Abstract
Recent emphasis on translational research (TR) is highlighting the role of epidemiology in translating scientific discoveries into population health impact. The authors present applications of epidemiology in TR through 4 phases designated T1–T4, illustrated by examples from human genomics. In T1, epidemiology explores the role of a basic scientific discovery (e.g., a disease risk factor or biomarker) in developing a “candidate application” for use in practice (e.g., a test used to guide interventions). In T2, epidemiology can help to evaluate the efficacy of a candidate application by using observational studies and randomized controlled trials. In T3, epidemiology can help to assess facilitators and barriers for uptake and implementation of candidate applications in practice. In T4, epidemiology can help to assess the impact of using candidate applications on population health outcomes. Epidemiology also has a leading role in knowledge synthesis, especially using quantitative methods (e.g., meta-analysis). To explore the emergence of TR in epidemiology, the authors compared articles published in selected issues of the Journal in 1999 and 2009. The proportion of articles identified as translational doubled from 16% (11/69) in 1999 to 33% (22/66) in 2009 (P = 0.02). Epidemiology is increasingly recognized as an important component of TR. By quantifying and integrating knowledge across disciplines, epidemiology provides crucial methods and tools for TR.
Keywords: epidemiology, genomics, medicine, public health, translational research
In a recent editorial launching the new journal Science Translational Medicine, Dr. Elias Zerhouni, former director of the National Institutes of Health, remarked that despite decades of advances in our understanding of human biology and the emergence of powerful new technologies, such as genomics, the transformation of scientific discoveries into effective health interventions continues to elude us (2). There is daunting complexity when applying basic discoveries and experimental approaches to treating and preventing human disease, requiring a strong translational research (TR) agenda. He stressed the need for “more and better TR, both for the sake of our patients and because much of the research funding … comes from the primary expectation of the public that such scientific investigations will reduce the burden of disease” (2, p. 1). Translation of promising scientific discoveries into day-to-day practice is slow and uncertain (3), with only a few scientists willing to venture into the translation gap, sometimes referred to as the “valley of death” (4, p. 840). Perhaps no field has generated higher expectations, deeper frustrations, and more “translation anxiety” than advances in human genomics. In 2003, Dr. Claude Lenfant, former director of the US National Heart, Lung, and Blood Institute, remarked that the gap between what we know about diseases and what we do to prevent and treat them will likely grow wider as new technologies emerge, as there is plenty of evidence that even established interventions have been “lost in translation” (5, p. 868). He asked, “Let's be realistic: If we didn't do it with aspirin, how can we expect to do it with DNA?” (5, p. 874). In public health, even with decades-long knowledge, cigarette smoking, physical inactivity, and poor diet are the main contributors to the global burden of common chronic diseases, such as diabetes and coronary heart disease. Yet, progress has been slow in translating this knowledge into a reduction of the burden of these diseases (6). This may be due to many forces other than the lack of data and scientific evidence, including political, social, legal, and economical. In this paper, we present a framework for epidemiology as a fundamental building block of the TR enterprise, using illustrations from the emerging field of human genomics. The emerging field of genomics offers a nice example, because it has the advantage of showing the potential contribution of epidemiology to all phases of TR. Moreover, it exemplifies the difficulties of translation (despite high expectations) and the relatively steep decrease in the number of studies as one moves from T1 to T4. However, the framework can be applicable to all areas of health besides genomics.
Although epidemiologic methods have helped to discover the underlying causes of many human diseases, they may be even more crucial for providing the data for translating these discoveries into population health benefits. We discuss translational epidemiology (TE) along a multidisciplinary research continuum (Table 1; Figure 1) and highlight the function of knowledge synthesis at each stage. Over the past few years, the term “translational research” has been used in the context of clinical and other enterprises in the United States and globally (7–9); however, to our knowledge, this is the first paper to present an inclusive perspective of TE. Our main objective is to describe the overall contributions of epidemiology to TR. We hope that the framework will support further discussion of the contributions of epidemiology to translational research and spur the integration of epidemiologic concepts into training curricula for clinical and public health practice to meet the increasing challenge of translating scientific discoveries into population health benefits.
Table 1.
Epidemiology and the Phases of Translation and Knowledge Synthesis—From Discovery to Population Health Impact
| Phase | Details | Role of Epidemiology | Examples From Genomics |
| T0 | Description and discovery | Describing patterns of health outcomes by place, time, and person; finding determinants of health outcomes by use of observational studies | Describing patterns of health outcomes in relation to inbreeding, migration, and family history to generate hypotheses about genetic factors; genome-wide association studies as a tool for gene discovery |
| T1 | From discovery to health applications (tests, interventions) | Characterizing discovery and assessing potential health applications by using clinical and population studies | Assessing prevalence, associations, interactions, sensitivity, specificity, and predictive value of testing for genetic risk factors |
| T2 | From health application to evidence guidelines | Assessing the efficacy of interventions to improve health and prevent disease by using observational and experimental studies | Assessing the clinical utility of genetic risk factors in improving health outcomes |
| T3 | From guidelines to health practice | Assessing the implementation and dissemination of guidelines into practice | Assessing the factors associated with implementation of BRCA testing in practice |
| T4 | From health practice to population health outcomes | Assessing the effectiveness of interventions on health outcomes | Assessing the effectiveness of newborn screening programs |
| Knowledge synthesis | Systematic review of what we know and what we do not know and how we know it | Knowledge synthesis applies to all phases of translation by use of evidence synthesis and systematic reviews. | T1—evaluating the credibility of genetic associations and assessing the genetic effects and interactions (through HuGENet) |
| T2—systematic reviews on the clinical validity and utility of genomic applications for specific intended uses (through EGAPP appraisal) |
Abbreviations: EGAPP, Evaluation of Genomic Applications in Practice and Prevention; HuGENet, Human Genome Epidemiology Network; T0–T4, designated phases of translational research.
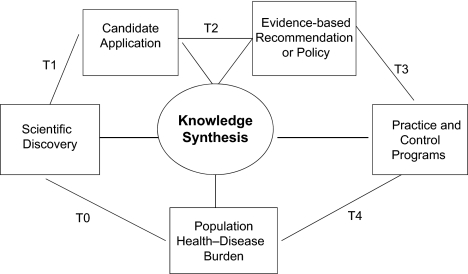
Figure 1.
Epidemiology and the phases of translational research: T0, scientific discovery research; T1, translational research from discovery to candidate application; T2, translational research from candidate application to evidence-based recommendation or policy; T3, translational research from recommendation to practice and control programs; T4, translational research from practice to population health impact.
THE CONTRIBUTION OF EPIDEMIOLOGY TO SCIENTIFIC DISCOVERIES
The schema in Figure 1 combines both clinical and public health approaches to translating scientific discoveries into effective, evidence-based approaches to treatment, prevention, and control of human disease in populations. A “scientific discovery” (designated T0) refers to new knowledge and insight into the causes, pathobiology, or natural history of disease. T0 research can come from laboratory sciences, as well as clinical and public health disciplines. In fact, epidemiology has long had an important role in describing disease occurrence in populations (descriptive epidemiology) and understanding the determinants of such occurrence (analytical or risk factor epidemiology). For example, epidemiologic studies helped to establish cigarette smoking as a major risk factor for lung cancer and folic acid as protective against the occurrence of neural tube defects (10). Because of the multilevel determinants of human disease (from molecular to social and ecologic factors), team or transdisciplinary science is increasingly recognized as the key to new discoveries (6). As remarked recently by Bob Hiatt, “… epidemiology has a central role in team science, no matter what the health issue at hand” (11, p. 859). In human genomics, consortia and networks have rapidly emerged as important infrastructure for the discovery of genetic risk factors for various common diseases, especially for genome-wide association studies, which require very large sample sizes for discovery and replication (12, 13).
TRANSLATIONAL RESEARCH: FROM SCIENTIFIC DISCOVERY TO POPULATION HEALTH IMPACT
A central dilemma in medicine and public health is the translation of basic discoveries into clinical practice and population health. A familiar concept in TR refers to the translation of basic laboratory discoveries to useful clinical applications as “bench to bedside” research (1, p. 211; 11, p. 859), as the “effective translation of the new knowledge, mechanisms, and techniques generated by advances in basic science research into new approaches for prevention, diagnosis, and treatment of disease” (1, p. 211). In 2006, the National Institutes of Health launched the Clinical and Translational Science Award program as a stimulus for TR, which it defined as “the process of applying discoveries generated during research in the laboratory, and in preclinical studies, to the development of trials and studies in humans” (14). Many laboratory research scientists view TR as a linear, unidirectional process for using laboratory discoveries to develop therapeutics for study in human clinical trials; however, even this form of TR is iterative, including feedback loops from bedside to bench (15). These concepts apply to the development of both therapeutic agents and biomarkers that could be used for diagnosis and early detection (e.g., prostate-specific antigen testing for early detection of prostate cancer).
On the other hand, public health and health services researchers tend to view TR as a process for developing evidence-based interventions and implementing them in practice, thus “ensuring that new treatments and research knowledge actually reach the patients or populations for whom they are intended and are implemented correctly” (1, p. 211). The production of a new drug, an end point for “bench-to-bedside” TR, is only the starting point for this second phase of translation. The Institute of Medicine’s Clinical Research Roundtable (16) reconciled these 2 views of TR by defining 2 research gaps: T1 is “the transfer of new understandings of disease mechanisms gained in the laboratory into the development of new methods for diagnosis, therapy, and prevention and their first testing in humans” (16, p. 1279); and T2 is “the translation of results from clinical studies into everyday clinical practice and health decision making” (16, p. 1279). One example is the evidence for whether or not prostate-specific antigen testing can be proven to improve morbidity and mortality when used in intervention trials.
Westfall et al. (17) proposed further dividing the second phase of TR, defining T2 as research to develop evidence-based recommendations and policies and T3 as research on implementing and disseminating evidence-based interventions in practice. For example, in the case of prostate-specific antigen, T2 would conduct trials to evaluate the benefits and harms of using prostate-specific antigen screening to detect prostate cancer, while T3 would implement research to get prostate-specific antigen screening in practice. Khoury et al. (18) recently adapted the TR framework to genomic medicine and added a final phase, T4, which focuses on evaluating the population-level health impact of interventions. In the case of prostate-specific antigen, T4 research would involve looking at the health impact of prostate screening in unselected populations (or real world practice). Figure 1 represents the relations among the 4 phases of TR research, with knowledge synthesis (discussed below) playing a central role. This schema is consistent with the comprehensive view of TR, which is not limited to drug development but includes behavioral, social, and policy interventions to address individual or collective determinants of health, recently published by Ogilvie et al. (19). It is noteworthy that Figure 1 depicts a logical progression from T1 through T4 research. However, the process is not necessarily linear. T3 and T4 research can be done on a candidate application that is not used in practice even before an evidence-based recommendation is made on its appropriate use (refer to the example of personal genomics tests below).
HOW DOES EPIDEMIOLOGY CONTRIBUTE TO TRANSLATIONAL RESEARCH?
Using the proposed T1–T4 translational research framework (Table 1; Figure 1), we examine the role of epidemiology in providing data to help translate initial basic discoveries into clinical and public health applications. We use the term “translational epidemiology” (TE) to describe the application of epidemiologic methods in all phases of TR (i.e., the study of disease distribution and determinants in populations). TE overlaps with and encompasses such terms as “applied epidemiology” (20), “clinical epidemiology” (21), and “field epidemiology” (22)—all referring to post-discovery research but used in various ways, depending on the context of applications of epidemiologic research, according to the purpose of the research, the phase of its application, and the determinants and outcomes of interest. TE is at the intersection of clinical and population-based research, including observational studies and randomized controlled trials (sometimes called experimental epidemiology) (23), assessing how basic discoveries can be used to improve health.
T1 epidemiologic research assesses the potential of a discovery for developing a “candidate application” for use in clinical or public health practice. Candidate applications are not limited to diagnostics or therapeutics; they can be considered more broadly to include behavioral, social, and policy interventions (e.g., tobacco regulations to help control smoking-related morbidity and mortality; physical activity to prevent type 2 diabetes).
In human genomics research, T1 epidemiologic research is needed to replicate and characterize genetic associations discovered by candidate gene studies and genome-wide association studies (24). For example, characterizing the prevalence of genetic risk factors in different populations and ethnic groups, assessing their contributions to disease burden, and evaluating gene-gene and gene-environment interactions are all questions requiring additional epidemiologic research (25). The importance of T1 epidemiologic research in human genomics cannot be underestimated. Recently, several companies have begun marketing so-called personal genomic tests directly to consumers; these tests are derived from discovery research and are offered with the implicit purpose of guiding health risk assessment and disease prevention (26). However, many associations included in personal genomic tests have not been replicated (27), and the risks calculated by different companies are often inconsistent (28). Yang et al. (29) recently showed that, in general, the estimated lifetime risks of disease include a large amount of uncertainty depending on variation in disease incidence rates, temporal trends, risk ratios, prevalence of genetic risk factors, and interactions with other risk factors. T1 epidemiologic research can begin to look beyond odds ratios (30) to applied measures, such as the sensitivity, specificity, and predictive value of genetic variants, singly or in combination, in the context of other disease risk factors (such as environmental exposures). For example, T1 epidemiologic research in human genomics includes the analysis of cohort studies to assess the value of adding genetic variants to conventional risk prediction models for coronary heart disease, some cancers, and type 2 diabetes (31). So far, these studies have shown that genetic risk factors add very little to the area under a receiver operating characteristic curve generated by well-established disease prediction algorithms (e.g., the Framingham score) (32).
T2 epidemiologic research is required to establish the clinical utility of a candidate application, concluding with a comprehensive assessment of the balance of benefits and harms of its use. Results of such research are the basis for evidence-based recommendations by professional groups or independent panels, such as the US Preventive Services Task Force (33). T2 epidemiologic research includes both observational studies and randomized controlled trials. Although phase III randomized controlled trials are routinely conducted to evaluate new drugs, behavioral interventions are also amenable to these trials. For example, the highly successful Diabetes Prevention Program recruited individuals at high risk for type 2 diabetes and randomized them prospectively to receive pharmacologic and behavioral interventions, including diet and physical activity (34).
In human genomics, T2 epidemiologic research assesses the value of genomic information in directing primary prevention, early detection, and treatment of disease (e.g., pharmacogenomics) (35). Principles of comparative effectiveness research are the basis for comparing the results of gene-directed interventions with those of standard interventions (36). So far, very few genomic applications have been evaluated for clinical utility (18, 37). In general, most discovered common genetic variants have low clinical validity and, furthermore, information from single or small sets of genetic markers discovered in genome-wide association studies is unlikely to have clinical utility (38). Even when a strong genetic association exists (e.g., factor V Leiden with recurrent venous thromboembolism), genetic testing may not improve clinical outcomes (39). In general, few randomized controlled trials have been conducted to evaluate the clinical utility of genomic applications in practice. One of the few examples of a genomic application evaluated by an ongoing randomized controlled trial is the use of breast cancer gene expression profiles to direct chemotherapy to women at high risk of recurrence (40).
T3 epidemiologic research addresses the major challenge of translating candidate applications into health-care practice and disease prevention programs. The Institute of Medicine's report, Crossing the Quality Chasm: A New Health System for the 21st Century, summarized the difficulty of effective implementation and diffusion of proven health-care interventions (41). According to McGlynn et al. (42), patients in the United States receive only half of the preventive services for which evidence-based recommendations exist. The overuse of inefficient or potentially harmful interventions is also an important concern (43). Discussing global health issues, Madon et al. recently remarked that “many evidence-based innovations fail to produce results when transferred to communities, largely because their implementation is untested, unsuitable, or incomplete” (44, p. 1728). Because health delivery schemes are difficult to study with randomized controlled trials, other epidemiologic approaches contribute to the “implementation sciences” for assessing facilitators and barriers to uptake and implementation of evidence-based recommendations. “Why do established programs lose effectiveness over days, weeks, or months? Why do tested programs sometimes exhibit unintended effects when transferred to a new setting? How can multiple interventions be effectively packaged to capture cost efficiencies and to reduce the splintering of health systems into disease specific programs? Answering questions like these will require analysis of biological, social, and environmental factors that impact implementation” (44, p. 1728).
In human genomics, few applications are currently ready for implementation in clinical practice. A notable exception is the breast cancer susceptibility gene (BRCA) mutation testing for assessing risk of breast and ovarian cancer. In 2005, the US Preventive Services Task Force issued an evidence-based recommendation that “women whose family history is associated with an increased risk for deleterious mutations in BRCA1 or BRCA2 genes should be referred for genetic counseling and evaluation for BRCA testing” (45). Very few studies to date have evaluated the extent and determinants of uptake of such recommendations in various clinical and population settings. For the personal genomic tests available (with no evidence-based recommendations), Kolor et al. (46) recently reported on 2 epidemiologic surveys of the population and primary care providers in the United States, and they have documented that a substantial fraction of the population and providers are aware of these tests. Determinants of awareness include gender (female), age (older groups), education (higher education), and race (whites). Although only a small proportion (0.3%) of respondents have used these tests, providers are getting questions about such tests from their patients and are likely to change some aspect of their practice as a result of these tests, even without the requisite evidence base around their use in practice. Simple T3 epidemiologic data such as these help to document the mismatch between scientific evidence and everyday practice, and they highlight the need for more education and oversight of such products (46).
T4 epidemiologic research evaluates the real world effectiveness of a candidate application in terms of population-level outcomes, such as morbidity, mortality, and disability, at the population or health-care-system level. As Ogilvie et al. (19) point out, the true end point of TR is not simply institutionalizing effective interventions but improving population health. Established epidemiologic methods for surveillance can be applied to risk factors (e.g., monitoring the prevalence of obesity and cigarette smoking by using the state-based Behavioral Risk Factor Surveillance System) (47) or disease occurrence: For example, cancer surveillance data have been used to model the impact of mammography screening on breast cancer mortality (48). This view of T4 research encompasses the whole spectrum of determinants of health, from the individual to the collective level, along with a corresponding spectrum of interventions.
In human genomics, very few applications have been evaluated by T4 research. Perhaps the most notable example is newborn screening for inherited metabolic disorders. Although mandated public health newborn screening programs have been in place for decades, they have only recently integrated new technologies (particularly tandem mass spectrometry) for identifying an expanded number of disorders (49). Surveillance and outcomes research are being used to document the real world effectiveness and potential harms of these new tests (50).
KNOWLEDGE SYNTHESIS: AN ENGINE FOR TRANSLATIONAL EPIDEMIOLOGY
Knowledge synthesis is a systematic approach to reviewing the evidence on what we know and what we do not know, and how we know it. We have indicated the essential role for knowledge synthesis in all phases of TR by placing it at the center of Figure 1. Knowledge synthesis methods, such as meta-analysis, are becoming standard in developing evidence-based recommendations for practice (T2 research). The Cochrane Collaboration (51), an international not-for-profit and independent organization founded in 1993, produces and disseminates systematic reviews of health-care interventions and promotes the search for evidence in the form of clinical trials and other studies of interventions. It continues to play a pivotal role in developing and promoting quantitative synthesis of evidence of what works and what does not work in health-care interventions. Increasingly, other independent groups, such as the US Preventive Services Task Force, are adopting similar methods (33, 52).
In human genomics, knowledge synthesis plays a key role in T1 epidemiologic research. Since 1998, the Human Genome Epidemiology Network (HuGENet) (53) has sought to synthesize information on gene-disease associations through human genome epidemiology (HuGE) reviews and meta-analyses. The Journal has been an active partner with HuGENet in publishing numerous HuGE reviews over the past decade (54). HuGENet has also drafted guidelines for assessing the credibility of genetic associations based on the amount of evidence, the extent of replication, and the degree of protection from bias (55). It has become the norm for publications reporting a discovery from genome-wide association studies to include a meta-analysis of replication data sets (56, 57). As genomics research begins to yield candidate applications for clinical and public health practice, knowledge synthesis will have an increasingly important role in T2 research. For example, the Centers for Disease Control and Prevention-supported initiative, Evaluation of Genomic Applications in Practice and Prevention, known as EGAPP, is a rigorous process for evaluating the evidence on analytical and clinical validity and the clinical utility of genomic applications for clinical and public health practice in the United States (58, 59). An independent EGAPP Working Group selects topics, oversees systematic reviews of evidence, and makes evidence-based recommendations; 5 systematic reviews and 4 evidence recommendations had been published by 2009 (58).
TRANSLATIONAL EPIDEMIOLOGY IN THE AMERICAN JOURNAL OF EPIDEMIOLOGY
Finally, we thought it would be instructive to describe the trends and types of translational research articles published in the Journal as a snapshot of the field. Although analysis of any one journal is not necessarily generalizable to the whole field, we were curious to see how much scientific discovery and translational research are published in a premier epidemiology journal. We compared the contents of 5 issues of the American Journal of Epidemiology published in 2009 with the corresponding issues published in 1999. We read the titles and abstracts of all the articles and classified them according to our TE framework, from T0 through T4 and knowledge synthesis. We assessed all the papers in each selected issue and used a simple classification of the article by purpose of the study. We applied the definitions of T0–T4 described in Table 1. If a paper addressed multiple phases of translation, we coded the highest phase of translation. For illustration, Table 2 shows examples (60–65) of each type of paper in 1999 and in 2009. The proportion of articles that could be classified as TE increased from 16% (11/69) in 1999 to 33% (22/66) in 2009 (P = 0.02 by chi-square test; Table 2). Although this exercise is by no means an exhaustive or representative analysis of the epidemiology literature, it suggests a positive trend. We are planning additional analyses of the published literature in epidemiology, and clinical and public health journals could help to clarify the trends in translational research and the contributions of epidemiology to the field. It is possible that each journal has its own preferred niche in the T1–T4 continuum, and an evaluation of many journals could give a wider perspective about the interface of translational research and epidemiology.
Table 2.
Articles in the American Journal of Epidemiology by Translation Phase, 1999 and 2009
| Issue and Year | Descriptive/Discovery Epidemiology, no. | Translational Epidemiology, no. | Total, no. |
| January 1 | |||
| 2009 | 9 | 3 (1 T1, 1 T4, 1 KS) | 12 |
| 1999 | 18 | 2 (1 T1, 1 KS) | 20 |
| March 1 | |||
| 2009 | 8 | 5 (2 T1, 2 T4, 1 KS) | 13 |
| 1999 | 9 | 1 (1 KS) | 10 |
| July 1 | |||
| 2009 | 10 | 4 (2 T1, 1 T4, 1 KS) | 14 |
| 1999 | 10 | 3 (3 T1) | 13 |
| October 1 | |||
| 2009 | 10 | 4 (3 T1, 1 T4) | 14 |
| 1999 | 11 | 2 (1 T1, 1 T4) | 13 |
| December 1 | |||
| 2009 | 7 | 6 (1 T1, 2 T4, 3 KS) | 13 |
| 1999 | 10 | 3 (1 T1, 2 T4) | 13 |
| Total | |||
| 2009 | 44 | 22 (9 T1, 7 T4, 6 KS)a | 66 |
| 1999 | 58 | 11 (6 T1, 3 T4, 2 KS)b | 69 |
Abbreviations: KS, knowledge synthesis; T0–T4, designated phases of translational research.
Examples of translational epidemiology in 2009 include T1 (reference 60), T4 (reference 61), and KS (reference 62).
Examples of translational epidemiology in 1999 include T1 (reference 63), T4 (reference 64), and KS (reference 65).
CONCLUDING REMARKS
In summary, we have presented a framework for epidemiology as a fundamental science for translating basic discoveries into population health benefits. The TE framework combines clinical and public health approaches to disease treatment, prevention, and control. In combination with basic, clinical, and other population sciences, TE provides the key data needed to document what we know and what we do not know, and what works and what does not work, thus influencing further research, practice, and policy development. The framework encompasses previously described areas of epidemiology, such as clinical and applied epidemiology, and unifies various concepts according to the purpose of the research, the phase of its application, and the determinants and outcomes of interest. We hope this framework supports further discussion of the field's potential contributions to translational research and spurs the continued integration of epidemiologic concepts into training curricula for clinical and public health practice. Finally, it is important to acknowledge that actual translation is much more complicated as different forces—such as private investments in research and development, policy and legal frameworks, oversight and regulation, product marketing, coverage and reimbursements, consumer advocacy, provider awareness, and consumer access—can accelerate or impede the translation process. These factors can often operate independently from research evidence as we recently explored in genomic medicine (66). Nevertheless, TE is a necessary ingredient to move specific discoveries from research into practice in an evidence-based fashion. In an era of health reform and comparative effectiveness research, epidemiologic data are fundamental to informed decision-making by health-care providers, policy makers, and citizens.
Acknowledgments
Author affiliations: Office of Public Health Genomics, Centers for Disease Control and Prevention, Atlanta, Georgia (Muin J. Khoury, Marta Gwinn); Division of Cancer Control and Population Sciences, National Cancer Institute, Bethesda, Maryland (Muin J. Khoury); Department of Hygiene and Epidemiology, University of Ioannina School of Medicine and Biomedical Research Institute, Ioannina, Greece (John P. A. Ioannidis); Institute for Clinical Research and Health Policy Studies and Tufts Clinical and Translational Science Institute, Tufts Medical Center and Tufts University School of Medicine, Boston, Massachusetts (John P. A. Ioannidis); and Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts (John P. A. Ioannidis).
The opinions in this paper are those of the authors and do not necessarily reflect those of the US Department of Health and Human Services.
Conflict of interest: none declared.
Glossary
Abbreviations
- HuGE
human genome epidemiology
- HuGENet
Human Genome Epidemiology Network
- TE
translational epidemiology
- TR
translational research
References
- 1.Woolf SH. The meaning of translational research and why it matters. JAMA. 2008;299(2):211–213. doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- 2.Zerhouni EA. Space for the cures: science launches a new journal dedicated to translational research in biomedicine. Sci Trans Med. 2009;1(1):1–2. doi: 10.1126/scitranslmed.3000341. [DOI] [PubMed] [Google Scholar]
- 3.Contopoulos-Ioannidis DG, Alexiou GA, Gouvias TC, et al. Life cycle of translational research for medical interventions. Science. 2008;321(5894):1298–1299. doi: 10.1126/science.1160622. [DOI] [PubMed] [Google Scholar]
- 4.Butler D. Translational research: crossing the valley of death. Nature. 2008;453(7197):840–842. doi: 10.1038/453840a. [DOI] [PubMed] [Google Scholar]
- 5.Lenfant C. Shattuck lecture—clinical research to clinical practice—lost in translation? N Engl J Med. 2003;349(9):868–874. doi: 10.1056/NEJMsa035507. [DOI] [PubMed] [Google Scholar]
- 6.Collins JL, Marks JS, Koplan JP. Chronic disease prevention and control: coming of age at the Centers for Disease Control and Prevention [editorial] Prev Chronic Dis. 2010;6(3):A81. [PMC free article] [PubMed] [Google Scholar]
- 7.MRC Center for causal analyses in translational epidemiology (CAITE) Bristol, United Kingdom: University of Bristol; 2010. University of Bristol, United Kingdom. ( http://www.bristol.ac.uk/caite/). (Accessed January 1, 2010) [Google Scholar]
- 8.Mayo Clinic. Rochester, MN: Mayo Clinic; 2010. Multidisciplinary epidemiology and translational research in intensive care (METRIC) ( http://mayoresearch.mayo.edu/gajic_lab/). (Accessed January 1, 2010) [Google Scholar]
- 9.Wayne State University. Detroit, MI: Wayne State University; 2010. Translational research and clinical epidemiology (TRACE) ( http://trace.med.wayne.edu/). (Accessed January 1, 2010) [Google Scholar]
- 10.Ness RB, Andrews EB, Gaudino JA, et al. The future of epidemiology. Acad Med. 2009;84(11):1631–1637. doi: 10.1097/ACM.0b013e3181bbb4ed. [DOI] [PubMed] [Google Scholar]
- 11.Hiatt RA. Epidemiology: key to translational, team and transdisciplinary science. Ann Epidemiol. 2008;18(11):859–861. doi: 10.1016/j.annepidem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Seminara D, Khoury MJ, O'Brien TR, et al. The emergence of networks in human genome epidemiology: challenges and opportunities. Epidemiology. 2007;18(1):1–8. doi: 10.1097/01.ede.0000249540.17855.b7. [DOI] [PubMed] [Google Scholar]
- 13.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Institutes of Health. Bethesda, MD: National Institutes of Health; 2005. Institutional Clinical and Translational Science Award. ( http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-06-002.html). (Accessed January 1, 2010) [Google Scholar]
- 15.Canadian Institutes for Health Research. Ottawa, Canada: Canadian Institutes for Health Research; 2009. About knowledge translation. ( http://www.cihr-irsc.gc.ca/e/29418.html). (Accessed January 10, 2010) [Google Scholar]
- 16.Sung NS, Crowley WF, Jr, Genel M, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289(10):1278–1287. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 17.Westfall JM, Mold J, Faqnan L. Practice-based research—blue highways on the NIH road map. JAMA. 2007;297(4):403–406. doi: 10.1001/jama.297.4.403. [DOI] [PubMed] [Google Scholar]
- 18.Khoury MJ, Gwinn M, Yoon PW, et al. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9(10):665–674. doi: 10.1097/GIM.0b013e31815699d0. [DOI] [PubMed] [Google Scholar]
- 19.Ogilvie D, Craig P, Griffin S, et al. A translational framework for public health research. BMC Public Health. 2009;9:116. doi: 10.1186/1471-2458-9-116. (doi: 10.1186/1471-2458-9-116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thacker SB, Buffington J. Applied epidemiology for the 21st century. Int J Epidemiol. 2001;30(2):320–325. doi: 10.1093/ije/30.2.320. [DOI] [PubMed] [Google Scholar]
- 21.Sackett DL. Clinical epidemiology. What, who, and whither. J Clin Epidemiol. 2002;55(12):1161–1166. doi: 10.1016/s0895-4356(02)00521-8. [DOI] [PubMed] [Google Scholar]
- 22.Gregg MB. Field Epidemiology. 2nd ed. New York, NY: Oxford University Press; 2002. [Google Scholar]
- 23.Ahrens W, Krickeberg K, Pigeot I. Introduction to epidemiology. In: Ahrens W, Pigeot I, editors. Handbook of Epidemiology. Berlin, Germany: Springer; 2005. pp. 1–42. [Google Scholar]
- 24.Wellcome Trust Case Control Consortium. Genomewide association study in 14000 cases of common diseases and 3000 controls. Nature. 2007;447(7145):661–667. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoury MJ, Bedrosian S, Gwinn M, et al., editors. Chapter 1 Human genome epidemiology—the road map revisited. In: Human Genome Epidemiology: Building the Evidence for Using Genetic Information to Improve Health and Prevent Disease. 2nd ed. New York, NY: Oxford University Press; 2010. pp. 3–12. [Google Scholar]
- 26.Khoury MJ, McBride CM, Schully SD, et al. The scientific foundation for personal genomics: recommendations from a National Institutes of Health–Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009;11(8):559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssens AC, Gwinn M, Bradley LA, et al. A critical appraisal of the scientific basis of commercial genomic profiles used to assess health risks and personalize health interventions. Am J Hum Genet. 2008;82(3):593–595. doi: 10.1016/j.ajhg.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng PC, Murray SS, Levy S, et al. An agenda for personalized medicine. Nature. 2009;461(7265):724–726. doi: 10.1038/461724a. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, Flanders WD, Moonesinghe R, et al. Using lifetime risk estimates in personal genomic profiles: estimation of uncertainty. Am J Hum Genet. 2009;85(6):786–800. doi: 10.1016/j.ajhg.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraft P, Wacholder S, Cornelis MC, et al. Beyond odds ratios—communicating disease risk based on genetic profiles. Nat Rev Genet. 2009;10(4):264–269. doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 31.Janssens AC, van Duijn CM. Genome-based prediction of common diseases: methodological considerations for future research. Genome Med. 2009;1(2):20. doi: 10.1186/gm20. (doi:10.1186/gm20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzoulaki I, Liberopoulos G, Ioannidis JP. Assessment of claims of improved prediction beyond the Framingham risk score. JAMA. 2009;302(21):2345–2352. doi: 10.1001/jama.2009.1757. [DOI] [PubMed] [Google Scholar]
- 33.Washington, DC: US Preventive Services Task Force; 2010. Agency for HealthCare Research Quality, US Preventive Services Task Force (USPSTF) ( http://www.ahrq.gov/CLINIC/uspstfix.htm). (Accessed January 1, 2010) [Google Scholar]
- 34.National Institute of Diabetes and Digestive and Kidney Diseases. Bethesda, MD: National Institute of Diabetes and Digestive and Kidney Diseases; 2010. Diabetes Prevention Program Outcomes Study. ( http://www.niddk.nih.gov/patient/dpp/dppos.htm). (Accessed January 1, 2010) [Google Scholar]
- 35.Limdi NA, Veenstra DL. Expectations, validity, and reality in pharmacogenetics. [published online ahead of print December 7, 2009]. J Clin Epidemiol. (doi:10.1016/j.jclinepi.2009.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khoury MJ, Rich EC, Randhawa G, et al. Comparative effectiveness research and genomic medicine: an evolving partnership for 21st century medicine. Genet Med. 2009;11(10):707–711. doi: 10.1097/GIM.0b013e3181b99b90. [DOI] [PubMed] [Google Scholar]
- 37.Schully SD, Benedicto CB, Gillanders EM, et al. Translational research in cancer genetics: the road less traveled. [published online ahead of print December 29, 2009]. Public Health Genomics. (doi: 10.1159/000272897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal JB, Brotman DJ, Necochea AJ, et al. Predictive value of factor V Leiden and prothrombin G20210A in adults with venous thromboembolism and in family members of those with a mutation: a systematic review. JAMA. 2009;301(23):2472–2485. doi: 10.1001/jama.2009.853. [DOI] [PubMed] [Google Scholar]
- 39.Ioannidis JP. Personalized genetic prediction: too limited, too expensive, or too soon? Ann Intern Med. 2009;150(2):139–141. doi: 10.7326/0003-4819-150-2-200901200-00012. [DOI] [PubMed] [Google Scholar]
- 40.National Cancer Institute. Bethesda, MD: National Cancer Institute; 2010. TailorX: testing personalized treatment for breast cancer. ( http://www.cancer.gov/clinicaltrials/digestpage/TAILORx). (Accessed January 1, 2010) [Google Scholar]
- 41.Institute of Medicine. Crossing the Quality Chasm: A New Health System for the 21st Century. Washington, DC: National Academy Press; 2001. [PubMed] [Google Scholar]
- 42.McGlynn EA, Asch SM, Adams J, et al. The quality of health care delivered to adults in the United States. N Engl J Med. 2003;348(26):2635–2645. doi: 10.1056/NEJMsa022615. [DOI] [PubMed] [Google Scholar]
- 43.Chalmers I, Matthews R. What are the implications of optimism bias in clinical research? Lancet. 2006;367(9509):449–450. doi: 10.1016/S0140-6736(06)68153-1. [DOI] [PubMed] [Google Scholar]
- 44.Madon T, Hofman KJ, Kupfer L, et al. Public health. Implementation science. Science. 2007;318(5857):1728–1729. doi: 10.1126/science.1150009. [DOI] [PubMed] [Google Scholar]
- 45.US Preventive Services Task Force. Washington, DC: US Preventive Services Task Force; 2005. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility. ( http://www.ahrq.gov/clinic/uspstf/uspsbrgen.htm). (Accessed January 1, 2010) [Google Scholar]
- 46.Kolor KK, Liu T, St Pierre J, et al. Health care provider and consumer awareness, perceptions, and use of direct-to-consumer personal genomic tests, United States, 2008 [letter] Genet Med. 2009;11(8):595. doi: 10.1097/GIM.0b013e3181b1cc2c. [DOI] [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention. Atlanta, GA: Centers for Disease Control and Prevention; 2010. The Behavioral Risk Factor Surveillance System (BRFSSS) ( http://www.cdc.gov/brfss/). (Accessed January 1, 2010) [Google Scholar]
- 48.Mandelblatt JS, Cronin KA, Bailey S, et al. Effects of mammography screening under different screening schedules: model estimates of potential benefits and harms. Ann Intern Med. 2009;151(10):738–747. doi: 10.1059/0003-4819-151-10-200911170-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCabe LL, McCabe ER. Expanded newborn screening: implications for genomic medicine. Annu Rev Med. 2008;59:163–175. doi: 10.1146/annurev.med.59.110106.132016. [DOI] [PubMed] [Google Scholar]
- 50.Grosse SD, Rogowski WH, Ross LF, et al. Population screening for genetic disorders in the 21st century: evidence, economics, and ethics. Public Health Genomics. 2010;13(2):106–115. doi: 10.1159/000226594. [DOI] [PubMed] [Google Scholar]
- 51.The Cochrane Collaboration. Ottawa, Canada: The Cochrane Collaboration; 2010. ( http://www.cochrane.org/). (Accessed January 1, 2010) [Google Scholar]
- 52.Agency for Healthcare Quality Research. Washington, DC: Agency for Healthcare Quality Research; 2010. Evidence-based practice centers. ( http://www.ahrq.gov/clinic/epc/). (Accessed January 1, 2010) [Google Scholar]
- 53.Centers for Disease Control and Prevention. Atlanta, GA: Centers for Disease Control and Prevention; 2010. The Human Genome Epidemiology Network (HuGENet) ( http://www.cdc.gov/genomics/hugenet/default.htm). (Accessed January 1, 2010) [Google Scholar]
- 54.Higgins JP, Little J, Ioannidis JP, et al. Turning the pump handle: evolving methods for integrating the evidence on gene-disease association. Am J Epidemiol. 2007;166(8):863–866. doi: 10.1093/aje/kwm248. [DOI] [PubMed] [Google Scholar]
- 55.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 56.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–368. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 57.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atlanta, GA: Office of Public Health Genomics, Centers for Disease Control and Prevention; 2010. Evaluation of Genomic Applications in Practice and Prevention (EGAPP) ( http://www.egappreviews.org/). (Accessed January 1, 2010) [Google Scholar]
- 59.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11(1):3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei EK, Colditz GA, Giovannucci EL, et al. Cumulative risk of colon cancer up to age 70 years by risk factor status using data from the Nurses’ Health Study. Am J Epidemiol. 2009;170(7):863–872. doi: 10.1093/aje/kwp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mosley BS, Cleves MA, Siega-Riz AM, et al. Neural tube defects and maternal folate intake among pregnancies conceived after folic acid fortification in the United States. National Birth Defects Prevention Study. Am J Epidemiol. 2009;169(1):9–17. doi: 10.1093/aje/kwn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zintzaras E, Zdoukopoulos N. A field synopsis and meta-analysis of genetic association studies in peripheral arterial disease: the CUMAGAS-PAD database. Am J Epidemiol. 2009;170(1):1–11. doi: 10.1093/aje/kwp094. [DOI] [PubMed] [Google Scholar]
- 63.Chen W, Srinivasan SR, Elkasabany A, et al. Cardiovascular risk factors clustering features of insulin resistance syndrome (syndrome X) in a biracial (black-white) population of children, adolescents, and young adults: the Bogalusa Heart Study. Am J Epidemiol. 1999;150(7):667–674. doi: 10.1093/oxfordjournals.aje.a010069. [DOI] [PubMed] [Google Scholar]
- 64.Mossong J, Nokes DJ, Edmunds WJ, et al. Modeling the impact of subclinical measles transmission in vaccinated populations with waning immunity. Am J Epidemiol. 1999;150(11):1238–1249. doi: 10.1093/oxfordjournals.aje.a009951. [DOI] [PubMed] [Google Scholar]
- 65.Katerndahl DA, Lawler WR. Variability in meta-analytic results concerning the value of cholesterol reduction in coronary heart disease: a meta-meta-analysis. Am J Epidemiol. 1999;149(5):429–441. doi: 10.1093/oxfordjournals.aje.a009830. [DOI] [PubMed] [Google Scholar]
- 66.Khoury MJ. Dealing with the evidence dilemma in genomics and personalized medicine. Clin Pharm Ther. 2010;87(6):635–638. doi: 10.1038/clpt.2010.4. [DOI] [PubMed] [Google Scholar]