Abstract
Type I interferons play an important role in the early defense against viral and other pathogens. These innate responses are also critically important in shaping the subsequent adaptive response. Thus, a more thorough knowledge of innate response types and mechanisms will improve our understanding of pathogenesis and guide the development of new therapeutics.
Interferon alpha is used clinically in the treatment of HIV and Hepatitis C infections. The majority of IFA-α therapies is based on a single IFN-α subtype, IFN-α2. However, IFN-α comprises a family of multiple subtypes. The biological functions of the distinct subtypes and how they relate to disease are poorly understood.
The current study developed the tools to distinguish and measure multiple IFN-α subtypes on the mRNA level in rhesus macaques that are used widely as an important animal model for human diseases. We were able to identify and measure nine distinct rhesus IFN-α subtypes.
Further, we could demonstrate that in response to oral pathogenic SIV infection, several IFN-α subtypes are rapidly induced in lymphoid but not at oral and gastrointestinal mucosal surfaces. While each IFN-α subtype was induced at distinct levels, their relative expression patterns were identical in all lymphoid tissues examined.
Keywords: IFN alpha subtypes, SIV infection, lymphoid and mucosal tissues
Introduction
Type I interferons have been known for more than 50 years when it was first discovered that virally infected cells secrete a substance that prevents the infection of other cells with virus [1]. We now know that this “substance”, called interferon, in fact is comprised of a mixture of multiple subtypes that include alpha, beta, omega, kappa, epsilon, tau and zeta interferon. In humans, IFN-α represents a family of 21 IFN-α subtypes. While some IFN-α genes are pseudogenes, the majority of them are expressed at the protein level, and all bind to the same receptor.
The antiviral activity of IFN-α and IFN-β has been extensively studied, but many open questions remain. Why do multiple IFN-α subtypes exist? What regulates the biological activity of all the diverse type I interferons when they all bind and signal through the same receptor? Different pathogens can induce distinct IFN-α subtypes in vitro [2–5], and IFN-α subtypes can differ in their antiviral activity. However, only few in vivo studies have assessed the functional activity of individual IFN-α subtypes and their roles during infection. Mice expressing an IFN-α1 transgene had significantly lower virus titers after murine CMV (MCMV) infection than mice transfected with the IFN-α4 or IFN-α9 gene [6]. In another murine study, acute CMV-induced myocarditis was most efficiently reduced with IFN-α9 or IFN-β [7]. Murine IFN-α1 was also highly effective in reducing HSV-induced ocular infection [8]. In contrast, IFN-α1 exerted less antiviral activity in a murine model of influenza infection when compared to IFN-α5 and IFN-α6 [9]. Recently, it was demonstrated that IFN-α1, 4 and 9, but not IFN-α6 were able to reduce virus replication in the murine model of Friends retrovirus infection [10]. How these results translate to human diseases is unknown.
Current IFN-α based clinical treatments like pegylated interferon (e.g. Pegasys) and RoferonA, often used to combat Hepatitis C infection, are based on the application of a single IFN-α subtype, IFN-α2. An exception is the use of Multiferon™. This is despite the fact that in human patients infected with Hepatitis C, IFN-α5 appears to be the most prominent IFN-α subtype expressed in the liver [4]. Based on the few, but striking results demonstrating differences in antiviral activity by various IFN-α subtypes, and considering the central role of type I interferons in the early immune defense and in directing T and B cell differentiation, a more comprehensive understanding of IFN-α subtypes in different disease settings is needed to determine whether distinct IFN-α subtype treatments might be beneficial in targeting distinct types of infections.
We previously showed that IFN-α mRNA is rapidly induced after oral SIV infection in infant rhesus macaques [11]. Despite similar high levels of virus replication in lymphoid and mucosal tissues, we observed significantly higher increases in IFN-α mRNA levels in lymphoid compared to mucosal tissues [11]. As the early antiviral response at entry sites of the virus is critically important in the early control of virus replication, a thorough analysis of the various IFN-α subtypes and their expression patterns in mucosal compared to lymphoid tissues is warranted.
Therefore, we conducted the study presented here, in which we developed the tools to distinguish and measure multiple IFN-α subtypes at the mRNA level in rhesus macaques, an animal species used as a model system for many human diseases. We then determined which IFN-α subtypes were induced in response to pathogenic SIV infection and/or whether distinct tissues show unique IFN-α expression patterns.
The results show that following SIV infection of infant rhesus macaques several IFN-α subtypes are rapidly induced in lymphoid but not at oral and gastrointestinal mucosal surfaces. While each IFN-α subtype was induced at distinct levels, their relative expression patterns were identical in all lymphoid tissues examined.
Materials and Methods
Animals and SIV infection
Newborn rhesus macaques (Macaca mulatta) were housed and hand-reared in a primate nursery in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care Standards at the California National Primate Research Center (CNPRC). Animal procedures were approved by the UC Davis Institutional Animal Use and Care Committee (IACUC). All animals were born to rhesus macaques of the CNPRC colony and were negative for HIV-2, SIV, type D retrovirus, and simian T-cell lymphotropic virus type 1. Six infant macaques were exposed to SIVmac251 (obtained from the CNPRC Analytical Core) using a previously described repeated oral exposure model [11, 12]. The infected animals were euthanized 3 days after the last SIVmac251 exposure, i.e. day 8 after the first virus exposure. The time of euthanasia is subsequently referred to as 1 week post-infection (p.i.). Six age-matched SIV-naive macaques served as controls.
Tissue collection and cell isolation
At the time of euthanasia, blood, gingiva, tonsil, retropharyngeal and submandibular lymph nodes (LN), jejunum, colon, mesenteric LN, and axillary LN samples were collected. Tissue samples were stored in RNAlater (Ambion, Austin, TX) at −20°C until RNA preparation. In addition, cell suspensions for functional assays were prepared as described [11]. Cell isolation from intestinal tissues was performed according to previously described methods [13]. Briefly, ~2 inch pieces of the ileum and colon were rinsed with PBS and then minced using sterile scalpels. The tissue suspensions were placed in a shaking waterbath in RPMI 1640 containing 7.5% FBS and collagenase type II (0.5 mg/ml) for 30 minutes at 37°C. After the digestion, the single cell suspension was passed through a 100 μm filter, spun down and resuspended in 10% FBS in RPMI 1640. The remaining undigested tissue was resuspended in collagenase-media and the digestion step was repeated a total of 3 times. Mucosal lymphocytes were then isolated from the obtained single cell suspension by performing a 35%/60% Percoll (Sigma) gradient centrifugation. Intestinal lymphocytes were collected from the 35%/60% interface and washed twice with PBS before being resuspended in 10% FBS in RPMI 1640.
RNA isolation and cDNA preparation
Prior to total RNA isolation, using Trizol (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol, the tissue samples were homogenized using a Power Homogenizer (PowerGen 7 mm × 195 mm; Fisher Scientific). RNA was used to determine tissue viral RNA (vRNA) levels and for gene expression analysis. RNA samples were DNase-treated with DNA-free (Ambion) for 1 hr. at 37°C. Complementary DNA (cDNA) was prepared using random hexamer primers (Amersham-Pharmacia Biotech, Inc., Piscataway, NJ) and M-MLV-Reverse Transcriptase (Invitrogen). Due to the low expression levels of some IFN-α subtype mRNA levels in tissues of SIV-naïve control animals, cDNA pre-amplification was performed using the ABI TaqMan Pre-Amp Master Mix Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions.
Interferon–α subtype cloning and primer/probe design
At the initiation of the study, the rhesus macaque genome was not fully sequenced. However, we recently developed a nested PCR strategy to clone the human IFN-α subtypes [14]. Based on the strategy applied to human IFN-α subtype cloning and the available sequence information listed on the Baylor College of Medicine Rhesus Macaque Sequencing Project website, rhesus IFN-α subytpe primers located in the 5′ and 3′ UTR regions were designed (see Results). The PCR was carried out using the Stratagene Easy A Hi Fidelity PCR cloning enzyme under the following conditions: 1 min. at 94°C, 30 cycles at 94°C for 15 sec, 55°C for 15 sec, 70°C for 45 sec, followed by 70°C for 5 min. The obtained PCR products were cloned using Invitrogen’s TOPO TA cloning kit (TOPO TA Cloning® Kit with One Shot® MAX Efficiency™ DH5α-T1R E. coli). For each IFN-α subtype, a minimum of three clones were sequenced and compared for sequence homology using BioEdit Software [15]. Once the sequence was confirmed to be IFN-α subtype specific, inner primer/probe sets were designed using the previous described criteria for the amplification of human IFN-α subtypes [14]. The specificity of IFN-α subtype specific primer-probe sets was confirmed by using 0.01 ng DNA of each cloned IFN-α subtype (plasmids) for amplification with matched and all non-matched primer/probe sets.
Real-Time (RT) PCR was performed as previously described [11, 16]. Briefly, samples were tested in duplicate, and the RT-PCR for the housekeeping gene GAPDH and the target gene from each sample were run in parallel on the same 96 well Optical Plate (Applied Biosystems, Foster City, CA) in a 25 μl reaction volume containing 5 μl cDNA + 20 μl Mastermix (Applied Biosystems). All sequences were amplified using the 7900 default amplification program: 2 min. at 50°C, 10 min. at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min. at 60°C. Results were analyzed with the SDS 7900 system software, version 2.1. (Applied Biosystems). In this analysis, the Ct value for the housekeeping gene (GAPDH) was subtracted from the Ct value of the target gene (delta, ΔCt) to normalize for mRNA input. The average ΔCt value for the tissue sample from all uninfected animals was then subtracted from the delta Ct value of the corresponding tissue sample from each of the SIV infected monkey (ΔΔCt). Since the target gene (cytokine) and the reference gene (GAPDH) are amplified with the same efficiency, and GAPDH values did not differ depending on the infection status of animals (data not shown), the increase in cytokine mRNA levels in tissue samples of SIV infected monkeys compared to tissue samples of uninfected animals could then be calculated as: Increase = 2−ΔΔCt (User Bulletin #2, ABI Prism 7700 Sequence Detection System (Applied Biosystems).
Immunochistochemistry (IHC) for IFN-α and MxA protein detection
Tissue section slides were stained using the DAKO Autostainer as follows: Formalin-fixed, paraffin-embedded tissue sections were cleared and rehydrated with xylene (3x), followed by a graded series of ethanol incubations (100%, 95%, 80%, and 50%), brief rinsing with water and then with Tris-buffered saline containing Tween 20 (TBST; Dako). For IFN-α detection, slides underwent antigen retrieval with AR10 (Biogenex, diluted 1:10) in a pressure cooker, followed by a rinse with water and TBST. Slides for both IFN-α and MxA detection were blocked with Protein Block (Dako) for 10 minutes, and then incubated in a humidified chamber with rabbit anti-human IFN-α antibody (US Biological) or mouse anti-human MxA antibody (kindly provided by Dr. O. Haller, University of Freiburg, Department of Virology, Germany) for 1 hr. All incubations were done at RT. Slides were washed with TBST (2x), incubated for 20 minutes with Ready-to-Use Peroxidase Quenching solution (DAKO) and rinsed with TBST (2x). Next, slides for IFN-α detection were treated with Advance Link (Dako), and slides for MxA staining with anti-mouse HRP polymer (Dako) for 15 and 30 minutes, respectively, followed by another wash with TBST (2x). An additional incubation (15 minutes) with Advance Enzyme (Dako) was performed for IFN-α detection. Then Substrate-DAB+ (Dako) was added for 10 minutes, slides were washed with running water for 5 minutes and counterstained with Hematoxylin (Dako) for 5 minutes. Finally, slides were dehydrated with 90% EtOH (3 × 1 minute) and 100% xylene (3 × 2 minutes).
Tissue sections from age-matched SIV-naïve animals were included as negative controls and tissue sections from SIV-infected animals that had been previously confirmed to be IFN-α and/or MxA positive served as positive controls.
Slides were examined with a Zeiss Axioskop microscope using Axiovision Software (Zeiss). Images were acquired at 200x magnification. Relative quantitation of IFN-α or MxA positive cells per tissue was performed using two different methods. First, tissue sections were analyzed using Image ProPlus software (Media Cybernetics). Briefly, for each tissue 3–5 images per slides were acquired and analyzed. IFN-α or MxA positive cells showed a dark brown color compared to IFN-α or MxA negative cells only showing the blue/purple hematoxylin staining. Then, the T cell area within a lymph node section was outlined, and based on the assigned color parameters for positive and negative staining, the software generated area values for positive and negative staining within the determined T cell area. Thus, a relative assessment of positive and negative staining patterns was obtained, but an exact quantitation of positive cells per tissue mm2 was not performed. To confirm the results, IFN-α or MxA positive cells were manually counted in each of the T cell areas examined (3–5 fields per tissue). Depending on the number of positive cells present per field, a score was assigned. A score of “1” was assigned when a particular field contained less than 50 positive IFN-α positive cells, a score of “2” for >50<75, and a score of “3” for fields with >75 IFN-α positive cells. Microscopic fields with MxA positive cells were assigned a score of “1” for >3<10 positive cells/field, a score of “2” for >10<25, and a score of “3” if >25 MxA positive cells were counted. Fields negative for IFN-α or MxA positive cells were assigned a score of “0”. Both methods resulted in a similar ranking of tissues with respect to the number of IFN-α and/or MxA positive cells, and therefore only the data from the latter scoring analysis are reported.
Analysis of pDC frequencies and function
The frequencies and ability of pDC to produce IFN-α were determined essentially as described previously [17]. Briefly, PBMC or tissue cell suspensions were prepared and stimulated with HSV-2 for 6 hours at 37°C, 5%CO2. Brefeldin A (10 μg/ml) was added for the last 5 hours of incubation. After in-vitro stimulation, pDC were analyzed by flow cytometry. pDC were identified as lineage (CD3, CD20, CD14, CD16) negative, HLA-DR and CD123 positive. Generally, 300,000 events were acquired on a FACS ARIA. Data were analyzed with FlowJo (TreeStar, Ashland, OR). Frequencies of pDC were calculated as percent of mononuclear cells, and frequencies of IFN-α and TNF-α producing cells as percentage of pDC.
In-vitro PBMC stimulation to determine IFN-α subtype mRNA expression
Rhesus PBMC were purified and stimulated at 1×10^6 cells/ml in RPMI1640 (supplemented with 10%FBS and penicillin/streptomycin) with HSV-2 (MOI=1), 50 μg of CpG ODN 2336 (InVivoGen) or 10 μg ssRNA40/LyoVec (InVivoGen) for 5 hours. RNA was purified as described [11, 16, 18] to determine mRNA levels of the various IFN-α subtypes by RT-PCR.
Virus load measurement
Plasma and tissue RNA samples were analyzed for viral RNA as described previously [11].
Statistical analysis
Results were analyzed using ANOVA One Way Analysis of Variance or the Student t-test using the GraphPad Prism and InStat software programs, version 4 (Graph Pad Software, Inc., San Diego, CA). Correlations between two parameters were determined using regression analysis tools by GraphPad Prism software.
Results
Design of rhesus macaque-specific IFN-α subtype primer-probe sets
Our initial studies were based primarily on the available human IFÑα subtype sequence information. Assuming that sequences would be highly homologous between humans and rhesus macaques, we searched the Rhesus Macaque Sequencing Project Website at the Baylor College of Medicine for sequences with homology to human IFÑα subtypes. Based on these results, rhesus macaque specific IFÑα subtype sequences were amplified from rhesus genomic DNA using primer sequences located in the 5′ and 3′ UTR regions (Table 1a, [14]). The obtained PCR products were cloned, and for each expected rhesus IFN-α subtype, 3 clones were sequenced, aligned to each other and compared to the relevant human IFN-α subtype sequence. Using this strategy, we generated rhesus macaque specific clones for 8 out of the 13 known human IFN-α subtypes (IFN-α1/13, IFN–α2, IFÑα4, IFÑα6, IFÑα8, IFÑα14 and IFÑα21). Rhesus specific clones with sequence homology to human IFÑα 5, 7, 10, and 16 could not be generated with this initial approach. The clone obtained for rhesus IFÑα17 showed a similar degree of nucleotide homology to four human IFÑα subtype genes (IFÑα4, 7, 10, and 17), and was therefore not considered to represent a specific rhesus IFÑα subtype. All of obtained rhesus clones span the full coding sequence of the relative IFN-α subtype gene they encode, when compared to the rhesus IFN-α subtype gene listed in GenBank (Table 2), except for the IFN-α8 clone that misses the last 25 base pairs. The sequence homology of each IFN-α subtype clone (beginning from the start codon) to the coding sequence of the rhesus IFN-α subtype gene in GenBank ranges from 98–100%. Note that, as in humans, the rhesus macaque sequences for IFÑα1 and IFÑα13 are almost identical, and therefore were amplified by the same primer sets and are represented by a single clone.
Table 1a.
PCR primer sequences for the Cloning of Rhesus IFN-α Subtypes
| Gene | Forward Primer (5′-3′)* | Reverse Primer (5′-3′) |
|---|---|---|
| IFN-α1/13 | GGCTCTAAACTCATGTAAAGAGTGCAT | AGCATGGTCATAGTTATAGCAGGG |
| IFN-α2 | TAAAtGAaTATGTTCCCTATTTAAGGCTAG | AAAGGTGAGCTGGtgtattag |
| IFN-α4 | CAAAAACAGAGATAGAAAGTACAACTAGGGA | TGCCTGCACAGGTATACACCAA |
| IFN-α6 | AACTAGTATGTTCCCTATTTAAGACCTACACAT | CGTTTCTGTGTTGGATCAGGTCTT |
| IFN-α8 | cccaaagttaaggtcatccat | tgcaagttgattgataaagagaagg |
| IFN-α14 | CCAGCAGCATCTTCGGGATT | GTATTAGTCAATGAGAATCATTTCCATGA |
| IFN-α21 | AGTTTTGAGTGCAGGGGAAAAAT | AAGTGTGGACTGGTGTATTAGTCATACA |
Note: Lower case letters indicate that sequences differ from the human cloning primers by Szubin et al., 2008
Table 2.
Human and rhesus macaque IFN-α subtype comparison
| Human IFN-α Subtype | Rhesus Macaque | ||||
|---|---|---|---|---|---|
| Gene | GenBank No. | Gene | GenBank No. | Identity w. Human nt Sequence | Identity w. Human aa Sequence |
| IFN-α1 | NM024013 | IFN-α1/13 | XM001099165 | 96% | 95% |
| IFN-α13 | XM001117329 | 97% | |||
| IFN-α13 | XM001082177 | 97% | |||
| IFN-α13 | NM006900 | IFN-α13 | XM001117329 | 97% | |
| IFN-α13 | XM001082177 | 97% | |||
| IFN-α1/13 | XM001099165 | 97% | 95% | ||
| IFN-α2 | NM000605 | IFN-α2 | XM001107516 | 94% | 92% |
| IFN-α4 | NM021068 | IFN-α2 | XM001107635 | 93% | 90% |
| IFN-α5* | NM002169 | IFN-α6 | XM001099374 | 91% | |
| IFN-α6 | NM021002 | IFN-α6 | XM001099374 | 97% | 95% |
| IFN-α7* | NM021057 | IFN-α4 | XM001107635 | 92% | |
| IFN-α8 | NM002170 | IFN-α8 | XM001107458 | 95% | 89% |
| IFN-α10* | NM002171 | IFN-α4 | XM001107635 | 92% | |
| IFN-α14 | BC104159 | IFN-α14 | XM001107576 | 95% | 90% |
| IFN-α16* | NM002173 | IFN-α4 | XM001107635 | 93% | |
| IFN-α17 | NM021268 | IFN-α17 | XM001107999 | 92% | 91% |
| IFN-α4 | XM001107635 | 93% | |||
| IFN-α21 | NM002175 | IFN-α21 | XM001108051 | 91% | 89% |
Blast search of the human against the rhesus sequences did not yield a predicted rhesus sequence of the same, but instead a related IFN-α subtype gene
Coincidentally, while we were pursuing the human [14] and rhesus studies, the rhesus macaque genome was published [19]. A search of all human IFÑα subtype sequences against the now annotated rhesus macaque genome database revealed the predicted rhesus IFÑα17 sequence. This rhesus IFÑα17 sequence was used in the subsequent real-time PCR assay design. Thus, while there are 13 expressed human IFÑα subtypes, we could identify and design rhesus-specific molecular assays to distinguish between 9 IFÑα subtypes (IFÑα1/13, IFÑα2, IFÑα4, IFÑα6, IFÑα8, IFÑα14, IFÑα17, and IFÑα21).
Consistent with our failure to obtain rhesus IFÑα7, IFÑα10 and IFÑα16 clones or sequences, the human IFÑα7, IFÑα10 and IFÑα16 sequences were most similar to the predicted sequence for rhesus IFÑα4. In addition, a rhesus counterpart for the human IFÑα5 gene was not found. Instead, the human IFÑα5 sequence appeared to be most closely related to the predicted sequence for rhesus IFÑα6.
Table 2 presents the summary of the Genbank annotation numbers for the human IFÑα subtypes, their rhesus counterparts and their nucleotide sequence homology. Sequence differences for human and rhesus macaque IFN-α subtypes extended to the amino acid level (Figure 1), thus, demonstrating the divergent evolution of the rhesus macaque and human IFÑα subtype genes [20].
Figure 1.
Sequence comparison of rhesus macaque and human IFN-α2. Panel A: Nucleotide sequence alignment of rhesus macaque (underlined) and human IFN-α2. Panel B: Amino acid alignment of rhesus macaque (underlined) and human IFN-α2. Sequence differences are indicated by bold italics letters.
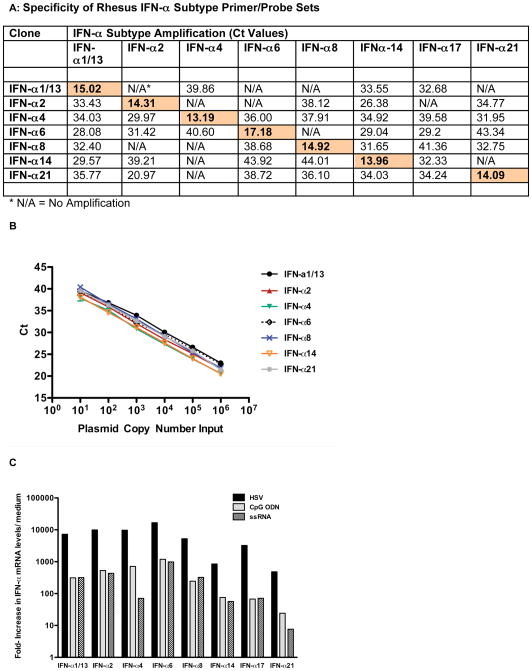
Next, primer-probe sets for single-round real time PCR amplification for all confirmed rhesus IFÑα subtype sequences were designed. Towards this goal, previously developed real-time PCR primer/probe sets for a subset of human IFÑα subtypes (IFÑα1/13, IFÑα2, IFÑα6 and IFÑα8) [5] were compared to rhesus IFÑα subtype sequences and mismatched nucleotides between the human and rhesus sequence were optimized for the rhesus specific sequence (Figure 2, Table 1b and 1c). All other primer-probe sets were designed using the ABI Primer Design Software. Despite the extensive nucleotide homology between all rhesus IFÑα subtypes (Figure 2), each subtype-specific primer-probe set preferentially amplified the unique IFÑα subtype it was matched to (Figure 3A). After confirming the specificity of the designed rhesus-specific IFÑα subtype primer-probe sets, PCR amplification of all cloned rhesus IFÑα subtypes was performed over a range from 10 to 1×10^6 copies plasmid DNA to ensure that all IFÑα subtype sequences were amplified with comparable efficiency. The similar slopes and the partial overlap of the amplification curves of the various IFÑα subtypes (Figure 3B) suggest similar efficiency in the PCR amplification.
Figure 2.
Rhesus macaque IFN-α subtype sequence alignment. Rhesus macaque IFN-α subtype sequences were aligned using ClustalW2 at www.ebi.ac.uk. Grey shaded regions with star symbols denote regions of nucleotide homology between all rhesus IFN-α subtypes. The forward and reverse primer sequences are indicated in bold black and italic letters, respectively, while probe sequences are in underlined.
Table 1b.
Real Time PCR Primer-Probe Sets for IFN-α Subtype Amplification
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Probe (5′-3′) |
|---|---|---|---|
| IFN-α1/13 | CTTCAACCTCTTTACCACAAAAGAcTC | TGCTGGTAGAGTTCAGcGCA | TGCTTGGGATGAGGACCTCCTAGACA |
| IFN-α2 | CTTGAAGGACAGACATGACTTTGGA | GGATGGTTTGAGCCTTTTGGA | TTCCCCAGGAGGAGTTTGGCAACC |
| IFN-α4 | TCCTGCCTGAAGGACAGgCAT | TGGATCATCTCATGGaGGA | ACTTTGCATTCCCCCAGGAGGA |
| IFN-α6 | TCCATGAGGTGATTCAGCAGAC | GCTGCTGGTAAAGTTCAGTATAGAGTTT | CTGTTGCTTGGGATGAGAGGCTTCTAGAC |
| IFN-α8 | CCTTCTAGATGAATTCTACATCGAACTTG | ACTCTATCACCCCCACTTCCTG | CAGCTGAATGACCTGGAGTCCTGTGTG |
| IFN-α14 | GCCCTGGTGGTGCTTAGCTA | TGTGCCATGAGCATCAAAGTC | AGTCAAGCTGCTCTCTGGGCTG |
| IFN-α17 | TCTGGGCTGTGATCCTCCC | TCCTTCAGACAGGAGAAAGGAGAGTT | TCCTTGATACTCCTGGCACAAATGGGAA |
| IFN-α21 | ACAAGGCTCAAGCCATCTCT | AGGATGCAGTCCACATTCGT | TACAGGAGGTTGGAGTGGAAGAGACT |
Table 1c.
Location of Rhesus IFN-α Subtype Primers and Probes within their respective Rhesus Gene
| Gene | Genbank No | Forward Primer Sequence Location | Reverse Primer Sequence Location | Probe Sequence Location |
|---|---|---|---|---|
| IFN-α1/13 | XM001099165 | 132–158 | 196–215 | 165–190 |
| IFN-α2 | XM001107516 | 225–247 (1) | 277–297 (1) | 250–273 |
| IFN-α4 | XM001107635 | 293–313 | 378–396 | 315–336 |
| IFN-α6 | XM001099374 | 272–293 | 352–379 | 323–351 |
| IFN-α8 | XM001107458 | 352–380 | 416–437 (2) | 386–412 |
| IFN-α14 | XM001107576 | 85–113 | 164–184 | 116–127 |
| IFN-α17 | XM001107999 | 205–223 | 280–306 | 251–278 |
| IFN-α21 | XM001108051 | 356–375 | 544–563 | 512–537 |
(1) Number in parentheses represent the number of mismatches between the primer or probe sequence and the actual gene sequence
Figure 3.
Specificity and amplification of rhesus macaque IFN-α subtype primer/probe sets. Panel A: Specificity of the rhesus macaque IFN-α subtype primer/probe sets tested by PCR using IFN–α subtype-specific cDNA clones. Shown are Ct values for amplification of each distinct rhesus IFN-α subtype cDNA clone by each of the various IFN-α subtype primer/probe sets. Shaded boxes indicate IFN-α subtype specific amplification of the relevant IFN-α subtype cDNA clone. Panel B: PCR efficiency of rhesus IFN-α subtype amplification. Shown are the Ct values per input copy number for each of the IFN-α subtype cDNA clones. Amplification was linear for all rhesus IFN-α subtypes over the tested range from 10 to 10^6 copies. Panel C: IFN-α subtype induction in rhesus PBMC. Rhesus PBMC were stimulated with HSV, CpG ODN or ssRNA for 6 hours and then mRNA levels for various rhesus IFN-α subtypes were determined by real time PCR. Shown are the relative increases for each IFN-α subtype mRNA in comparison to the mRNA level of the same IFN-α subtype in medium control cultures. One representative example out of 3 experiments is shown.
IFÑα mRNA subtype measurement following in vitro stimulation
To determine the extent to which the various identified rhesus IFÑα subtypes can be measured with the developed assay, rhesus PBMC were stimulated for 6 hours with either HSV, a virus that induces strong IFÑα production in human and rhesus PBMC [17, 21], the TLR9 ligand CpG ODN 2236, or the TLR7/8 ligand ssRNA40/LyoVec. The results demonstrate that multiple IFÑα subtypes can be induced in vitro in rhesus macaque PBMC (Figure 3C). The magnitude of individual IFÑα subtype mRNA levels was dependent on the stimulus type. HSV induced at least 10-fold higher IFÑα subtype mRNA levels compared to CpG ODN and ssRNA. Generally, IFÑα14, IFÑα17 and IFÑα21 were less induced than IFÑα1/13, IFÑα2, IFÑα4, IFÑα6, and IFÑα8. Consistent with earlier studies in humans [14], while the magnitude of induction of the various IFÑα subtypes differed dependent on the stimulus used, there was no apparent preferential induction of any IFÑα subtype by any one treatment.
Taken together, these results demonstrated that this highly sensitive and specific assay could be used to measure the induction of individual IFN-α subtypes in rhesus macaques during the course of in vivo infection with SIV or other pathogens.
IFÑα subtype expression patterns in lymphoid tissues after oral SIVmac251 infection in infant macaques
To determine the levels and IFN-a subtype responses to SIV infection in infants, we utilized available tissue samples from 6 infant macaques that were infected orally at 4 weeks of age by repeated SIVmac251 inoculation. Of those 6 macaques, all but one animal tested positive for plasma viral RNA one week after first SIV exposure [11]. The one that tested negative (# 36444) had detectable viral RNA in the submandibular lymph node and the colon (76 and 139 SIV RNA copies per μg tissue RNA, respectively), indicating infection of all 6 SIV-exposed macaques.
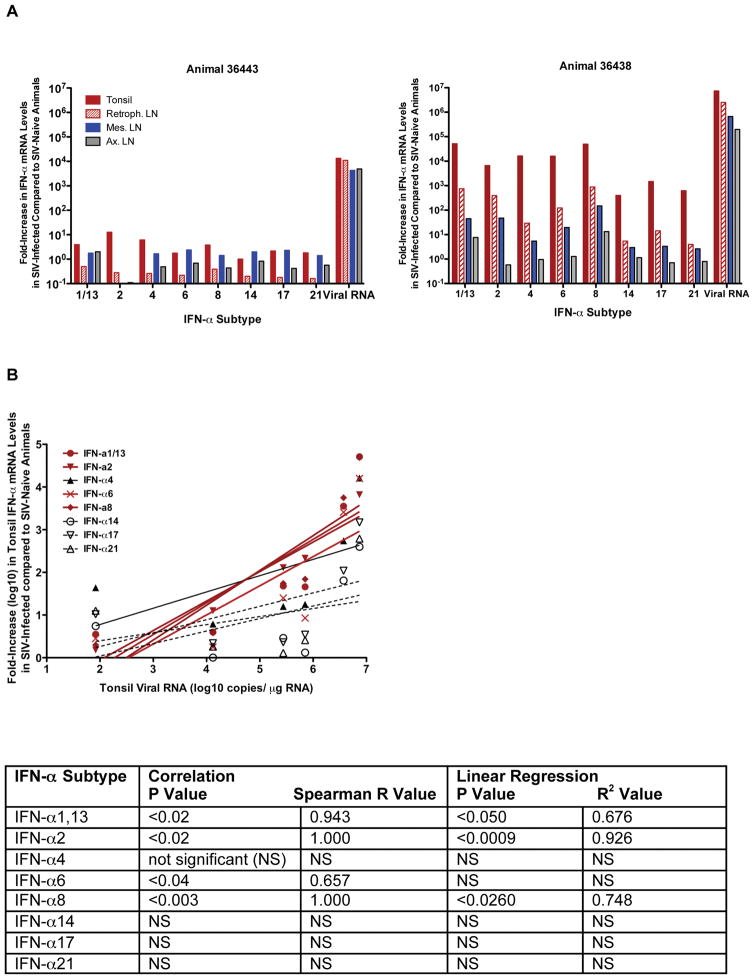
SIV infection resulted in the increase of multiple IFN-α subtypes in various tissues of all animals (Figure 4 and data not shown). Similar to the in vitro studies (Figure 3C), the magnitude of mRNA induction differed between the individual IFN-α subtypes with the highest increases seen for IFÑα1/13, IFÑα2, IFÑα6 and IFÑα8. The relative expression levels between the various IFÑα subtypes in the different tissues of an individual animal, however, were remarkably similar. Thus, IFÑα subtypes did not show any tissue-specific expression patterns in orally SIV-infected infant macaques.
Figure 4.
IFN-α subtype mRNA levels in tissues at 1 week after oral SIV infection. Panel A: The relative increase in IFN-α subtype mRNA levels in tissues of infant macaques at 1 week after oral SIV exposure in comparison to average IFN-α subtypes in the same tissues of age-matched infant macaques are shown. Shown are the data for the animals with the lowest (#36443) and highest level (#36438) of SIV replication in plasma and tissues. The x-axis denotes the IFN-α subtype and virus replication (copies per μg total tissue RNA). The different tissues are indicated by different colors (tonsil-filled red bars, retropharyngeal LN- empty red bars, mesenteric LN- blue bars, axillary LN- grey bars). Panel B: Relationship between SIV RNA and IFN-α subtype mRNA expression levels in the tonsil of SIV-infected infant macaques. The SIV replication levels, expressed as SIV RNA copies/μg tonsil RNA, of all six infant macaques are plotted against the log10 increase in IFN-α subtype mRNA levels in the tonsil. A positive relationship between SIV replication and distinct IFN-α subtype mRNA levels is indicated by red lines.The table below the figure lists the P and Spearman R values for the correlation, and the P and R2 values for the linear regression analysis between SIV replication and IFN-α subtype mRNA levels.
Generally, the overall magnitude of IFÑα subtype induction seemed to be correlated with the level of virus replication in individual animals. The monkey with the highest plasma SIV RNA levels (36438) also showed the highest increases in IFÑα mRNA levels for all the IFÑα subtypes tested (Figure 4A and data not shown). Furthermore, in the tonsil, but not in the axillary or mesenteric LN, the mRNA expression levels of the most strongly upregulated IFN-α subtypes, IFÑα 1/13, IFÑα2, IFÑα6, and IFÑα8, positively correlated with the levels of virus replication in the same tissue (Figure 4B). Consistent with the pattern of virus dissemination after oral SIV exposure [11], IFÑα subtype mRNA expression levels were generally highest in the sites closest to the exposure site, i.e. the tonsil, and lowest in the distant mesenteric LN. In fact, IFÑα mRNA levels of animal 36438 were significantly higher (P<0.05) in the tonsil compared to the retropharyngeal, the axillary and the mesenteric lymph node, and the retropharyngeal lymph node had higher gene expression of all IFÑα subtypes than the mesenteric lymph node (P<0.05).
IFÑα protein production in lymphoid tissues after oral SIV infection
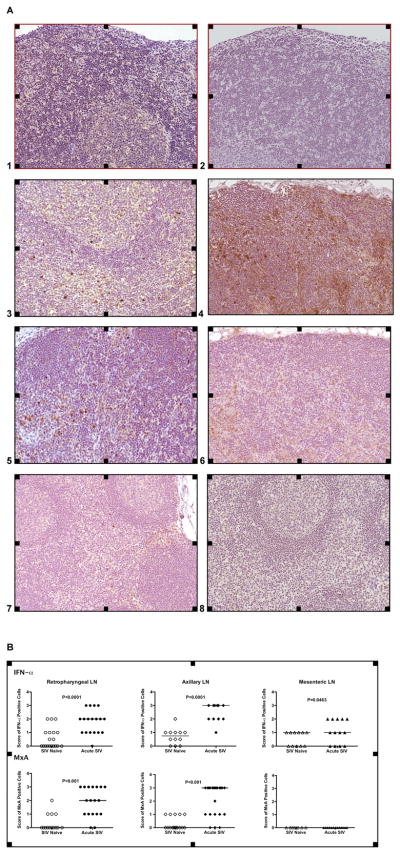
To assess whether IFÑα subtype mRNA expression increases correlated with enhanced IFÑα protein production, we performed immunohistochemistry in various lymph nodes. The majority of IFÑα positive cells were localized in the T cell areas of the lymph node (Figure 5A). Relative frequencies of IFÑα positive cells per T cell area for each lymph node correlated with the IFÑα subtype mRNA expression data. The highest frequencies of IFÑα positive cells were found predominantly in lymph nodes draining the oral cavity (retropharyngeal LN) and the tonsil, but less so in the more distal mesenteric LN at one week after oral SIV exposure (Figures 5A and 5B). The co-localization of an interferon-inducible protein, MxA, with IFN-α in the same tissues was evidence that the IFN-α was biologically active (Figures 5A and 5B). Consistent with low frequencies of IFN-α positive cells in mesenteric lymph nodes, MxA positive cells were not or only at low frequencies detectable in the mesenteric lymph nodes (Figure 5). Thus, at the earliest time points post SIV infection, the expression of MxA is likely related to the total frequencies of IFN-α secreting cells and to the time IFN-α is present in the tissue to result in the induction of IFN-α –inducible genes like MxA.
Figure 5.
Detection of IFN-α positive cells by IHC. Panel A: IFN-α (Images 1, 3, 5, and 7) and MxA (Images 2, 4, 6, and 8) positive cells (Magnification: 20x). Images 1 and 2 show IFN-α and MxA staining in retropharyngeal LN of SIV-negative age-matched infants (negative control). IFN-α and MxA positive cells of SIV-infected infants in the retropharyngeal lymph node (Images 3, 4), the axillary (Images 5, 6) and the mesenteric LN (Images 7, 8) show a brown staining pattern. Panel B: Relative frequencies of IFN-α and MxA positive cells. The individual scores of the relative frequencies of IFN-α and MxA positive cells in each analyzed microscopic field within the T cell area in the tissues of SIV-negative and SIV-infected infants. The scores were determined as described in Materials and Methods.
Plasmacytoid dendritic cells as source of IFÑα
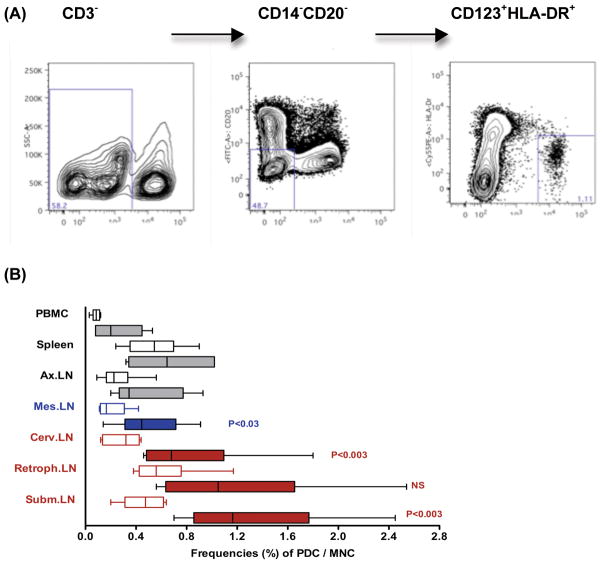
Plasmacytoid dendritic cells (pDC) have been identified as the main source of IFN–α during acute viral infections. To determine whether the increases in IFÑα mRNA and protein were due to increases in pDC frequencies and/or their activation state, we compared frequencies in peripheral blood and lymphoid tissues of 4-week old SIV-naïve to SIV-infected infant macaques at 1 week after SIV exposure using flow cytometry (Figure 6). The submandibular lymph node is the main draining LN in the oral cavity [22], and indeed, pDC frequencies were significantly increased in the submandibular and cervical LN of SIV-infected compared to SIV-naïve animals. For the retropharyngeal LN, a similar trend was observed, but did not reach statistical significance, likely due to the relatively high animal-to-animal variation in pDC frequencies. The low cell yields from infant tonsils did not allow the pDC analysis for tonsils. Importantly though the tissues with the highest numbers of pDC at 1 week after oral SIV infection, e.g. the submandibular and retropharyngeal LN, also showed the highest increases in IFÑα mRNA levels (Figure 4).
Figure 6.
Frequencies of pDC in tissues of infant macaques. Panel A shows the gating strategies applied to the flow cytometric analysis. The percentage of pDC among mononuclear cells isolated from various tissues of 1 SIV-infected (n=6, filled bars) and age-matched SIV-naive (n=6, empty bars) infant macaques is shown in Panel B. Statistically significant differences between SIV-naïve and SIV-infected infants in specific tissues are indicated by P values.
Thus, oral SIV exposure in infant macaques results in (i) the rapid and simultaneous induction of multiple IFÑα subtypes in various tissues, (ii) the appearance of IFÑα producing cells in lymph nodes, and (iii) increased pDC frequencies in various lymph nodes draining the oral cavity. The magnitude of the type I interferon response was highest in the tonsil and in lymph nodes draining the oral entry site of SIV, thus following the pattern of virus dissemination.
Mucosal IFÑα responses
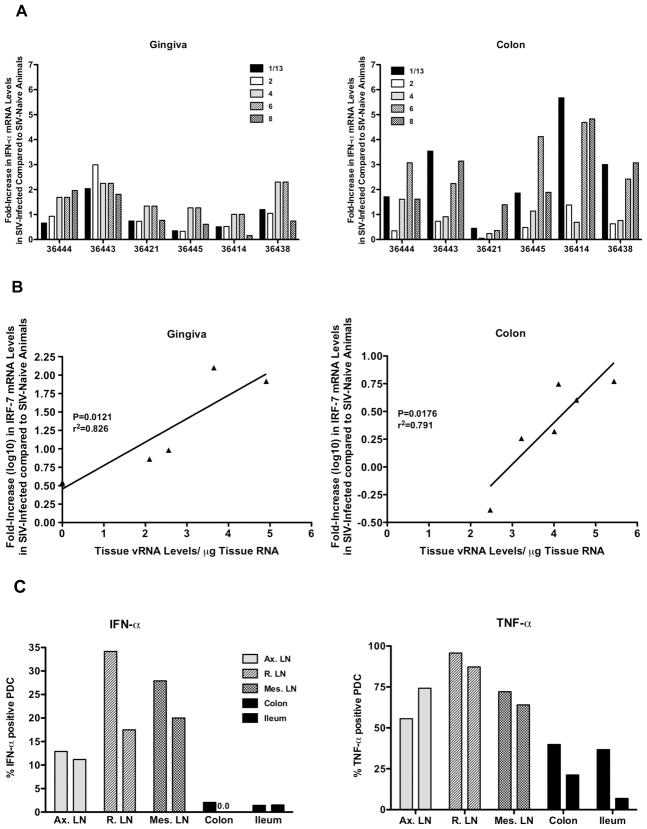
In contrast to the strong induction in lymphoid tissues, individual IFÑα subtype mRNA levels increased only slightly (2- to maximal 6-fold) in the gingiva and the colon (Figure 7A). This is consistent with our previous data generated with a primer/probe set that preferentially amplified IFÑα2 [11, 18]. This was not due to the induction of type I interferon genes other than IFN-α. In fact, elevated IFN-β mRNA levels were only observed in the tonsil, and only in the two animals (36438, 36414) with the highest virus replication overall (data not shown). Similarly, IFN-κ mRNA levels in the gingiva of SIV-infected animals were not increased compared to SIV-naïve animals (data not shown), despite the fact that interferon kappa is preferentially produced by epidermal keratinocytes and can be induced in the gingiva [23]. Despite our failure to detect a strong induction of any type I interferon in the mucosa, IRF-7 mRNA levels were strongly expressed in the gingiva and the colon compared to the controls, and positively correlated with the level of SIV replication in the same tissue (Figure 7B).
Figure 7.
IFN-α responses in mucosal tissues. Panel A shows the relative increases in IFN-α subtype mRNA levels in the gingiva (left panel) and colon (right panel) of SIV-infected infants compared to IFN-α mRNA levels of SIV-naïve infants. Panel B demonstrates the relationship between tissue SIV RNA levels and relative log10 increases in IRF-7 mRNA levels in the gingiva (left panel) and colon (right panel) of the same SIV-infected infant macaques. Panel C shows the frequencies of IFN-α and TNF-α producing pDC in lymphoid and mucosal tissues after in –vitro stimulation with Imiquimod. Cell suspensions from tissues of SIV-naïve, 6-month old macaques were stimulated for 6 hours with Imiquimod and then analyzed for the frequencies of IFN-α (left panel) and TNF-α (right panel) producing pDC using multiparameter flow cytometry. Note that frequencies were lower in the colon and ileum compared to cell suspensions prepared from lymph nodes. The two bars per tissue are representative of two different animals.
To determine whether the differences in IFÑα induction between lymph nodes and mucosal sites were due to quantitative or qualitative differences in pDC’s, we compared IFÑα responses by pDC in cell suspensions from lymphoid and mucosal tissues of 6 months old macaques after in vitro stimulation with Imiquimod, a TLR7/8 agonist. Consistent with the ex vivo IFÑα expression analysis, few pDC from the intestinal tissues produced IFÑα (Figure 7C). TNF-α production by mucosal pDC was also reduced, albeit to a lesser extent.
Thus, despite the increase of IRF-7 mRNA levels in the gingiva and the colon after SIV infection, and in contrast to the strong induction of IFÑα subtypes in lymphoid tissues, mucosal tissues are relatively devoid of IFÑα producing cells during acute SIV infection. The in-vitro TLR7/8 stimulation studies indicated a lower and/or altered responsiveness of pDC from intestinal tissues compared to lymph node pDC within an individual animal.
Discussion
Innate responses are critically important in the early control of pathogen replication and thereby prevention of dissemination from the local entry to distal sites. Innate responses also determine the nature of the subsequent adaptive response and are involved in the maintenance of the pathogen-specific response. Therefore, a more detailed knowledge of the breadth of innate responses will be beneficial in our overall understanding of pathogenesis and in the design of more efficacious intervention strategies.
Type I interferons have long been known as a first line defense mechanism in viral infections. In fact, IFÑα is used clinically in the treatment of patients infected with hepatitis or HIV. However, while IFÑα treatment is successful in some patients, it shows no effect in others. Multiple factors could contribute to treatment failure [24, 25]. Knowing that IFÑα comprises a family of multiple subtypes, it is imperative that we gain insight into the regulation and function of these various IFÑα subtypes in human diseases.
The current study developed IFÑα subtype specific primer/probe sets as a means to dissect the IFÑα response in rhesus macaques (Table 1, Figures 2 and 3). As expected, the rhesus IFÑα subtype clones revealed a high degree of homology to the human IFÑα subtype genes (Table 2). Interestingly though, nucleotide alignments between the human and macaque sequences demonstrated that not all human IFÑα subtypes necessarily correspond to the same numeric IFÑα subtype in rhesus macaques and vice versa. The comparative analysis demonstrated that the human sequences for IFÑα7, IFÑα10 and IFÑα16 had no apparent rhesus gene homologue, but showed a high degree of nucleotide homology to rhesus IFÑα4. Similarly, we were unable to clone rhesus IFÑα5 and the alignment of the human IFÑα5 sequence against predicted rhesus IFÑα subtype sequences identified rhesus IFÑα6 as having the highest homology to human IFÑα5. In this context, it is noteworthy that human IFÑα6 has been proposed to be a pseudogene [26, 27]. While we recently showed that IFÑα6 could be induced at low levels after in vitro TLR stimulation in human pDC [14], other human IFÑα subtypes, including IFÑα5, were induced at much higher levels. In contrast, in rhesus macaques, IFÑα6, but not IFÑα5, was strongly induced after in vitro TLR stimulation and in vivo SIV infection. Although one could therefore speculate that the functional equivalent of human IFÑα5 is rhesus IFÑα6, studies analyzing the genetic relatedness or divergence of human and rhesus IFÑα subtype sequences were beyond the scope of this study.
Genetic studies by Woelk et al. have suggested that IFN-α subtype genes in humans and chimpanzees are closely related [20]. In fact an alignment of all genic and intergenic regions of each of the IFN-α genes in humans shows the exact same location of the family locus compared to the chimpanzee IFN-α genes [20]. In contrast, only IFN-α1, IFN-α2, IFN-α8, IFN-α13 and IFN-α6 are preserved in the locus between humans and rhesus macaques. Thus, the authors proposed that the most recent common ancestor contained only a subset of the IFN-α subtype genes, but then the human/chimpanzee and the rhesus IFN-α subtype genes diverged separately [20]. Both, gene duplication and gene conversion contributed to the diversity of the IFN-α subytpe genes observed in the different species [20].
As rhesus macaques are used as model systems for multiple human diseases, it is important to identify the similarities and differences in immune responses between rhesus monkeys and humans. In HIV-1 infected patients, increased serum levels of IFÑα have been associated with disease progression [28, 29]. Recently, it was demonstrated that, dependent on the stage of infection, the expression of IFÑα subtypes in PBMC of HIV-1 infected patients varies [30], a finding that has potential implications for improved diagnostics and treatment. The current study in the rhesus macaque model of SIV infection focuses on the early IFÑα subtype responses as they relate to virus dissemination and anatomic compartment.
The data demonstrate that multiple IFÑα subtypes are induced in acutely SIV infected infant macaques (Figure 4). Consistent with IFÑα1 as the first IFÑα subtype induced following stimulation and necessary for the induction of all other IFÑα subtypes in humans, we observed a strong increase of IFÑα1/13 mRNA levels in lymphoid tissue of rhesus macaques at 1 week after SIV infection. A significant increase of IFÑα mRNA levels was also observed for IFÑα2, IFÑα4, IFÑα6, and IFÑα8. Why gene expression levels were less elevated for IFN-α14, IFÑα17 and IFÑα21 could not be determined. A recent study showed that the induction of distinct IFN-α subtypes is regulated by the differential recruitment of the transcription factors IRF-3 and IRF-7 to the promoter regions of the individual IFN-α subtype genes [31]. Thus, in future studies, it would be interesting to determine how IRF-3 and IRF-7 expression and activation levels are altered by SIV infection in future studies. Importantly, if it could be demonstrated that different IFN-α subtypes differ in their anti-SIV activity, the understanding of regulation of these subtypes would be critical. Similarly, it would be important to determine whether SIV proteins could induce or inhibit IFN-α directly. In vitro studies have shown that some HIV-1 proteins, e.g. gp120, can directly induce IFN-α in human pDC[32]. At the same time, through the recognition of a common epitope described for HIV-1 gp41and IFN-α, antibodies against IFN-α could interfere with HIV-1 gp41 binding to cells, and vice versa, HIV gp41-specific antibodies could inhibit IFN-α function [33, 34].
Interestingly though, while the various IFÑα subtypes were induced with different magnitude, the overall expression pattern was similar in each of the lymphoid tissues tested in any individual animal. Thus, there was no tissue-specific expression of distinct IFÑα subtypes and their induction occurred with similar kinetics. Furthermore, we provide evidence that increased mRNA levels of IFÑα translated into the production of IFÑα protein and also the production of the IFN-α-inducible protein MxA (Figure 5).
The increased pDC frequencies observed in several lymph nodes close to the virus entry site at one week after oral SIV exposure, and the established positive correlation between IFÑα mRNA levels and virus replication in the tonsil, suggested that this early IFÑα response in infant macaques was predominantly exerted by pDC (Figure 6). This conclusion is supported by recent studies in adult macaques showing the fast influx of pDC into lymph nodes in acute SIV infection [35–37] and by demonstrating that the role of pDC in the early IFÑα response [38]. We cannot exclude the possibility though that differences in IFN-α production between various tissues could be due to different frequencies of functionally impaired and/or dying pDC. Studies in SIV infected macaques have shown that pDC undergo rapid apoptotic cell death after migration into lymph nodes [35].
It is noteworthy that infant macaques had significantly higher frequencies of pDC in lymph nodes draining the oral cavity compared to pDC frequencies observed in the same tissues of six adult macaques (data not shown). One could speculate that the exploration of new things by mouth in infants might result in increased innate effector mechanisms at sites of oral exposure. In general, in both, infant and adult macaques, pDC frequencies were highest in the spleen, followed by lymph nodes draining the oral cavity, and lowest in the mesenteric lymph nodes and in PBMC (data not shown).
In contrast to lymphoid tissues, mucosal tissues appeared less able to induce potent type I interferon responses (Figure 7). Although SIV infection induced slightly elevated mRNA levels for a number of IFÑα subtypes, within an individual animal the magnitude was considerably lower compared to the increase of IFÑα subtype mRNA levels observed in lymphoid tissues. Consistent with the low IFÑα subtype mRNA induction in the gingiva and colon, IFÑα positive cells could rarely be detected by IHC (data not shown).
Assuming that pDC are responsible for the early IFÑα response in SIV infection, pDC frequencies and/or function must have differed between lymphoid and mucosal tissues of these animals. In fact, we present in-vitro data demonstrating that TLR stimulation resulted in lower frequencies of cytokine-producing pDC in intestinal compared to PBMC and lymph node cell suspensions. This finding is consistent with data obtained in mouse studies [39–43]. Considering that mucosal sites present the main entry sites for SIV and HIV, future studies need to address this question more thoroughly by comparing the functional ability of dendritic cells isolated from effector versus inductive sites within mucosal tissues and comparing them to dendritic cell function in lymphoid tissues of the same animal.
Acknowledgments
We would like to thank Dr. Otto Hahn (University of Freiburg, Germany) for providing the MxA antibody, and appreciate the technical assistance by Mathieu Lemieux and by Joyce Lee and Kathy Lantz of the CNPRC Analytical Core. The work was supported by the funding from the NIH/NIDCR to K.A. (1R21DE016541) and N.B. (1P01 DE016839), and by the CNPRC Base Grant (P51 RR00169-41).
References
- 1.Isaacs A, Lindenmann J. Virus Interference I. The interferon Proc R Soc Ser B, London. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Brandt ER, Linnane AW, Devenish RJ. Expression of IFN A genes in subpopulations of peripheral blood cells. Br J Haematol. 1994;86(4):717–25. doi: 10.1111/j.1365-2141.1994.tb04820.x. [DOI] [PubMed] [Google Scholar]
- 3.Foster GR, Finter NB. Are all type I human interferons equivalent? J Viral Hepat. 1998;5(3):143–52. doi: 10.1046/j.1365-2893.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 4.Larrea E, Alberdi A, Castelruiz Y, Boya P, Civeira MP, Prieto J. Expression of interferon-alpha subtypes in peripheral mononuclear cells from patients with chronic hepatitis C: a role for interferon-alpha5. J Viral Hepat. 2001;8(2):103–10. doi: 10.1046/j.1365-2893.2001.00273.x. [DOI] [PubMed] [Google Scholar]
- 5.Loseke S, Grage-Griebenow E, Wagner A, Gehlhar K, Bufe A. Differential expression of IFN-alpha subtypes in human PBMC: evaluation of novel real-time PCR assays. J Immunol Methods. 2003;276(1–2):207–22. doi: 10.1016/s0022-1759(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 6.Yeow WS, Lawson CM, Beilharz MW. Antiviral activities of individual murine IFN-alpha subtypes in vivo: intramuscular injection of IFN expression constructs reduces cytomegalovirus replication. J Immunol. 1998;160(6):2932–9. [PubMed] [Google Scholar]
- 7.Cull VS, Bartlett EJ, James CM. Type I interferon gene therapy protects against cytomegalovirus-induced myocarditis. Immunology. 2002;106(3):428–37. doi: 10.1046/j.1365-2567.2002.01423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr DJ, Noisakran S. The antiviral efficacy of the murine alpha-1 interferon transgene against ocular herpes simplex virus type 1 requires the presence of CD4(+), alpha/beta T-cell receptor-positive T lymphocytes with the capacity to produce gamma interferon. J Virol. 2002;76(18):9398–406. doi: 10.1128/JVI.76.18.9398-9406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James CM, Abdad MY, Mansfield JP, et al. Differential activities of alpha/beta IFN subtypes against influenza virus in vivo and enhancement of specific immune responses in DNA vaccinated mice expressing haemagglutinin and nucleoprotein. Vaccine. 2007;25(10):1856–67. doi: 10.1016/j.vaccine.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 10.Gerlach N, Gibbert K, Alter C, et al. Anti-retroviral effects of type I IFN subtypes in vivo. Eur J Immunol. 2009;39(1):136–46. doi: 10.1002/eji.200838311. [DOI] [PubMed] [Google Scholar]
- 11.Abel K, Pahar B, Van Rompay KK, et al. Rapid virus dissemination in infant macaques after oral simian immunodeficiency virus exposure in the presence of local innate immune responses. J Virol. 2006;80(13):6357–67. doi: 10.1128/JVI.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rompay KK, Schmidt KA, Lawson JR, Singh R, Bischofberger N, Marthas ML. Topical administration of low-dose tenofovir disoproxil fumarate to protect infant macaques against multiple oral exposures of low doses of simian immunodeficiency virus. J Infect Dis. 2002;186(10):1508–13. doi: 10.1086/344360. [DOI] [PubMed] [Google Scholar]
- 13.Shacklett BL, Yang O, Hausner MA, et al. Optimization of methods to assess human mucosal T-cell responses to HIV infection. J Immunol Methods. 2003;279(1–2):17–31. doi: 10.1016/s0022-1759(03)00255-2. [DOI] [PubMed] [Google Scholar]
- 14.Szubin R, Chang WL, Greasby T, Beckett L, Baumgarth N. Rigid interferon-alpha subtype responses of human plasmacytoid dendritic cells. J Interferon Cytokine Res. 2008;28(12):749–63. doi: 10.1089/jir.2008.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucelic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 16.Abel K, Rocke DM, Chohan B, Fritts L, Miller CJ. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J Virol. 2005;79(19):12164–72. doi: 10.1128/JVI.79.19.12164-12172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abel K, Wang Y, Fritts L, et al. Deoxycytidyl-deoxyguanosine oligonucleotide classes A, B, and C induce distinct cytokine gene expression patterns in rhesus monkey peripheral blood mononuclear cells and distinct alpha interferon responses in TLR9-expressing rhesus monkey plasmacytoid dendritic cells. Clin Diagn Lab Immunol. 2005;12(5):606–21. doi: 10.1128/CDLI.12.5.606-621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel K, Alegria-Hartman MJ, Rothaeusler K, Marthas M, Miller CJ. The relationship between simian immunodeficiency virus RNA levels and the mRNA levels of alpha/beta interferons (IFN-alpha/beta) and IFN-alpha/beta-inducible Mx in lymphoid tissues of rhesus macaques during acute and chronic infection. J Virol. 2002;76(16):8433–45. doi: 10.1128/JVI.76.16.8433-8445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 20.Woelk CH, Frost SD, Richman DD, Higley PE, Kosakovsky Pond SL. Evolution of the interferon alpha gene family in eutherian mammals. Gene. 2007;397(1–2):38–50. doi: 10.1016/j.gene.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung E, Amrute SB, Abel K, et al. Characterization of virus-responsive plasmacytoid dendritic cells in the rhesus macaque. Clin Diagn Lab Immunol. 2005;12(3):426–35. doi: 10.1128/CDLI.12.3.426-435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner JA. The lymph vessel system of the mouth cavity and pharynx. Laryngorhinootologie. 1995;74(10):622–8. doi: 10.1055/s-2007-997814. [DOI] [PubMed] [Google Scholar]
- 23.LaFleur DW, Nardelli B, Tsareva T, et al. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J Biol Chem. 2001;276(43):39765–71. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- 24.Jorns C, Holzinger D, Thimme R, et al. Rapid and simple detection of IFN-neutralizing antibodies in chronic hepatitis C non-responsive to IFN-alpha. J Med Virol. 2006;78(1):74–82. doi: 10.1002/jmv.20506. [DOI] [PubMed] [Google Scholar]
- 25.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105(19):7034–9. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiscott J, Cantell K, Weissmann C. Differential expression of human interferon genes. Nucleic Acids Res. 1984;12(9):3727–46. doi: 10.1093/nar/12.9.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez S, Reeves R, Island ML, et al. Silencer activity in the interferon-A gene promoters. J Biol Chem. 1997;272(36):22788–99. doi: 10.1074/jbc.272.36.22788. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs D, Shearer GM, Boswell RN, et al. Negative correlation between blood cell counts and serum neopterin concentration in patients with HIV-1 infection. AIDS. 1991;5(2):209–12. doi: 10.1097/00002030-199102000-00012. [DOI] [PubMed] [Google Scholar]
- 29.von Sydow M, Sonnerborg A, Gaines H, Strannegard O. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res Hum Retroviruses. 1991;7(4):375–80. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann C, Taubert D, Jung N, et al. Preferential upregulation of interferon-alpha subtype 2 expression in HIV-1 patients. AIDS Res Hum Retroviruses. 2009;25(6):577–81. doi: 10.1089/aid.2008.0238. [DOI] [PubMed] [Google Scholar]
- 31.Genin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20(4):283–95. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Del Corno M, Gauzzi MC, Penna G, Belardelli F, Adorini L, Gessani S. Human immunodeficiency virus type 1 gp120 and other activation stimuli are highly effective in triggering alpha interferon and CC chemokine production in circulating plasmacytoid but not myeloid dendritic cells. J Virol. 2005;79(19):12597–601. doi: 10.1128/JVI.79.19.12597-12601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YH, Xiao Y, Dierich MP. HIV-1 gp41 and type I interferon: sequence homology and biological as well as clinical implications. Immunol Res. 2000;22(1):61–6. doi: 10.1385/ir:22:1:61. [DOI] [PubMed] [Google Scholar]
- 34.Chen YH, Wu W, Yang J, Sui SF, Sun J, Dierich MP. Antibodies against human IFN-alpha and -beta recognized the immunosuppressive domain of HIV-1 gp41 and inhibit gp41-binding to the putative cellular receptor protein p45. Immunol Lett. 1999;69(2):253–7. doi: 10.1016/s0165-2478(99)00098-x. [DOI] [PubMed] [Google Scholar]
- 35.Brown KN, Wijewardana V, Liu X, Barratt-Boyes SM. Rapid influx and death of plasmacytoid dendritic cells in lymph nodes mediate depletion in acute simian immunodeficiency virus infection. PLoS Pathog. 2009;5(5):e1000413. doi: 10.1371/journal.ppat.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malleret B, Karlsson I, Maneglier B, et al. Effect of SIVmac infection on plasmacytoid and CD1c+ myeloid dendritic cells in cynomolgus macaques. Immunology. 2008;124(2):223–33. doi: 10.1111/j.1365-2567.2007.02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malleret B, Maneglier B, Karlsson I, et al. Primary infection with simian immunodeficiency virus: plasmacytoid dendritic cell homing to lymph nodes, type I interferon, and immune suppression. Blood. 2008;112(12):4598–608. doi: 10.1182/blood-2008-06-162651. [DOI] [PubMed] [Google Scholar]
- 38.Mandl JN, Barry AP, Vanderford TH, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med. 2008;14(10):1077–87. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- 39.Alpan O, Rudomen G, Matzinger P. The role of dendritic cells, B cells, and M cells in gut-oriented immune responses. J Immunol. 2001;166(8):4843–52. doi: 10.4049/jimmunol.166.8.4843. [DOI] [PubMed] [Google Scholar]
- 40.Chirdo FG, Millington OR, Beacock-Sharp H, Mowat AM. Immunomodulatory dendritic cells in intestinal lamina propria. Eur J Immunol. 2005;35(6):1831–40. doi: 10.1002/eji.200425882. [DOI] [PubMed] [Google Scholar]
- 41.Iwasaki A, Kelsall BL. Freshly isolated Peyer’s patch, but not spleen, dendritic cells produce interleukin 10 and induce the differentiation of T helper type 2 cells. J Exp Med. 1999;190(2):229–39. doi: 10.1084/jem.190.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwasaki A, Kelsall BL. Mucosal immunity and inflammation. I. Mucosal dendritic cells: their specialized role in initiating T cell responses. Am J Physiol. 1999;276(5 Pt 1):G1074–8. doi: 10.1152/ajpgi.1999.276.5.G1074. [DOI] [PubMed] [Google Scholar]
- 43.Iwasaki A, Kelsall BL. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J Exp Med. 2000;191(8):1381–94. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]