Abstract
Self-fertilization is a common mating system in plants and is known to reduce genetic diversity, increase genetic structure and potentially put populations at greater risk of extinction. In this study, we measured the genetic diversity and structure of two cedar glade endemic species, Leavenworthia alabamica and L. crassa. These species have self-incompatible (SI) and self-compatible (SC) populations and are therefore ideal for understanding how the mating system affects genetic diversity and structure. We found that L. alabamica and L. crassa had high species-level genetic diversity (He=0.229 and 0.183, respectively) and high genetic structure among their populations (FST=0.45 and 0.36, respectively), but that mean genetic diversity was significantly lower in SC compared with SI populations (SC vs SI, He for L. alabamica was 0.065 vs 0.206 and for L. crassa was 0.084 vs 0.189). We also found significant genetic structure using maximum-likelihood clustering methods. These data indicate that the loss of SI leads to the loss of genetic diversity within populations. In addition, we examined genetic distance relationships between SI and SC populations to analyze possible population history and origins of self-compatibility. We find there may have been multiple origins of self-compatibility in L. alabamica and L. crassa. However, further work is required to test this hypothesis. Finally, given their high genetic structure and that individual populations harbor unique alleles, conservation strategies seeking to maximize species-level genetic diversity for these or similar species should protect multiple populations.
Keywords: Leavenworthia, self-incompatibility, selfing, genetic diversity, mating system
Introduction
The evolution of self-fertilization from a predominantly outcrossing mating system is a common form of mating system evolution in plants (Stebbins, 1970) and has also occurred repeatedly in hermaphroditic animals (Jarne and Auld, 2006). Shifting to a self-fertilizing mating system increases homozygosity, reduces effective population sizes (Pollak, 1987; Schoen and Brown, 1991), and may reduce genetic diversity (Jarne and Städler, 1995; Hamrick and Godt, 1996; Nybom, 2004; Glémin et al., 2006). Given that the rate of evolution is a function of the level of genetic variance (Fisher, 1930), populations or species engaging in substantial inbreeding may be more likely to go extinct than those that outcross.
Inbreeding also changes the partitioning of genetic variation among populations thereby increasing genetic structure (Wright, 1951, 1965). In plants, species with mixed-mating or predominantly selfing mating systems have greater genetic differentiation among populations compared with outcrossers (Hamrick and Godt, 1996; Nybom, 2004). Selfing species may also experience recurrent extinction and recolonization events, which have important implications for species conservation because these metapopulation dynamics will further reduce genetic diversity and increase genetic structure (Ingvarsson, 2002). Furthermore, inbreeding may also have more pronounced negative effects in endemic plant species, which often have low genetic diversity and high structure compared with widespread congeners (Cole, 2003). It is therefore especially critical to understand the effects of mating system variation on the genetic diversity and structure of endemic, selfing plants.
A majority of the studies of the effect of mating system on population genetics have used a comparative approach; choosing closely related taxa differing only in mating system (for example, Fenster and Ritland, 1992; Charlesworth and Charlesworth, 1995; Williams et al., 2001). In this study, we take advantage of a mating system polymorphism in two highly endemic sister taxa: Leavenworthia alabamica and L. crassa to explicitly test the effects of mating system on genetic diversity and structure.
Leavenworthia has long been a genus of historical importance in plant evolutionary genetics with the work of Reed Rollins and David Lloyd. Within both L. alabamica and L. crassa there are self-incompatible (SI) and self-compatible (SC) populations (Rollins, 1963; Lloyd, 1965). The ancestral mating system in the family (Bateman, 1955) and genus (Lloyd, 1967; Beck et al., 2006) is sporophytic self-incompatibility, and its genetic control is likely the same as in other mustard genera (Busch et al., 2008). Within Leavenworthia, a small genus of eight annual species, self-fertilization has evolved at least four times and perhaps more if there were multiple origins of SC within L. alabamica and L. crassa (Rollins, 1963; Beck et al., 2006).
SC populations of L. alabamica and L. crassa set seed autogamously and have distinctive floral morphology from their SI counterparts (Lloyd, 1965; Lyons and Antonovics, 1991; Busch, 2005). Lloyd (1965) found that SC populations had smaller flowers, reduced anther-stigma distance, and shorter styles relative to SI populations, as well as anthers that dehisced toward rather than away from the stigma. On the basis of a combination of autogamy rates, floral traits and vegetative morphology, Lloyd (1965) divided populations of L. alabamica into 6 land races (1 SI and 5 SC), and L. crassa into 15 (4 SI and 11 SC). In addition to well-documented SI and SC in both species, L. alabamica and L. crassa are sister species (Beck et al., 2006) that share the same life-history, mode of pollination and method of seed dispersal (Rollins, 1963; Lloyd, 1965), making it possible to compare mating system effects without the confounding influence of these other factors. Thus, these sister species offer a unique opportunity to examine the parallel effects of mating system evolution on genetic diversity and structure of populations.
Although there have been previous studies of mating system and its effect on genetic diversity and structure in L. alabamica and L. crassa (Solbrig, 1972; Solbrig and Rollins, 1977; Charlesworth and Yang, 1998; Liu et al., 1998, 1999; Busch, 2005), their results may have been biased because of limited sampling both within and among populations. In this study, we use significantly more loci and larger population sample sizes to address the following questions: (1) Do SI and SC populations differ in genetic diversity? (2) How are populations structured genetically? (3) What are the genetic relationships between SI and SC populations? We then discuss these data with respect to the evolution of self-compatibility within L. alabamica and L. crassa and their conservation.
Materials and methods
Study species and population sampling
L. alabamica and L. crassa are limestone glade endemics within the Brassicaceae (Rollins, 1963; Beck et al., 2006). Limestone glades are characterized by thin calcareous soil over a dolomitic limestone base (Baskin et al., 1995). L. alabamica and L. crassa are winter annuals, growing during winter months when the soil is extremely wet. They germinate in the fall (September–November), with flowering and fruiting occurring mid-March to early May (Rollins, 1963; Lloyd, 1965). Both are pollinated by generalist bee species (Lloyd, 1965).
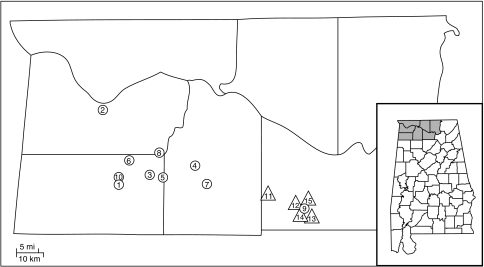
Glades of L. alabamica and L. crassa are patchily distributed within the Moulton and Tennessee Valleys of northwestern Alabama (Rollins, 1963). L. crassa occurs within two counties in the eastern Moulton Valley, whereas L. alabamica occurs in four counties, one of which (Colbert Co.) is located in the Tennessee Valley (Figure 1). Their ranges overlap in Morgan Co., where possible hybrids have been observed (Lloyd, 1965). We collected seeds from 10 populations of L. alabamica and 5 of L. crassa (Figure 1). Fewer populations were collected for L. crassa because of habitat destruction.
Figure 1.
Geographic distribution of sampled populations of L. alabamica and L. crassa within northwestern Alabama. L. alabamica populations (circles) are numbered 1–10: Isbell, Tuscumbia, Speck, Hatton, Winchell, Russell, Landers, StCross, CPCC and CR46, respectively. L. crassa populations (triangles) are numbered 11–15: CR203, CR55, Quarry, Bramlett and NCP, respectively. The inset (lower right) shows the location of the map area (shaded) within Alabama. L. alabamica's range extends from population 2 to 9, and L. crassa's from population 11 to 13.
Seeds were collected from individuals every few meters along transects spanning each population. The number of individuals sampled varied with population size (estimated visually). Populations tended to be either large (>1000 individuals) or small (<500 individuals). GPS coordinates were recorded and geographic distances calculated between populations using the software GENEPOP (Rousset, 2008).
We used the race maps from Lloyd (1965) to determine the breeding system of each population. For L. alabamica, races sampled were: a1=SI; a2=SC; a3=SC; Tuscumbia=SC; and Russellville=SC (Table 1). For L. crassa, we sampled four races: c1=SI; c3=SI; c5=SC; and c15=SC (Table 2). It is important to note that race c5 was in close geographic proximity to race c3 and that the two overlapped in some areas of Lloyd's map. Thus, it is possible that CR55 is actually race c3 because self-pollinations were not performed.
Table 1. Genetic diversity for Leavenworthia alabamica populations.
| Population | N | Population size | SI status | Lloyd's racea | Pp (%) | Total Ap | APp | Aep | Hop | Hep | Mean I |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Isbell | 30 | L | SI | a1 | 57.9 | 40 | 2.9 | 1.45±0.121 | 0.182±0.053 | 0.234±0.056 | 0.9295 |
| Speck | 23 | L | SI | a1 | 68.4 | 40 | 2.6 | 1.45±0.116 | 0.145±0.046 | 0.246±0.051 | 0.938 |
| Hatton | 112 | L | SI | a1 | 68.4 | 42 | 2.8 | 1.34±0.129 | 0.114±0.041 | 0.167±0.053 | 0.9191 |
| Winchell | 24 | L | SI | a1 | 57.9 | 38 | 2.7 | 1.42±0.124 | 0.137±0.042 | 0.222±0.054 | 0.9432 |
| StCross | 90 | L | SI | a1 | 63.2 | 37 | 2.5 | 1.24±0.070 | 0.110±0.031 | 0.156±0.039 | 0.9206 |
| CR46 | 65 | S | SI | a1 | 63.2 | 38 | 2.6 | 1.38±0.048 | 0.165±0.050 | 0.214±0.051 | 0.9291 |
| Landers | 177 | L | SC | a2 | 63.2 | 40 | 2.8 | 1.20±0.078 | 0.065±0.022 | 0.125±0.039 | 0.8309 |
| CPCC | 104 | S | SC | a3 | 52.6 | 31 | 2.2 | 1.07±0.056 | 0.055±0.050 | 0.038±0.027 | 0.909 |
| Tuscumbia | 46 | S | SC | Tuscumbia | 42.1 | 27 | 2 | 1.07±0.052 | 0.057±0.051 | 0.041±0.026 | 0.9114 |
| Russell | 32 | S | SC | Russellville | 31.6 | 25 | 2 | 1.09±0.053 | 0.059±0.049 | 0.057±0.029 | 0.9032 |
Abbreviations: Aep, mean effective number of alleles per locus; APp, mean number of alleles per polymorphic locus; Hep, expected heterozygosity (genetic diversity); Hop, observed heterozygosity; I, Nei's genetic identity; Pp, percent polymorphic loci; SC, self-compatible; SI, self-incompatible; total Ap, total number of alleles per population (including monomorphic loci).
Populations are ordered by size and self-incompatibility status. Standard errors are shown where available.
Table 2. Genetic diversity statistics for Leavenworthia crassa populations.
| Population | N | Population size | SI status | Lloyd's racea | Pp (%) | Total Ap | APp | Aep | Hop | Hep | Mean I |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCP | 255 | L | SI | c3 | 68.7 | 37 | 2.91 | 1.23±0.090 | 0.102±0.035 | 0.150±0.047 | 0.935 |
| CR203 | 57 | S | SI | c1 | 62.5 | 31 | 2.5 | 1.42±0.114 | 0.158±0.058 | 0.229±0.058 | 0.922 |
| CR55 | 148 | S | SC | c5 | 62.5 | 32 | 2.6 | 1.16±0.063 | 0.075±0.027 | 0.109±0.036 | 0.937 |
| Bramlett | 19 | S | SC | c15 | 25 | 21 | 2.25 | 1.09±0.057 | 0.010±0.005 | 0.061±0.035 | 0.874 |
| Quarry | 116 | S | SC | c15 | 25 | 22 | 2.5 | 1.14±0.080 | 0.039±0.031 | 0.082±0.042 | 0.863 |
Abbreviations: Aep, mean effective number of alleles per locus; App, mean number of alleles per polymorphic locus; Hep, expected heterozygosity (genetic diversity); Hop, observed heterozygosity; I, Nei's genetic identity; Pp, percent polymorphic loci; SC, self-compatible; SI, self-incompatible; total Ap, total number of alleles per population (including monomorphic loci).
Populations are ordered by size and self-incompatibility status. Standard errors are shown where available.
Genetic analyses
Seeds from each population were grown in The University of Georgia greenhouses. Seeds were planted in Fafard (Syngenta Group Co., Agawam, MA, USA) potting soil mixed with lime (approximately 128 g lime per bag), and rosettes were transplanted into 15 cm pots. Plants were watered daily and fertilized weekly with Peter's 20-10-20 Peat Lite Special at ∼250 p.p.m. Once rosettes were large enough, leaf tissue was snap frozen in liquid nitrogen and stored in a −80 °C freezer until extraction of allozyme proteins. Owing to variation in germination, sample sizes varied among populations (see Tables 1 and 2).
Each individual was crushed with a mortar and pestle, and an extraction buffer (Wendel and Parks, 1982) was added to solubilize and stabilize the enzymes. Enzyme extracts were absorbed onto chromatography paper wicks and stored at −80 °C until starch gel (11%) electrophoresis. We resolved the following enzymes for L. crassa: diaphorase (DIA), fluorescent esterase (FE), leucine-amino peptidase (LAP), malate dehydrogenase, malic enzyme, 6-phosphogluconate dehydrogenase, phosphoglucoisomerase (PGI), shikimate dehydrogenase, triose-phosphate isomerase (TPI) and UTP-glucose-1-phosphate (UGPP). These enzymes were also resolved for L. alabamica, including one additional enzyme: colorimetric esterase). For L. alabamica, we scored 19 loci on 11% starch gels using the following gel-electrode buffer combinations: buffer 34: (colorimetric esterase-1, colorimetric esterase-2, DIA-1, DIA-2, LAP-1, LAP-2, PGI-1, PGI-2, TPI-1, TPI-2 and UGPP); buffer 8–: FE-1, FE-3, FE-4, and malic enzyme); buffer 11: malate dehydrogenase, 6-phosphogluconate dehydrogenase-1, shikimate dehydrogenase-2, and UGPP). For L. crassa, we used four gel-electrode buffer combinations to score 16 loci: buffer 34: (DIA-1, DIA-2, LAP-1, LAP-2, PGI-1, PGI-2, TPI-1, TPI-2); buffer 4: (6-phosphogluconate dehydrogenase-1 and shikimate dehydrogenase-2); buffer 8–: (FE-1, FE-3, FE-4 and malic enzyme); buffer 11: (malate dehydrogenase and UGPP). Three additional loci were scored for L. alabamica that could not be resolved sufficiently for L. crassa. All loci were numbered sequentially, with the most anodal locus given the number one. The stain recipe for DIA is given in Cheliak and Pitel (1984), and for UGPP is given in Manchenko (1994). Recipes for all other stains and buffers are found in Soltis et al. (1983).
Statistical analyses
Measures of genetic diversity were calculated for L. alabamica and L. crassa (Hamrick and Godt, 1989), and for SI and SC populations (Hedrick, 2005) using GenAlEx 6.3 (Peakall and Smouse, 2006): percentage of polymorphic loci (P), mean number of alleles per locus (A) and per polymorphic locus (AP), effective number of alleles (Ae), and observed (Ho) and expected (He) heterozygosity. Population values (Tables 1 and 2) are given subscript ‘p'. We report the unbiased estimator of He (Nei and Chesser, 1983) in all tables. Following Schoen and Brown (1991), we also calculated the coefficient of variation for mean Hep of each species, and for mean Hep of SI and SC populations of each species.
We compared mean population values of L. alabamica and L. crassa for each genetic diversity measure using Welch's t-tests performed in JMP 6.0 (SAS Institute, Cary, NC, USA, 2005) as in Mable and Adam (2007). We also performed t-tests to examine mean differences in genetic diversity between SI and SC populations of each species. In addition, we used FSTAT (Goudet, 2001) to compare mean Hep values of SI and SC populations of each species. We used 1000 permutations of the data to compute the variance around Hep. As population size was highly correlated with SI status (Tables 1 and 2), its affect on genetic diversity was not examined separately. In addition, we tested deviations from Hardy–Weinberg equilibrium at each polymorphic locus, using GENEPOP (Rousset, 2008) to calculate the fixation index (F; Wright, 1922) and conduct Fisher's global exact tests (Guo and Thompson, 1992).
We computed Nei's genetic identity (I) and distance (D) values for each pair of populations (Nei, 1972) within species using GenAlEx 6.1 (Peakall and Smouse, 2006). We then used Nei's D to construct a UPGMA phenogram in PHYLIP 3.68 (Felsenstein, 2005) for each species to examine genetic distance relationships between populations. In addition, we tested isolation by distance within each species using Rousset's (1997) measure of genetic distance, FST/(1-FST), and calculated by GENEPOP (Rousset, 2008). A Mantel test was used to determine if there was a significant positive correlation between genetic and log-transformed geographic distance (Mantel, 1967).
To estimate the proportion of total genetic diversity found between populations at each polymorphic locus, we calculated between-population genetic structure (FST) of both species using AMOVA (Excoffier et al., 1992; Michalakis and Excoffier, 1996) in GenAlEx 6.1 (Peakall and Smouse, 2006). This was carried out for all populations of each species, as well as for SI and SC populations separately. Calculations were based on 16 and 12 polymorphic loci for L. alabamica and L. crassa, respectively. FST values were tested for significance against 9999 random permutations of the data.
We also examined genetic differentiation of L. alabamica and L. crassa populations using the clustering method of Pritchard et al. (2000) implemented in Structure 2.3.2. This program probabilistically assigns each individual to k clusters or populations, and individuals can be assigned to one or more populations as an indication of admixture. We ran the program using admixture and independent allele frequency models from k=1 to 20 for L. alabamica and k=1 to 10 for L. crassa, with 10 replicates at each k, and 100 000 MCMC reps after a burn-in period of 10 000. Log-likelihood values are assigned to each k, and we used the method described in the Structure manual to determine optimal k (that is, the optimal k has the smallest ln-likelihood value with the least variance between reps of the same k). We also used the alternative method of Evanno et al. (2005), which determines k using first- and second-order derivatives of the –ln probability of the data, as well as the second-order derivative divided by the s.d. For each species' optimal k, the program Distruct (Rosenberg, 2004) was used to show the clustering results. We then compared the results from Structure to those of Instruct (Gao et al., 2007), which assumes non-random mating and was designed for species with significant self-fertilization.
Finally, Filatov and Charlesworth (1999) found evidence that balancing selection maintains DNA nucleotide polymorphism at the cytosolic PGI locus in the closely related L. stylosa. Three polymorphisms conferred amino acid charge changes, although none were correlated with the allozyme mobility classes documented by Charlesworth and Yang (1998). Much lower levels of nucleotide polymorphism were observed in L. alabamica (Busch, 2005) and L. crassa (Liu et al., 1999) at this locus, but we cannot rule out the possibility that PGI is under balancing selection. To make sure that our results were not dependent on a locus experiencing selection, we re-ran all analyses with PGI excluded. Our results did not change, so we report only analyses with all loci included.
Results
Genetic diversity within species
For L. alabamica, 16 of 19 loci were polymorphic (84.2%) in at least one population, whereas L. crassa had a slightly lower number, 12 of 16 loci (75.0% Table 3). L. alabamica had AP=3.38, and L. crassa had AP=3.17. There were also differences in A (3.00 vs 2.63), Ae (1.47 vs 1.32), and He (0.229 vs 0.183). The species did not differ significantly in their mean population values for any measures of genetic diversity. The range of population values was larger in L. crassa for P, A and AP, but smaller for Ae and He. Range differences are likely due to the number of populations sampled for each species. For all species-level measures, L. alabamica had more genetic diversity than L. crassa.
Table 3. Comparisons of species-level diversity and genetic structure for Leavenworthia alabamica and L. crassa.
| P (%) | AP | A | Ae | He | FST | |
|---|---|---|---|---|---|---|
| L. alabamica | 84.2 | 3.38 | 3 | 1.47 | 0.229 | 0.45 |
| Mean population values | 56.8±3.75 | 2.51±0.104 | 1.88±0.070 | 1.27±0.050 | 0.150±0.015 | — |
| Range | 31.6–68.4 | 2.00–2.91 | 1.32–2.21 | 1.07–1.45 | 0.038–0.246 | — |
| L. crassa | 75 | 3.17 | 2.63 | 1.32 | 0.183 | 0.36 |
| Mean population values | 48.7±9.76 | 2.55±0.106 | 1.79±0.106 | 1.21±0.057 | 0.126±0.020 | — |
| Range | 25.0–68.7 | 2.25–2.91 | 1.31–2.31 | 1.09–1.42 | 0.061–0.229 | — |
Abbreviations: A, mean number of alleles per locus (including monomorphic loci); Ae, mean effective number of alleles per locus; AP, mean number of alleles per polymorphic locus; FST, among population genetic structure; He, mean expected heterozygosity (gene diversity); P, percent of polymorphic loci.
Standard errors are shown where available.
Genetic diversity within populations
We found that genetic diversity within populations depended on their SI status (Tables 1 and 2). For L. alabamica, populations designated by Lloyd (1965) as SC had significantly lower (P<0.05) Ae (1.11 vs 1.38), Hop (0.059 vs 0.142) and Hep (0.065 vs 0.206), but not P (47.37 vs 63.17), A (30.75 vs 39.17) or AP (2.24 vs 2.68) (Table 4). Hep was significantly different in both the JMP (t-test) and FSTAT (permutation) analysis. In line with expectations, the coefficient of variation of mean Hep was higher for SC populations than SI populations (62.37 vs 17.76%).
Table 4. Comparison of mean genetic diversity statistics for SI and SC populations of Leavenworthia alabamica and L. crassa.
| N | Pp (%) | Total Ap | APp | Aep | Hop | Hep | |
|---|---|---|---|---|---|---|---|
| L. alabamica | |||||||
| SI | 6 | 63.17±1.92 | 39.17±0.749 | 2.68±0.060 | 1.38±0.033 | 0.142±0.011 | 0.206±0.015 |
| SC | 4 | 47.37±6.80 | 30.75±3.31 | 2.24±0.177 | 1.11±0.062 | 0.059±0.002 | 0.065±0.020 |
| L. crassa | |||||||
| SI | 2 | 65.6±3.10 | 34.00±3.00 | 2.70±0.205 | 1.32±0.095 | 0.173±0.054 | 0.189±0.039 |
| SC | 3 | 37.5±12.50 | 25.00±3.51 | 2.45±0.104 | 1.13±0.021 | 0.055±0.043 | 0.084±0.014 |
Abbreviations: Aep, mean effective number of alleles per locus; App, mean number of alleles per polymorphic locus; Hep, expected heterozygosity (genetic diversity); Hop, observed heterozygosity; Pp, percent polymorphic loci; SC, self-compatible; SI, self-incompatible; total Ap, total number of alleles per populations (including monomorphic loci).
Means are shown with standard errors. The number of populations (N) is shown in the first column.
For L. crassa, the permutation analysis showed that SC populations had significantly (P<0.05) lower mean Hep. However, SI and SC populations did not differ significantly for any genetic diversity measure in the t-test analysis, although the mean value of all measures was lower in SC populations (Table 4). It seems likely that other measures in addition to Hep would have been significantly different between SI and SC populations had more L. crassa populations been sampled.
Within L. alabamica, observed genotype frequencies conformed to Hardy–Weinberg expectations in all populations for only 12.5% of polymorphic loci. There were 68 cases (62.9%) of fixation indices significantly different from zero (P<0.05); 62 were cases of heterozygote deficiency and 6 were cases of heterozygote excess. Overall, FIS for L. alabamica was 0.290, and 0.320 and 0.285 for only SI and SC populations, respectively. Observed genotype frequencies of L. crassa conformed to Hardy–Weinberg expectations in all populations for only 16.7% of polymorphic loci. Fixation indices significantly differed from zero (P<0.05) in 28 cases (71.8%), with 25 cases of heterozygote deficiency and three cases of heterozygote excess. For L. crassa, overall FIS was 0.364, and 0.340 and 0.410 for only SI and SC populations, respectively. These data indicate that most loci show inbreeding in all populations of both species.
Nine (mean frequency=0.120) and six (mean frequency=0.193) private alleles were within five L. alabamica and four L. crassa populations, respectively. In L. alabamica, Hatton had four private alleles, followed by Landers (two), with Speck, StCross and CPCC each having one private allele. L. crassa populations with private alleles were CR55 (two), NCP (two), CR203 (one) and Bramlett (one). Interestingly, one private allele in CR55 is also only found in CPCC in L. alabamica. CPCC (Pop. 9) is an isolated L. alabamica population within close geographic proximity to populations of L. crassa (Figure 1). The presence of this allele in one L. alabamica and one L. crassa population may suggest gene flow has occurred.
Genetic structure and differentiation
The proportion of total genetic variation explained by differences between populations (FST) of L. alabamica was 0.450 (Table 3) and was significantly different from zero. This value drops to 0.174 and rises to 0.693 when considering only SI and SC populations, respectively. Mean genetic identity among populations of L. alabamica was 0.913±0.010, with a range from 0.831 to 0.943 (Table 1). Landers had the lowest mean genetic identity with other L. alabamica populations. Pairwise population estimates of FST ranged from 0.042 (Speck and Winchell) to 0.714 (Landers and CPCC). A significant positive correlation between genetic and geographic distance was detected (P=0.008), indicating that there is isolation by distance between L. alabamica populations.
For L. crassa, FST was 0.358 (Table 3), which was significantly different from zero. As for L. alabamica, this value drops to 0.235 and rises to 0.577 when considering only SI and SC populations, respectively. Mean genetic identity among populations of L. crassa was 0.906±0.016, and ranged from 0.863 to 0.937 (Table 2). Quarry and Bramlett had the lowest mean genetic identity with other populations. Pairwise FST values ranged from 0.046 (CR55 and NCP, populations in closest geographic proximity) to 0.758 (Quarry and Bramlett, both of the highly SC race c15). We did not detect a significant correlation between genetic and geographic distance in L. crassa (P>0.1), suggesting only weak isolation by distance. However, this may be an artifact of the limited number of populations sampled for L. crassa.
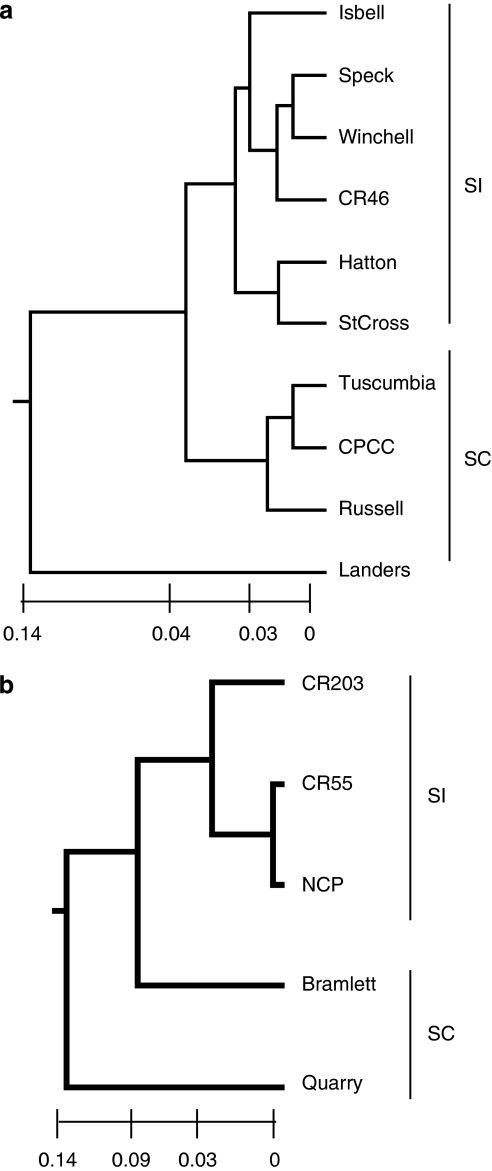
We also examined population genetic differentiation using Nei's I and D, and used the D values to construct a UPGMA phenogram for each species (Figure 2). Mean I was 0.924±0.009 and 0.922±0.021 for L. alabamica and L. crassa populations, respectively.
Figure 2.
UPGMA phenograms of 10 L. alabamica and 5 L. crassa populations based on Nei's (1972) genetic distance values: (a) L. alabamica populations, (b) L. crassa populations. Branch lengths represent genetic distances (see scale). SI=self-incompatible, SC=self-compatible.
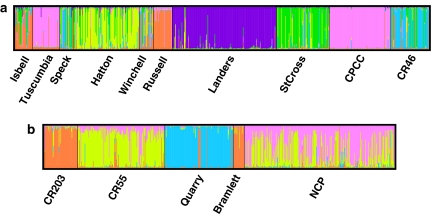
The Structure analysis yielded optimal k=6 and k=4 for L. alabamica and L. crassa, respectively (Figure 3). The Evanno et al. (2005) methods did not show clear peaks for our data (results not shown), but did show the greatest rate of change in the likelihood values at k=6 and k=4, and these results were also obtained by using the Structure manual method. In L. alabamica the first cluster was formed by Isbell, Winchell and Russell, the second by Tuscumbia and CPCC, the third by Speck and CR46, the fourth by Hatton, the fifth by Landers and the sixth by StCross. In L. crassa the first cluster was formed by CR203 and Bramlett, the second by CR55, the third by Quarry and the fourth by NCP. Instruct (Gao et al., 2007) identified the same clusters at these k (results not shown).
Figure 3.
Bar plot of assignment of individual genotypes to each of k sub-populations using the clustering method of the program Structure: (a) L. alabamica individuals were assigned to six sub-populations, (b) L. crassa individuals were assigned to four sub-populations. The different colors in each plot represent the k sub-populations. Physical populations sampled for each species are also shown.
Discussion
Inbred populations generally have less genetic diversity (Jarne and Städler, 1995; Hamrick and Godt, 1996; Nybom, 2004; Glémin et al., 2006), potentially putting populations at increased risk of extinction. Increased population genetic structure is also associated with inbreeding (Hamrick and Godt, 1996; Nybom, 2004) and that population extinction could cause the loss of genetic variation within species. To better understand the genetic consequences of mating system change on endemic plants, we examined its population genetic effects in the cedar glade endemics L. alabamica and L. crassa. We also looked at the genetic relationship between SI and SC populations to make inferences regarding population history and the evolution of SC populations.
The genetic consequences of mating system change in Leavenworthia were examined previously, but with a number of limitations. Solbrig (1972) first compared genetic diversity in SI and SC species using allozymes. He reported a weak association between self-compatibility and lower allozyme diversity. However, he used few populations and an inaccurate measure of genetic diversity (Berg and Hamrick, 1997), reporting the number of homozygous individuals versus those showing some segregation in a population rather than identifying alleles and quantifying expected genetic diversity. Solbrig and Rollins (1977) sampled more populations, but used the same measure of genetic diversity as Solbrig (1972). It is therefore difficult to interpret the results of these studies.
Charlesworth and Yang (1998) also estimated genetic diversity in SI and SC species of Leavenworthia, but found unexpected results. The SI L. stylosa and SC L. uniflora and L. torulosa had, respectively, high and low measures of heterozygosity as expected, but selfing populations of L. alabamica and L. crassa had mainly high heterozygosity (ranging from ∼0.2 to 0.4); measures close to those of outcrossing populations (∼0.43–0.47). In addition, genetic structure was unexpectedly low for L. alabamica and L. crassa. However, sample sizes were limited.
In contrast, Liu et al. (1998, 1999) found low DNA sequence diversity in selfing L. crassa populations at three alcohol dehydrogenase loci and one locus of phosphoglucoisomerase, respectively. Busch (2005) analyzed the phosphoglucoisomerase locus of three populations of L. alabamica (one SI and two SC) and found significantly less sequence variation in the SC populations. Although they used only a few loci, the results were consistent with less genetic diversity occurring within selfing populations.
Genetic diversity within populations
We found less neutral genetic diversity in SC populations compared with SI populations for most measures (Table 4), a result consistent with inbreeding in SC populations. Our results differ from Charlesworth and Yang (1998) in that we found much lower expected heterozygosity in both SI and SC populations (Tables 1 and 2). This difference likely stems from the fact that we sampled more and different loci, and that we had larger sample sizes for most populations. Furthermore, our result agrees with low DNA sequence diversity in SC populations of L. crassa (Liu et al., 1998, 1999) and L. alabamica (Busch, 2005), and with Mable and Adam's (2007) study of genetic diversity in Arabidopsis lyrata, another mustard species with both SI and SC populations.
We found high fixation indices in both SI and SC populations, with most loci showing heterozygote deficiency and only a few cases of heterozygote excess. Charlesworth and Yang (1998) also found high fixation indices in SI and SC populations of L. alabamica and L. crassa, suggesting that SI populations may experience inbreeding high enough to reduce heterozygosity. This is possible given that low levels of autogamy and/or pseudo-compatibility are documented in SI races of L. alabamica and L. crassa (Lloyd, 1965, 1968; Busch et al., 2010). Pseudo-compatibility can fluctuate depending on environmental conditions (Levin, 1996); in some years SI populations may produce a significant number of progeny through self-fertilization.
Genetic diversity and structure within species
L. alabamica and L. crassa showed similarly high levels of genetic diversity (Table 3) and high genetic structure between populations (see FST values in Table 3 and clustering results in Figure 3). Our estimates of genetic structure are higher than those of Charlesworth and Yang (1998) and suggest that gene flow between populations is low. Given that limestone glades are geographically disjunct, the average distance between populations was approximately 10 km (Figure 1), and seeds of these species are gravity-dispersed, low gene flow between populations is expected. These genetic structure values also agree with those found in other annual species with gravity-dispersal and selfing (Hamrick and Godt, 1996). However, species-level genetic diversity for both L. alabamica and L. crassa was higher than most plant species with similar life-history traits. It seems likely that the high levels of genetic diversity observed are due to the maintenance of allelic variation in SI populations, variation that may then spread to SC populations through occasional gene flow events.
Genetic relationships between populations and the evolution of self-compatibility
Our evidence suggests that there may have been multiple origins of self-compatibility in L. alabamica and L. crassa. In Figure 2a, the L. alabamica selfing populations Tuscumbia, Russell and CPCC clustered together and Landers was the most genetically distant from all populations. In the Structure analysis (Figure 3a), Tuscumbia and CPCC formed a cluster, whereas Russell was more closely allied to SI populations (Isbell, Speck and Winchell) and Landers formed a distinct cluster. A large geographic distance separates Tuscumbia and CPCC (Figure 1), making their clustering unlikely to result from current gene flow. The two SC populations sampled in L. crassa were not genetically similar (Figures 2b and 3b). These results are consistent with multiple origins of self-compatibility in these species. Multiple origins of self-compatibility have also been found in Arabidopsis lyrata (Hoebe et al., 2009). However, the observed genetic distance between these populations may be due to local drift and/or selection post-establishment and does not reflect population phylogeny. As we cannot determine ancestral relationships from our allozyme data set, further work using markers suited to phylogeographic questions, such as cpDNA (Avise, 2000), is needed to truly test whether self-compatibility has evolved repeatedly within these two species.
Implications for conservation
In both L. alabamica and L. crassa, SI populations had high genetic diversity and SC populations low. In addition, populations were highly genetically structured and likely experience infrequent gene flow. However, private alleles were present in some populations, two of which were SC, showing that individual populations contain unique genetic variation. In addition, previous work has shown that SI and SC populations differ substantially in floral morphology (Lloyd, 1965; Lyons and Antonovics, 1991; Busch, 2005), suggesting that there are at least some quantitative genetic differences between populations with different mating systems. Thus, the overall genetic diversity of L. alabamica and L. crassa depends on the conservation of multiple populations of both mating types, and any efforts to conserve L. alabamica or L. crassa should consider these population genetic factors when designing a conservation strategy.
Acknowledgments
We thank the Bramlett family and the Nature Conservancy of Alabama for access to property, J Busch, T Haselkorn, S Scott, J Estill, L Mitchell and M Brown for help with seed collections, E Hedgepeth and J Wolf for aid with gel electrophoresis and G Schneider for map-making assistance. We also thank two anonymous reviewers for comments on the paper. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation. A Ruth L Kirschstein National Research Service Award from the National Institutes of Health to VAK funded this work.
The authors declare no conflict of interest.
References
- Avise JC. Phylogeography. Harvard University Press: Cambridge; 2000. [Google Scholar]
- Baskin JM, Webb DH, Baskin CC. A floristic plant ecology study of the limestone glades of northern Alabama. Bull Torrey Bot Club. 1995;122:226–242. [Google Scholar]
- Bateman AJ. Self-incompatibility systems in angiosperms III. Cruciferae. Heredity. 1955;9:53–68. [Google Scholar]
- Beck JB, Al-Shehbaz IA, Schaal BA. Leavenworthia (Brassicaceae) revisited: testing classic systematic and mating system hypotheses. Syst Bot. 2006;31:151–159. [Google Scholar]
- Berg EE, Hamrick JL. Quantification of genetic diversity at allozyme loci. Can J For Res. 1997;27:415–424. [Google Scholar]
- Busch JW. The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae) Am J Bot. 2005;92:1503–1512. doi: 10.3732/ajb.92.9.1503. [DOI] [PubMed] [Google Scholar]
- Busch JW, Joly S, Schoen DJ.2010Does mate limitation in self-incompatible species promote the evolution of selfing? The case of Leavenworthia alabamica Evolutionin press; 10.1111/j.1558-5646.2009.00925.x [DOI] [PubMed]
- Busch JW, Sharma J, Schoen DJ. Molecular characterization of Lal2, an SRK-like gene linked to the S-locus in the wild mustard Leavenworthia alabamica. Genetics. 2008;178:2055–2067. doi: 10.1534/genetics.107.083204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Quantitative genetics in plants: the effect of the breeding system on genetic variability. Evolution. 1995;49:911–920. doi: 10.1111/j.1558-5646.1995.tb02326.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth DB, Yang Z. Allozyme diversity in Leavenworthia populations with different inbreeding levels. Heredity. 1998;81:453–461. doi: 10.1046/j.1365-2540.1998.00415.x. [DOI] [PubMed] [Google Scholar]
- Cheliak WM, Pitel JA.1984Techniques for Starch Gel Electrophoresis of Enzymes from Forest Tree Species Petawawa National Forestry Institute, Canadian Forestry Service, Agriculture: Chalk River, Ontario; Information report P1-X-42. [Google Scholar]
- Cole CT. Genetic variation in rare and common plants. Annu Rev Ecol Evol S. 2003;34:213–237. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.2005PHYLIP (Phylogeny Inference Package) version 3.6Distributed by the author.Department of Genome Sciences, University of Washington: Seattle [Google Scholar]
- Fenster CB, Ritland K. Chloroplast DNA and isozyme diversity in two Mimulus species (Scrophulariaceae) with contrasting breeding systems. Am J Bot. 1992;79:1440–1447. [Google Scholar]
- Filatov DA, Charlesworth D. DNA polymorphism, haplotype structure and balancing selection in the Leavenworthia PgiC locus. Genetics. 1999;153:1423–1434. doi: 10.1093/genetics/153.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press: Oxford; 1930. [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov Chain Monte Carlo approach for joint Inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Bazin E, Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. P Roy Soc B-Biol Sci. 2006;273:3011–3019. doi: 10.1098/rspb.2006.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J.2001FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3)Available from http://www.unil.ch/izea/softwares/fstat.html .
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW.1989Allozyme diversity in plant speciesIn: AHD Brown, MT Clegg, AL Kahler, BS Weir (eds).Plant Population Genetics, Breeding and Genetic Resources Sinauer: Sunderland, MA; 43–63. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philos T Roy Soc B. 1996;351:1291–1298. [Google Scholar]
- Hedrick PW. Genetics of Populations. Jones and Bartlett: Boston; 2005. [Google Scholar]
- Hoebe PN, Stift M, Tedder A, Mable BK. Multiple losses of self-incompatibility in North-American Arabidopsis lyrata?: phylogeographic context and population genetic consequences. Mol Ecol. 2009;18:4924–4939. doi: 10.1111/j.1365-294X.2009.04400.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution. 2002;56:2368–2373. doi: 10.1111/j.0014-3820.2002.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Jarne P, Auld JR. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution. 2006;60:1816–1824. doi: 10.1554/06-246.1. [DOI] [PubMed] [Google Scholar]
- Jarne P, Städler T. Population genetic structure and mating system evolution in freshwater pulmonates. Cell Mol Life Sci. 1995;51:482–497. [Google Scholar]
- Levin DA. The evolutionary significance of pseudo-self-fertility. Am Nat. 1996;148:321–332. [Google Scholar]
- Liu F, Charlesworth DB, Kreitman M. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics. 1999;151:343–357. doi: 10.1093/genetics/151.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zhang L, Charlesworth DB. Genetic diversity in Leavenworthia populations with different inbreeding levels. P Roy Soc B-Biol Sci. 1998;265:293–301. doi: 10.1098/rspb.1998.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG. Evolution of self-compatibility and racial differentiation in Leavenworthia (Cruciferae) Contrib Gray Herb Harv. 1965;195:3–134. [Google Scholar]
- Lloyd DG. The genetics of self-incompatibility in Leavenworthia crassa Rollins (Cruciferae) Genetica. 1967;38:227–242. [Google Scholar]
- Lloyd DG. Pollen tube growth and seed set in self-incompatible and self-compatible Leavenworthia (Cruciferae) populations. New Phytol. 1968;67:179–195. [Google Scholar]
- Lyons EE, Antonovics J. Breeding system evolution in Leavenworthia: breeding system variation and reproductive success in natural populations of Leavenworthia crassa (Cruciferae) Am J Bot. 1991;78:270–287. [Google Scholar]
- Mable BK, Adam A. Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Mol Ecol. 2007;16:3565–3580. doi: 10.1111/j.1365-294X.2007.03416.x. [DOI] [PubMed] [Google Scholar]
- Manchenko GP. Handbook of Detection of Enzymes on Electrophoretic Gels. CRC Press: Ann Arbor; 1994. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Michalakis Y, Excoffier L. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996;142:1061–1064. doi: 10.1093/genetics/142.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. [Google Scholar]
- Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann Hum Genet. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics. 1987;117:353–360. doi: 10.1093/genetics/117.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RC. The evolution and systematics of Leavenworthia (Cruciferae) Contrib Gray Herb Harv. 1963;192:3–98. [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Brown AHD. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc Natl Acad Sci USA. 1991;88:4494–4497. doi: 10.1073/pnas.88.10.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbrig OT. Breeding system and genetic variation in Leavenworthia. Evolution. 1972;26:155–160. doi: 10.1111/j.1558-5646.1972.tb00182.x. [DOI] [PubMed] [Google Scholar]
- Solbrig OT, Rollins RC. Evolution of autogamy in species of the mustard genus Leavenworthia. Evolution. 1977;31:265–281. doi: 10.1111/j.1558-5646.1977.tb01007.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Haufler CH, Darrow DC, Gastony GJ. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. Am Fern J. 1983;73:9–27. [Google Scholar]
- Stebbins GL. Adaptive radiation of reproductive characteristics in angiosperms. I. Pollination mechanisms. Annu Rev Ecol Evol S. 1970;1:307–326. [Google Scholar]
- Wendel JF, Parks CR. Genetic control of isozyme variation in Camellia japonica L. J Hered. 1982;73:197–204. [Google Scholar]
- Williams CF, Ruvinsky J, Scott PE, Hews DK. Pollination, breeding system, and genetic structure in two sympatric Delphinium (Ranunculaceae) species. Am J Bot. 2001;88:1623–1633. [PubMed] [Google Scholar]
- Wright S. Coefficient of inbreeding and relationships. Am Nat. 1922;56:330–338. [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]