in the probe. Probes corresponding to portions of the zen VRE or sc promoter containing TAGteam elements yielded similar results (Supplemental Fig. S5B and data not shown). Higher levels of Grh(603–1032) were required to bind both TAGteam elements in the zen VRE probe than for the SxlPe or sc probes, as expected from our previous findings that Grh binds weakly to the TAGGTAG variant. The full-length rGrh protein showed similar binding behavior (Supplemental Fig. S6), suggesting that the DNA-binding and dimerization domains alone control binding-site specificity. Additionally, we note that attempts to co-immunoprecipitate full-length Grh and Zld from embryonic extracts or a mixture of purified epitope-tagged proteins were negative, indicating that interactions between the two full-length proteins through direct protein/protein interactions is not likely. Having shown that rGrh and Grh(603–1032) have similar binding profiles and binding of the truncated protein to oligonucleotide probes is easily distinguished from Zld binding, we used Grh(603–1032) in EMSAs to test for cooperativity or competition.
The minimal amount of rZld required to saturate binding and eliminate free probe was determined experimentally (data not shown). Reactions supplemented with increasing quantities of Grh(603–1032) showed reduced amounts of probe complexed with Zld and a concomitant increase in the amounts of probe bound by two Grh(603–1032) dimers (Fig. 5A and Supplemental Fig. S5B), demonstrating that Grh competes with Zld for TAGteam binding. As predicted from our previous results, Grh competed most weakly with Zld for binding to the zen VRE probe, which contains a TAGGTAG site to which Grh binds more weakly than Zld. Therefore Grh is capable of competing with the transcriptional activator Zld for binding to TAGteam sites, and these data suggest that Grh acts to repress transcription from TAGteam-containing promoters in the pre-CB embryo. Similarly, Grh has been shown to compete with an unidentified activator for binding to a TAGteam-related sequence in the dpp VRR (Huang et al., 1995). Thus, one possible mechanism for the previously reported Grh repression in the pre-CB embryo is competition with an activator for DNA binding (Huang et al., 1995; Liaw et al., 1995).
Given that at approximately equal molar amounts Grh and Zld compete for TAGteam binding, we determined the relative levels of each protein at different times during early embryonic development to learn whether they would have an opportunity to compete in the early embryo. Embryos were harvested at one-hour time intervals after egg laying (AEL), and levels of Grh and Zld were compared using quantitative immunoblots. Grh levels were constant in the early embryo (Fig. 5B). By contrast, levels of Zld were low in the 0–1 hour embryos and increased in the 1–2 hour embryos, when early gene expression initiates (Fig. 5B). To allow for a comparison between the relative amounts of each protein in the early embryo, approximate protein concentrations for Zld and Grh were determined by comparison of the immunoblot signals with the signal obtained from known amounts of recombinant protein. Each embryo contains approximately 1.8 × 109 molecules of Grh regardless of age, equating to ~9 × 108 molecules of Grh dimers with DNA-binding activity. This estimate is based on the fact that as determined by gel filtration chromatography little or no Grh protein exists as a monomer (Supplemental Fig. S7). Zld levels were ~7 × 108 molecules per embryo in the 0–1 hour embryo and increased to ~1.5 × 109 molecules per embryo in the 1–2 and 2–3 hour embryos. Thus, in the early embryo when there is no zygotic transcription, Grh levels are higher than Zld levels. These data, in combination with the fact that Grh may have a slightly higher affinity for CAGGTAG sites than Zld (compare EMSAs in Fig. 2C and 3C), suggest a model wherein Grh is likely bound to TAGteam elements and helps to maintain a transcriptionally silent state in the pre-CB embryo. At the time that early zygotic transcription initiates Zld levels have increased, raising the possibility that Zld now outcompetes Grh for TAGteam binding and thus helps drive gene expression.
Validating our suggestion that Grh is not an essential activator of early gene expression, maternal depletion of grh does not result in obvious defects in cellular blastoderm formation or viability (data not shown). Furthermore in situ hybridizations have not shown any obvious effects of maternal depletion or overexpression of Grh on the expression patterns of zen or tll in the stage 5 embryo (data not shown); it is unclear whether this is because grh expression is only being perturbed in the maternal germline. It is possible that premature expression of pre-CB genes resulting from the maternal depletion of grh will not result in a significant phenotypic consequence unless the embryo is subject to stress or Zld levels are perturbed, resulting in our failure to detect abnormal expression patterns for zen and tll. In addition, the extra maternally deposited Grh in the overexpression experiments may be overcome by the increase in Zld levels that occurs one hour after fertilization. Alternatively, the additional Grh binding sites in the pre-CB promoters might have other functions in regulating gene expression that confound these experiments where we observed the expression of the native genes. Importantly, we did not observe an expansion of tll expression in embryos maternally depleted for grh despite previously published reports to the contrary (Liaw et al., 1995). The only difference between our experiments and the published experiments were that we used the FLP-FRT system to generate embryos lacking maternal grh, while the previous work relied on X-ray induced mitotic recombination. Thus, we suggest that the expansion of tll observed by Liaw et al. (1995) may have been due to an unrelated defect caused by the irradiation.
We do note that overexpression of grh in the maternal germline leads to defects in nuclear division in the blastoderm embryo reminiscent of the defects observed in zld mutant embryos or when Zld levels are decreased by RNAi (Staudt et al., 2006), supporting our model that Grh acts as a transcriptional repressor by competing with Zld for DNA binding (Figs. 5D, E and Supplemental Fig S8). We detected anaphase bridges between dividing nuclei, aberrant cell divisions perpendicular to the normal plane of division, and a lack of synchronicity in cell divisions in about 50% of the blastoderm embryos generated from mothers of two different lines overexpressing grh in the maternal germline. None of these defects were noted in wild-type siblings. Interestingly, Grh overexpression in the pre-CB embryo resulted in ~50% reduction in hatching, indicating that these cell-division defects may ultimately decrease embryo viability (data not shown). These observations are consistent with the competition model suggested by our in vitro experiments. When Grh is overexpressed in the maternal germline the resulting abnormally high levels of Grh in pre-CB embryos may disrupt the ability of Zld to function properly in the very early embryo by competing for TAGteam-binding sites.
Conclusions
In summary, our data suggest that the concentrations of at least two TAGteam-binding factors (Grh and Zld), as well as the sequence variants of the TAGteam elements in the promoters, regulate gene expression in the pre-CB embryo, ensuring that transcription does not initiate prematurely. In its simplest form the model from our existing data is that Grh acts as to inhibit premature transcription in the pre-CB embryo during the first hour following fertilization by blocking the ability of Zld to bind to TAGteam sites and activate gene expression. As Zld levels increase during the second hour, Zld now successfully competes against the constant level of Grh for TAGteam binding and activates gene expression. This competition between Grh and Zld can ensure that despite minor fluctuations in Zld levels or other stochastic activating events, expression of pre-CB genes will not initiate prematurely. This model is supported by previous work showing that Grh binds to repressive elements in the tll and dpp promoters, mutation of the Grh binding site can cause an expansion of dpp expression, and Grh competes with an unidentified activator for binding to sites in the dpp promoter (Huang et al., 1995; Liaw et al., 1995). Furthermore, as Zld and Grh bind differentially to discrete TAGteam variants, activation at different promoters can be fine-tuned by the combination of TAGteam sequences present. This differential binding preference may explain, in part, how different pre-CB genes initiatetranscription at precise nuclear cycles. Thus Grh, Zld and the TAGteam elements could combinatorially regulate transcription in the pre-CB embryo, establishing the foundation for proper future embryonic development.
Supplementary Material
Acknowledgements
We are grateful to Jim Pesavento for performing the mass spectrometry and analysis. We thank R. Tjian and the Drosophila Genomic Resource Center for reagents. This work was supported by the National Institutes of Health grants GM23468 (to T.W.C.) and CA R37-30490 (to M.R.B.). M. M. H. was supported by the American Cancer Society Grant #PF-07-179-01-DDC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attardi LD, Tjian R. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 1993;7:1341–1353. doi: 10.1101/gad.7.7b.1341. [DOI] [PubMed] [Google Scholar]
- Attardi LD, Von Seggern D, Tjian R. Ectopic expression of wild-type or a dominant-negative mutant of transcription factor NTF-1 disrupts normal Drosophila development. Proc Natl Acad Sci U S A. 1993;90:10563–10567. doi: 10.1073/pnas.90.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Blastyak A, Mishra RK, Karch F, Gyurkovics H. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol Cell Biol. 2006;26:1434–1444. doi: 10.1128/MCB.26.4.1434-1444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ, Burke B, Brown NH, Hirsh J. Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 1989;3:1130–1145. doi: 10.1101/gad.3.8.1130. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Attardi LD, Admon A, Freeman M, Tjian R. Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 1989;3:1677–1688. doi: 10.1101/gad.3.11.1677. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Tamagishi M, Nishimoto Y, Taguchi O, Matsukage A, Yamaguchi M. A binding site for the transcription factor Grainyhead/Nuclear Transcription Factor-1 contributes to regulation of the Drosophila Proliferating Cell Nuclear Antigen gene promoter. J Biol Chem. 1999;274:35080–35088. doi: 10.1074/jbc.274.49.35080. [DOI] [PubMed] [Google Scholar]
- Huang JD, Dubnicoff T, Liaw GJ, Bai Y, Valentine SA, Shirokawa JM, Lengyel JA, Courey AJ. Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 1995;9:3177–3189. doi: 10.1101/gad.9.24.3177. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2009;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Rushlow CA, Zhou Q, Small S, Levine M. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. Embo J. 1992;11:3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Yamamoto KR, Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: evidence for a co-repressor in dorsal-mediated repression in Drosophila. Embo J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb MM, Laird CD. Increase in nuclear poly(A)-containing RNA at syncytial blastoderm in Drosophila melanogaster embryos. Dev Biol. 1976;52:31–42. doi: 10.1016/0012-1606(76)90004-x. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, Biggin MD. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw GJ, Rudolph KM, Huang JD, Dubnicoff T, Courey AJ, Lengyel JA. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308:381–385. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KA, Miller DF, Kaufman TC. Developmental distribution of RNA and protein products of the Drosophila alpha-tubulin gene family. Dev Biol. 1989;132:45–61. doi: 10.1016/0012-1606(89)90203-0. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OL., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Nakakura N, Miura T, Yamana K, Ito A, Shiokawa K. Synthesis of heterogeneous mRNA-like RNA and low-molecular-weight RNA before the midblastula transition in embryos of Xenopus laevis. Dev Biol. 1987;123:421–429. doi: 10.1016/0012-1606(87)90400-3. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Pilot F, Philippe JM, Lemmers C, Chauvin JP, Lecuit T. Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularisation. Development. 2006;133:711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell. 2006;17:2356–2365. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, Elias PM, Cunningham JM, Jane SM. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Tuckfield A, Clouston DR, Wilanowski TM, Zhao LL, Cunningham JM, Jane SM. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol Cell Biol. 2002;22:1936–1946. doi: 10.1128/MCB.22.6.1936-1946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uv AE, Harrison EJ, Bray SJ. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol Cell Biol. 1997;17:6727–6735. doi: 10.1128/mcb.17.11.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uv AE, Thompson CR, Bray SJ. The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol Cell Biol. 1994;14:4020–4031. doi: 10.1128/mcb.14.6.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 2003;31:4304–4316. doi: 10.1093/nar/gkg644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.