Abstract
DNA replication and the correct packaging of DNA into different states of chromatin are both essential processes in all eukaryotic cells. High-fidelity replication of DNA is essential for the transmission of genetic material to cells. Likewise the maintenance of the epigenetic chromatin states is essential to the faithful reproduction of the transcriptional state of the cell. It is becoming more apparent that these two processes are linked through interactions between DNA replication proteins and chromatin-associated proteins. In addition, more proteins are being discovered that have dual roles in both DNA replication and the maintenance of epigenetic states. We present an analysis of two Drosophila mutants in the conserved DNA replication protein Mcm10. A hypomorphic mutant demonstrates that Mcm10 has a role in heterochromatic silencing and chromosome condensation, while the analysis of a novel C-terminal truncation allele of Mcm10 suggests that an interaction with Mcm2 is not required for chromosome condensation and heterochromatic silencing but is important for DNA replication.
THE essential process of DNA replication does not occur in a vacuum; rather, it takes place within the context of the cell. More specifically, DNA replication occurs within the context of chromatin: an integrated network of DNA-associated proteins that have roles in packaging DNA, controlling transcription, and maintaining genome integrity. The maintenance and manipulation of these chromatin proteins are, like DNA replication, an essential process. The packaging of DNA has significant consequences for the transcriptional state of the underlying DNA. Repression or activation of different regions of the genome through packaging as open euchromatin or as repressive heterochromatin is cell type specific (Fraser et al. 2009; Minard et al. 2009). Moreover, these transcriptional states must be maintained and passed on to daughter cells during mitosis. If not passed on faithfully, genome instability and/or transcriptional misregulation can occur, both of which may lead to defects in cell proliferation, cancer, and other disease states (Jones et al. 2007; Hirst and Marra 2009).
By necessity, the process of DNA replication requires unencumbered access to the nitrogenous bases that make up the DNA strand. As a result, chromatin proteins must be removed. In the wake of the DNA replication fork this nascent DNA must be repackaged to recapitulate the previous chromatin state. While DNA replication benefits from complementary base pairing to build a DNA molecule through semiconservative replication, the reestablishment of epigenetic states occurs through more subtle and varied mechanisms (Groth et al. 2007). One central question in reconciling the processes of DNA replication and the establishment and/or maintenance of chromatin states is how are these processes linked? One model suggests that DNA replication proteins interact with separate chromatin establishment factors, thereby spatially linking the two processes. Supporting this model has been the discovery that a number of nonreplication proteins that associate with the DNA replication fork have been shown to have roles in the establishment of chromatin states (Groth et al. 2007). Another complementary model for the establishment of epigenetic states posits that DNA replication factors themselves have distinct roles in the establishment of different chromatin states. An excellent example of this has been the work on the origin recognition complex (ORC). The ORC has been shown to be a structural component of heterochromatin in yeast and has been shown in Drosophila to physically interact with Heterochromatin protein 1 (Hp1) and be involved in its correct localization (Pak et al. 1997; Huang et al. 1998; Shareef et al. 2001; Gerbi and Bielinsky 2002; Rusché et al. 2002). Finally, replication timing has been implicated in the establishment of chromatin structure with early S-phase replication being associated with euchromatin and late S-phase replication associated with heterochromatin (Hiratani and Gilbert 2009).
Enter into this, Mcm10. Mcm10 is a highly conserved protein that was identified in Saccharomyces cerevisiae in the same minichromosome maintenance assay that yielded the well-studied Mcm2–7 proteins that likely constitute the replicative helicase (Merchant et al. 1997; Tye and Tye 1999). Temperature-sensitive mcm10 mutants in yeast arrest in S phase with a 2C DNA content. At permissive temperatures these mutants are characterized by excessive pausing of replication forks at unfired origins of replication (Merchant et al. 1997). Further studies have firmly established a role for Mcm10 in replication. It has been shown to interact with members of the prereplication complex and elongation complex (Merchant et al. 1997; Homesley et al. 2000; Izumi et al. 2000; Christensen and Tye 2003; Lee et al. 2003; Das-Bradoo et al. 2006; Chattopadhyay and Bielinsky 2007; Zhu et al. 2007). Curiously, like the Mcm2–7 proteins, Mcm10 is exceptionally abundant in eukaryotic cells with nearly 40,000 molecules per haploid yeast cell (Kawasaki et al. 2000). A number of studies have suggested that only a subset of the Mcm10 present in the cell may be utilized in DNA replication processes. In S. cerevisiae a portion of the Mcm10 protein pool is diubiquitinated. This modified form of Mcm10 participates in an interaction with PCNA that is essential for cell proliferation (Das-Bradoo et al. 2006). Also suggesting that the majority of Mcm10 does not participate in essential processes is the observation that Drosophila tissue culture cells that are depleted of Mcm10 by RNAi continue to proliferate even with very low levels of Mcm10 (Christensen and Tye 2003). If, as some evidence suggests, only a portion of the Mcm10 pool is utilized for DNA replication, then in what other nonessential processes does Mcm10 play a role? Recently evidence has been uncovered that points to an involvement of Mcm10 in chromatin structure. Work using S. cerevisiae has demonstrated that Mcm10 is involved in transcriptional repression of the mating-type loci and links DNA replication proteins to heterochromatin formation (Douglas et al. 2005; Liachko and Tye 2005, 2009). Also pointing to a possible role in chromatin structure and chromosome condensation is that the depletion of Mcm10 in Drosophila tissue culture cells results in undercondensed metaphase chromosomes (Christensen and Tye 2003).
Here we add to the growing body of evidence for alternate roles for Mcm10 through the examination of two different mutant Mcm10 alleles in Drosophila melanogaster and the dissection of the domains of the Mcm10 protein related to key protein–protein interactions. A hypomorphic allele of Mcm10 displays defects consistent with a role for Mcm10 in DNA replication, chromosome condensation, and heterochromatin formation. On the other hand a C-terminal truncation allele of Mcm10 is attenuated for interaction with Mcm2 but does not display any defects in assays for chromosome condensation and heterochromatin formation. Taken together, the analysis of these two alleles suggests that Mcm10 may have separable roles in DNA replication, chromosome condensation, and heterochromatin formation.
MATERIALS AND METHODS
Fly husbandry/stocks:
Fly stocks (Mcm10Scim19 FlyBase ID, FBst0013070, y[1] w[67c23]; P{y[+mDint2] w[BR.E.BR]=SUPor-P}Mcm10[KG00233]; Mcm10d08029 FlyBase ID, FBst1011557, P{XP}Mcm10[d08029]; and Hp15 FlyBase ID, FBst0006234, In(1)w[m4h]; Su(var)205[5]/In(2L)Cy, In(2R)Cy, Cy[1]) were obtained from the Bloomington Fly Stock Center and the Exelixis Drosophila Stock Collection at Harvard Medical School. The dumpy variegating line (w; SM1/dp[w18] T(2:3) 3L^2R 2L^3L) and a Gla,dpov line (Gla, dp[Olv-12]/Cy, Roi) were kindly provided by R. MacIntyre (Cornell University). Mcm10 P-element insertions were confirmed by PCR (data not shown). The Mcm10 lines were both backcrossed more than seven times to w; Df(2L), b[82-2]/CyO to remove unwanted second-site mutations. Wild-type controls were generated through precise P-element excision from these respective lines and additional backcrosses to the deficiency line above. In all assays, both of these wild-type controls were identical and data for these wild-type controls were pooled. All fly stocks were maintained on Caltech media (U.S. Biological no. D9600-07) at room temperature.
Mcm10 transgene-containing flies were generated through germline transformation (BestGene Inc.). The transgenic construct was based on the Murphy vector pTWF where genomic Mcm10 was cloned into the Gateway system (Invitrogen, Carlsbad, CA). The 3.5-kb genomic MCM10 insert contains 1137 bp of the promoter region upstream of the coding sequence and has had the stop codon removed for C-terminal fusion of the FLAG tag in the pTWF vector. A transgenic fly line containing a nonlethal insertion into the third chromosome was identified and expression of the fusion protein was verified in early embryos by Western blot (anti-Flag) performed as described below (data not shown).
Antibodies/Western blots:
Antibodies and Western blots were performed as in Christensen and Tye (2003), except that starting tissues were early embryos from the genotypes indicated in the text. These were collected from well-fed females on grape agar plates after allowing 8 hr of oviposition. Tubulin loading controls were 1:4000, antitubulin DM1a, (Sigma, St. Louis).
Pupae size analysis:
Cleaned microscope slides were placed vertically in a fly food bottle containing the genotype indicated in the text. Third instar larvae were allowed to wander onto the slides and pupate. Slides were then removed and scanned using a flatbed scanner. Resulting images were analyzed using the Motic Plus imaging and analysis software package. Statistical analysis was performed using Minitab.
Hatch rates:
Recently eclosed, well fed, Drosophila were allowed to deposit eggs on yeast-dusted grape plates at 26° for 2 hr. Eggs were then counted. These grape plates were incubated for 24 hr at 26°, after which unhatched eggs were counted. A minimum of three independent trials were conducted for each genotype.
RNA extraction and RT–PCR:
Embryos (0–5 hr) were collected from each strain (Mcm10+, Mcm10Scim19, and Mcm10d08029) using grape agar plates, washed three times with 1-ml volumes of sterile distilled water, and stored in 1.5-ml tubes at −80° until RNA extractions were conducted. Total RNA was extracted from each strain using the TōTALLY RNA Kit (Ambion) from a 100-μl volume of embryos as directed in the manufacturer's protocol. The integrity of the extracted RNA was assayed by native agarose gel electrophoresis, using Ambion's supplied formaldehyde loading dye, and the concentration of the RNA preparations was quantified spectrophotometrically. First-strand cDNA synthesis was conducted using the SuperScript III First-Strand Synthesis System (Invitrogen), following manufacturer's protocol using 5 μg of total RNA extract from each strain.
RT–PCR was conducted using standard procedures (one 2-min denaturing step at 95°, 45-sec denaturing at 95°, 45-sec annealing at 60°, 1.5-min extension at 72°, and one final 5-min extension at 72°, for 20 cycles), using primers that amplified a portion of the Mcm10 gene common to both the full-length and the truncation allele: 5′-CACCATGGGTCCTGCTCA-3′ and 5′-TCAGACAGCGGGTGTGC-3′. The RP49 control was amplified as above except using primers 5′-CGGATCGATATGCTAAGCTGT-3′ and 5′-GCGCTTGTTCGATCCGTA-3′ for 17 cycles. For each RT–PCR reaction 2 μl of the appropriate first-strand synthesis reaction was used as a template to amplify each Mcm10 variant and the rp49 control, using a final concentration of 0.3 μm of appropriate primers and GoTaq DNA polymerase (Promega, Madison, WI). RT–PCR products were analyzed by mixing 5 μl of the Mcm10 variant reactions with 5 μl of their respective rp49 control reactions and assaying for band intensity using agarose gel electrophoresis and ethidium bromide staining (50 ng/ml). Gel imaging was conducted using the Gel Logic 100 Imaging System (Carestream Molecular Imaging), and densitometry was conducted with Kodak 1D Image Analysis Software using the asymmetric Gaussian fitting function (band sensitivity, 0; profile width, 80%).
Yeast two-hybrid system:
Yeast manipulation and growth were conducted using standard protocols as in Christensen and Tye (2003) and as found in manufacturer's protocols (Clontech; Matchmaker Yeast Two-Hybrid System). Yeast strain AH109 (Clontech) was used as the reporter strain. Plasmids used were pGBKT7 and pGADT7 (Clontech) except that both were converted to the Gateway cloning system (Invitrogen) by insertion of the Gateway cassette into the multiple cloning site. In addition, the KanR gene in pGBKT7 was disrupted by insertion of the AmpR gene to facilitate use in the Gateway system. Entry clones were all sequence verified prior to LR reactions with two-hybrid plasmids. Resulting two-hybrid clones were then sequence verified to ensure the proper reading frame was maintained and no ectopic mutations were introduced.
Polytene chromosomes and early embryos:
Third instar wandering larvae were harvested from age- and density-matched bottles and dissected in 1× PBS, pH 7.2, with 1% PEG 8000. Salivary glands were then transferred to a solution of 50% acetic acid, 2–3% lactic acid, and 3.7% formaldehyde and fixed for 2 min. Glands were transferred to a clean microscope slide and overlaid with a siliconized coverslip. Polytene chromosomes were spread using spiral tapping with a dull pencil. Spreading was monitored using phase-contrast microscopy. Once spread, the microscope slide and coverslip were sandwiched between filter paper and an additional microscope slide. This was then placed in a machinist's vise and pressure was applied using a torque wrench to 15 Newton meter. Following a 2-min incubation at this pressure, the slide and coverslip were removed and lowered into liquid nitrogen. Once equilibrated, the slide and coverslip were removed; the coverslip was popped off; and the slide was washed gently with 100% EtOH, allowed to air dry, and mounted with 7 μl of Vectashield with DAPI.
Embryos were collected, fixed with methanol/EGTA, prepared, and stained as in Kellum and Alberts (1995). Microscopy was performed using an Olympus IX81 motorized inverted microscope with a spinning disk confocal controlled by SlideBook software.
Larval brain squashes/mitotic index:
Third instar wandering larvae were harvested and dissected as for polytene chromosome preparations. Removed larval brains were transferred to hypotonic solution (0.5% sodium citrate) and incubated for 10 min. Brains were then fixed in acetic acid:methanol:water 11:11:2 for 30 sec. Brains were then transferred to a cleaned microscope slide and overlaid with a siliconized coverslip. These were then squashed, mounted, and visualized as for the polytene chromosome preparations above. Mitotic index determinations were performed on these squash preparations by selecting 10 random well-populated fields of view for each brain squash, using a 20× objective. Total nuclei were counted for each field and the total was divided by the total number of mitotic figures observed in each field to generate the fraction of cells in mitosis. To mitigate complications due to maternal loading Mcm10Scim19 homozygotes were used as females to generate Mcm10Scim19/+ and Mcm10Scim19/Df(2L) by crossing to Df(2L)/CyO, GFP. Mcm10Scim19 homozygotes were also used as females in the cross to Mcm10d08029/Mcm10d08029 to generate Mcm10Scim19/Mcm10d08029 larvae. Mcm10d08029/+ and Mcm10d08029/Df(2L) were generated as above except Mcm10d08029 homozygotes were used as females in the cross. Statistical analysis was performed using Minitabtm.
5-Ethynyl-2′-deoxyuridine incorporation assays:
S-phase detection in Drosophila neural tissue was ascertained using the Click-It reaction kit from Invitrogen (cat. no. C10337). Brains were dissected in fresh Grace's unsupplemented cell culture medium. An equal volume of 200 μm 5-ethynyl-2′-deoxyuridine (EdU) solution in DMSO was added to the well and brains from each strain were incubated for 30 min in the dark at room temperature. Following the incubation the liquid was removed from each well and the brains were rinsed three times with 1× PBS. Brains were fixed in fresh 3.7% formaldehyde in PBS, incubating at room temperature for 15 min in the dark. The liquid was then removed and the brains were rinsed two times with 1× PBS. Brains were then permeabilized using 0.1% Triton X-100 in PBS for 15 min at room temperature in the dark. The liquid was removed and the brains were rinsed two times with 1× PBS. Brains were then incubated in the Click-It reaction cocktail per the manufacturer's instructions for 30 min. The brains were rinsed two times with the reaction rinse buffer provided by the manufacturer. After removing the rinse buffer, Hoechst 33342 was prepared per the manufacturer's instructions for nuclear visualization for 10 min. The Hoechst solution was then removed and the brains were washed four times with 1× PBS. Brains were then mounted on Polylysine-coated slides with Vectashield.
Ovary dissection and visualization:
Wild-type, Mcm10Scim19, and Mcm10d08029 flies 3–7 days post-eclosion were fed with yeast for 2 days. Ovaries were extracted from female wild-type and mutant flies in PBS. Ovarioles were teased apart and then fixed in 4% formaldehyde with PBS + 0.1% Triton X-100 (PBX) for 20 min. After fixing, ovaries were stained for 5 min with 0.3 μg/ml DAPI in PBS. Ovaries were then washed three times for 5 min in PBX, followed by 1 hr PBX wash, and three times for 10 min in PBX washes. Finally, ovaries were mounted using Vectashield and imaged using confocal optical sectioning microscopy.
Position-effect variegation analysis:
The dpov allele was introgressed into the Mcm10 and Hp1 mutant lines using standard genetic crosses. Homozygous w; dpov, Mcm10mut flies and w; Hp15/CyO flies were then crossed to w; Gla, dpov/Cy. The resulting F1 flies with the respective genotypes w; dpov, Mcm10mut/Gla,dpov, and w; dpov, Hp15/Gla,dpov were crossed to the dumpy variegating line w; Cy / dpw18 T(2:3). This cross was incubated at 25° as dumpy variegation is temperature sensitive (R. MacIntyre, personal communication). The adult non-Glazed-eye progeny from these crosses were then scored for wing morphology (see text) and compared with Gla, dpov individuals acting as sibling controls. Greater than 500 flies from each genotype were scored.
RESULTS
Mcm10 is a conserved protein:
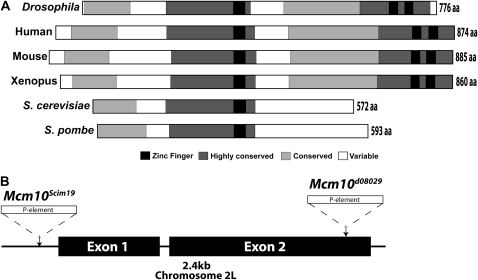
Mcm10 is found in eukaryotic organisms ranging from yeast to humans. Alignments from multiple species reveal that the Mcm10 protein contains several conserved regions (Figure 1A). The central highly conserved region of Mcm10 is present in all Mcm10 proteins and possesses an essential zinc-finger domain that has been implicated in protein–protein interactions (Robertson et al. 2008; Warren et al. 2008, 2009). In addition to this conserved core, higher eukaryotic Mcm10 proteins have expanded in length and contain a highly conserved C-terminal domain with two zinc-finger motifs (Robertson et al. 2008; Warren et al. 2008, 2009).
Figure 1.—
Alignment of multiple Mcm10 proteins and schematic of Mcm10 mutant alleles. (A) Alignment of Mcm10 proteins showing conserved zinc-finger motifs (solid bars), highly conserved regions (bars with dark shading, 23–40% similarity for metazoans), and moderately conserved regions (bars with light shading, 10–22% similarity for metazoans). (B) Schematic of Mcm10 gene region on chromosome 2L with P-element insertion sites indicated for the two Mcm10 alleles used in this study.
Genomic organization and viability of mcm10 mutant alleles:
Drosophila Mcm10 is located on the left arm of chromosome 2. P-element transposon-induced mutant alleles of Mcm10 have been identified (Figure 1B). The first Mcm10 allele identified was generated in a screen designed to identify genes responsible for the transmission of a centromere-attenuated minichromosome (Dobie et al. 2001). Mcm10Scim19 was found to be a P-element insertion 76 bp upstream of the translation start codon for Mcm10 that resulted in a dominant 17% reduction in transmission of the minichromosome (Dobie et al. 2001). Despite the close insertion of the P element to the start codon in Mcm10Scim19, the flies are homozygous viable. Chi-square analysis suggests that the allele is semilethal with the outcome of Mcm10Scim19/CyO X Mcm10Scim19/CyO demonstrating a significant (P < 0.0001) deviation from expected Mendelian ratios with Mcm10Scim19 homozygotes 42% reduced. The second allele of Mcm10 was identified in the Exelixis P-element insertion collection. Mcm10d08029 contains a P-element insertion in the second exon of Mcm10 and is predicted to truncate the C terminus of the Mcm10 protein by 85 amino acids (aa) (Gene Disruption Project and Exelixis 2005). The insertion of the P element in Mcm10d08029 adds the peptide DAEKRFS prior to encountering a stop codon within the P element. Despite this 85-aa truncation of the conserved C terminus of the Mcm10 protein, flies are homozygous viable for the Mcm10d08029 mutant allele. Like the Mcm10Scim19 allele, Mcm10d08029 homozygotes are significantly underrepresented (24% reduced, P < 0.05) in the outcomes of Mcm10d08029/CyO × Mcm10d08029/CyO, indicating that the allele is also semilethal. The semilethal nature of both Mcm10 alleles was rescued by a Mcm10 transgene (see materials and methods) inserted into the third chromosome (Mcm10Scim19/CyO or Mcm10d08029/CyO; p[Mcm10]/p[Mcm10]). Crosses including this transgene yielded phenotypic ratios not significantly different from those expected.
Putative hypomorphic allele of Mcm10:
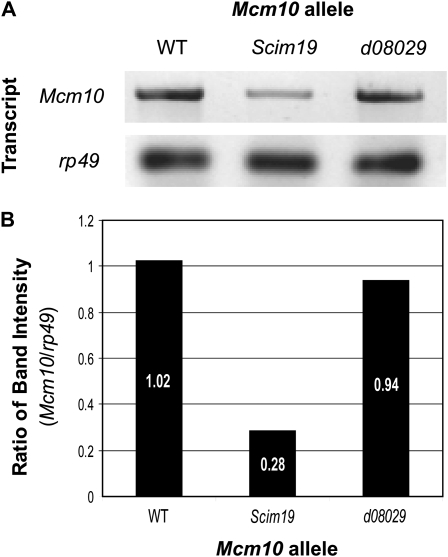
Due to the proximity of the P-element insertion to the start codon of Mcm10, the Mcm10Scim19 allele was predicted to be hypomorphic (Dobie et al. 2001). To test this we analyzed the transcription of Mcm10 by RT–PCR, using primers that anneal within the 5′ region of the transcript common to both Mcm10 alleles (Figure 2A). The Mcm10 transcript is maternally loaded and present at highest levels in the early embryo (Arbeitman et al. 2002; Gauhar et al. 2008). RT–PCR on mRNA extracted from early homozygous embryos (0–5 hr) deposited by homozygous mothers for the respective genotypes revealed a reduction in the levels of Mcm10 transcript in the Mcm10Scim19 background but not in the Mcm10d08029 background. When normalized to the rp49 loading control, the Mcm10Scim19 allele shows a 74% reduction in transcription while the Mcm10d08029 allele shows a negligible reduction compared to WT controls (Figure 2B).
Figure 2.—
RT–PCR measurement of relative Mcm10 transcript levels in the two Mcm10 alleles. (A) Visualization of transcript levels in the respective genotypes with rp49 loading control. (B) Bar graph of transcript levels as a ratio of rp49 control show that transcription of Mcm10 is significantly lower in the Mcm10Scim19 background.
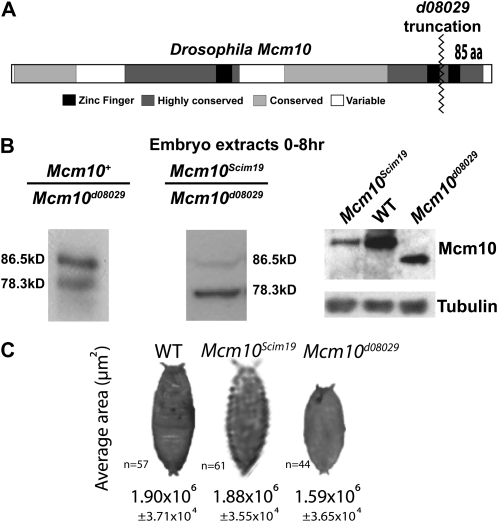
C-terminal truncation allele of Mcm10:
Truncating 85 aa from the C terminus of Mcm10 would remove one of the two conserved zinc-finger motifs and shifts the mass of the protein by 9.6 kDa (Figure 3A). To test this, Western blot analysis was performed on protein extracts from early embryos derived from Mcm10d08029/+ females crossed to males of the same genotype and Mcm10Scim19/Mcm10d08029 females crossed to males of the same genotype (Figure 3B). As predicted, the Mcm10d08029 P-element insertion results in a protein that migrates faster than wild-type protein with a molecular weight ∼8.2 kDa less than the native 86.5-kDa protein (Figure 3B). Despite being derived from heterozygous females, the band for the truncated Mcm10 appears lighter [80.3% of wild-type (wt) levels]. Given that transcription is only slightly affecting, as measured by RT–PCR, it is possible that the observed reductions of the truncated Mcm10 protein by Western blot are an artifact of the removal of antigenic residues from the C terminus of Mcm10. Alternatively the decrease in Mcm10 protein signal in the Mcm10d08029 background may be a reflection of protein stability differences. Western blot analysis was also performed to measure Mcm10 protein levels in embryos produced by Mcm10Scim19/Mcm10d08029 females (Figure 3B). Full-length Mcm10 protein levels are 28.8% of the truncation allele protein product. Since both Western blots contained the truncated protein, an estimate can be made of protein levels in the Mcm10Scim10 background: 23.1% of wild-type levels. Western blots of protein derived from the respective homozygous lines also demonstrated similar protein level reductions (Figure 3B). The levels of proteins observed in the Western blot analysis are in rough agreement with the reduction of 74% in the transcription of Mcm10 in the Mcm10Scim19 background (Figure 2).
Figure 3.—
Schematic of Mcm10d08029 truncation allele, shift in Mcm10 protein mobility, and reduction in pupae size. (A) The Mcm10d08029 allele is predicted to cut off 85 aa from the C terminus of the protein and remove a conserved zinc-finger domain. (B) Western blots probed with α-Mcm10 of protein extracts from early embryos laid by females with the genotypes +/Mcm10d08029 and Mcm10d08029/Mcm10Scim19, respectively. Left and right, Mcm10 protein mobility is altered in the Mcm10d08029-containing background, consistent with prediction from P-element insertion location. Right, Western blot showing reduced levels of full-length Mcm10 protein in Mcm10Scim19/Mcm10d08029-derived embryo extracts as compared to the truncation protein. Western blot of Mcm10 protein from homozygous fly lines shows similar protein levels and mobility shifts compared to the first two panels. (C) Measurement of average pupae size in wild type, Mcm10Scim19, and Mcm10d08029 shows that Mcm10d08029 pupae are on average 16% smaller than wild type (P < 0.00001).
Although viable, fly larvae homozygous for the Mcm10d08029 allele are, on average, smaller than wild-type controls. As a surrogate for size, the two-dimensional areas of pupae were measured in wild-type, Mcm10Scim19, and Mcm10d08029 homozygous strains (Figure 3C). T-tests revealed that Mcm10Scim19 pupae are not significantly different from wild type. On the other hand, Mcm10d08029 pupae are highly variable with respect to pupae size, as some pupae measure the same as wild type and others measure only half as big as wild type. Overall, Mcm10d08029 pupae were 16% smaller than wild-type controls (P < 0.00001). The majority of larval growth occurs through enlargement of polyploid cells that are the result of multiple rounds of DNA replication without mitosis. Another DNA replication mutation in mcm6 also results in smaller larvae, most likely due to its role in endoreplication (Schwed et al. 2002). Smaller size in larvae may also be due to defects in the proliferation of normal diploid imaginal disks as is the case with orc1 mutants (Park and Asano 2008).
Dissection of Mcm10 interaction domains:
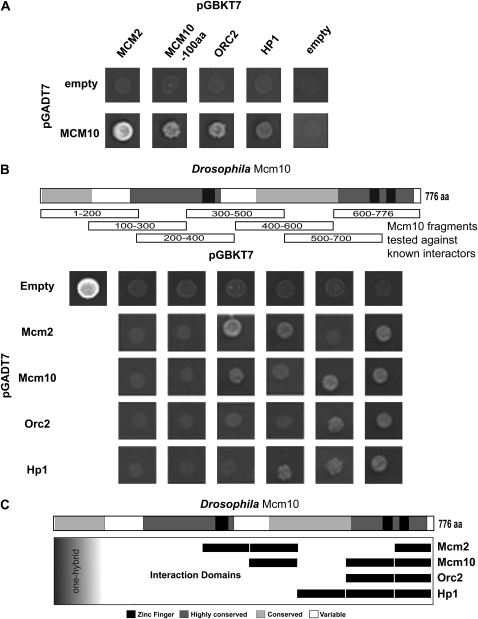
To delineate the regions of Drosophila Mcm10 responsible for protein interactions we performed two-hybrid analysis using overlapping fragments of the Mcm10 protein tested against known protein partners (Christensen and Tye 2003). To confirm previously reported Mcm10 interactions two-hybrid analysis was first performed using full-length Mcm10 fused to the Gal activation domain (Figure 4A). Mcm10 was fused to the activation domain due to the fact that fusion of Mcm10 to the Gal-binding domain results in weak one-hybrid activity (data not shown). Interactions were indicated by growth on media lacking histidine that was a result of transcription of the HIS3 reporter construct in the AH109 yeast two-hybrid strain. Growth occurred when GAD∷MCM10 was combined with different Drosophila proteins fused to the Gal-binding domain: GBK∷MCM2, GBK∷MCM10 (minus the first 100 aa), GBK∷ORC2, and GBK∷HP1, respectively. No growth was observed when GAD∷MCM10 was combined with empty vector. Nor was significant growth observed when the GBK fusion proteins were tested against GAD empty vector.
Figure 4.—
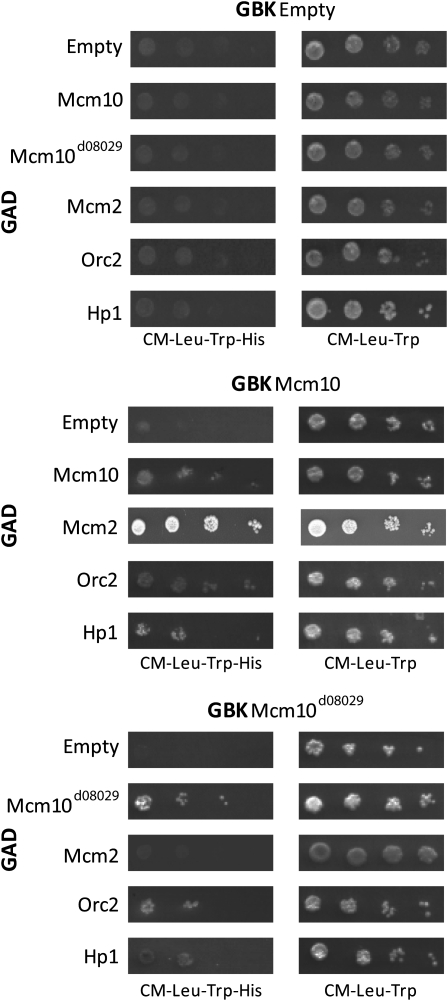
Two-hybrid dissection of Mcm10 interaction domains. (A) Two-hybrid interactions indicated by growth on media lacking histidine between Mcm10 and Orc2, Mcm2, and Hp1, respectively. Growth was not observed on empty controls. (B) Schematic representation of Mcm10 fragments tested against the interactions in A. Centered directly below each fragment are results for growth on media lacking histidine with growth indicating a positive interaction. Note that fragment 1–100 aa of Mcm10 demonstrated one-hybrid activity. (C) Representation of the results in B showing regions of Mcm10 interaction with the proteins tested.
To determine the regions of Mcm10 responsible for protein interactions and the Mcm10 one-hybrid activity we cloned portions of Mcm10 that corresponded to 200-aa fragments. Each of the successive fragments overlaps by 100 aa. This allows for a 100-aa resolution with respect to interaction domains. Each of these fragments was then fused to the Gal-binding domain by cloning into the pGBKT7 vector. These were tested for interaction against Gal activation domain fusions of Drosophila Mcm2, Mcm10, Orc2, Hp1, and empty, respectively, by transformation into the yeast HIS3 reporter strain and growth on media lacking histidine (Figure 4B).
The results revealed that Mcm10 interacts with Mcm2 via a central region that includes some of the highly conserved core and the central zinc finger and through the extreme C terminus of Mcm10 that includes one of the two conserved zinc-finger domains (Figure 4C). Mcm10 self-interaction occurs through a small central region and the C terminus including both conserved zinc-finger domains. The results also show that Mcm10 interaction with Orc2 is mediated though only the C-terminal domain including the two conserved zinc fingers. Finally, the interaction between Mcm10 and Hp1 occurs through an expanded portion of the C terminus including the zinc fingers and much of the higher eukaryotic conserved region.
The one-hybrid activity of Mcm10 was mapped to the first 100 aa (Figure 4B). To determine if this one-hybrid activity masked any protein interactions we swapped activation and binding fusion constructs for each of the proteins tested. However, no protein interactions were detected for the N-terminal portion of Mcm10 (data not shown). To eliminate the one-hybrid activity in further two-hybrid testing, clones were constructed that removed the first 100 aa of Mcm10 when fused to the Gal-binding domain.
Impact of Mcm10 C-terminal truncation on protein interactions:
Two intriguing observations concerning the C terminus of Mcm10 have been made. First, the C terminus of Mcm10 participates in interactions with Mcm2, Mcm10, Orc2, and Hp1. Second, the Mcm10d08029 allele results in a truncation of this C-terminal domain. These observations lead to the question: What is the impact of the Mcm10d08029 truncation on protein interactions? To address this question we performed semiquantitative yeast two-hybrid analysis with a truncation clone of Mcm10 that consisted of amino acids 101–691. Removal of the N-terminal 100 aa eliminated the one-hybrid activity and the removal of the C-terminal 85 aa was analogous to the Mcm10d08029 truncation. The truncation Mcm10d08029 clone and a Mcm10 clone missing only the first 100 aa were fused to the Gal-binding domain, respectively. These clones were tested by yeast two-hybrid analysis for interactions with Mcm10, Mcm2, Orc2, and Hp1 (Figure 5). The results indicate that self-interactions and interactions with Orc2 are attenuated by the removal of the last 85 aa compared to control. On the other hand, the relatively weak interaction with Hp1 is unaffected by the truncation. Finally, analysis also revealed that removing the last 85 aa of Mcm10 abolishes interaction with Mcm2.
Figure 5.—
Serial dilution yeast two-hybrid testing of the effect of the Mcm10d08029 allele on known protein interactions. The left column shows growth on media lacking histidine and the right column shows growth control on media with histidine. The top panel shows one-hybrid control showing no growth for empty vector controls. The middle panel shows the relative strength of two-hybrid interactions between Mcm10 and the proteins indicated. The bottom panel demonstrates the effect of the Mcm10d08029 85-aa C-terminal truncation on the relative strength of these same interactions.
These results suggest that, within the context of the native Mcm10 protein, the last 85 aa of Mcm10 are required for interaction with Mcm2. Indeed, when comparing the interactions of the proteins tested with only the C-terminal region of Mcm10, Mcm2 interacted only with the last 76 aa of Mcm10 (Figure 4C). The truncation of 85 aa completely removed this interaction domain. Unlike Mcm2, both Mcm10 self-interaction and Orc2 interaction occur over a larger 176-aa region at the C terminus of Mcm10. Both of these proteins demonstrated a reduction, but not elimination, of interaction with the truncated Mcm10. This is likely because some level of interaction is maintained through the remaining 91 aa. Finally, Hp1 interaction is unaffected by the removal of the last 85 aa of Mcm10. This is likely due to the fact that Hp1 interaction with Mcm10 occurs through a larger 276-aa region of the Mcm10 C terminus (Figure 4C). As a result 191 aa remain in the Mcm10 protein after the 85-aa truncation to participate in interactions with Hp1.
Cytological phenotypes of Mcm10 alleles:
The Mcm10Scim19 allele reduces transcription of an otherwise full-length transcript by 74%. On the other hand, the Mcm10d08029 allele is normal for transcription but is truncated at the C terminus by 85 aa. Given the previously reported roles for Mcm10 in DNA replication and chromosome condensation (Christensen and Tye 2003), the Mcm10Scim19 allele provides the opportunity to begin to understand the role of the bulk of Mcm10 in these processes, whereas the truncation allele of Mcm10 and its changes in protein interactions may provide a window into the significance of these interactions as they relate to both DNA replication and chromosome biology.
Polytene chromosomes:
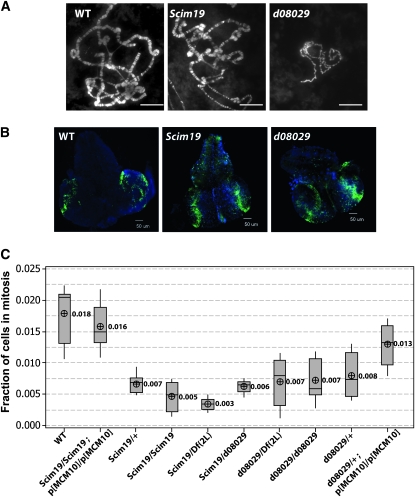
In Drosophila, different modes of DNA replication are developmentally regulated (Edgar and Orr-Weaver 2001; Asano 2009). DNA replication can occur without ensuing mitosis to generate polyploid tissues. The classic examples of this endoreplication are the polytene chromosomes found in the salivary gland tissues of wandering third instar larvae. Polytene chromosomes were examined in wild-type, Mcm10Scim19, and Mcm10d08029 homozygous backgrounds (Figure 6A). Micrographs revealed that a survey of Mcm10Scim19 polytene chromosomes appeared normal compared to that of wild-type controls. This suggests that normal levels of Mcm10 protein are not required for endoreplication. Conversely, the truncation of 85 aa from the C-terminal domain of Mcm10 in the Mcm10d08029 allele resulted in the underreplication of polytene chromosomes compared to wild type (Figure 6A). Underreplication of the salivary gland polytene chromosomes was observed more frequently in smaller Mcm10d08029 third instar wandering larvae than in larger larvae of the same genotype. This observation suggests that the variable larval size observed in the Mcm10d08029 background is likely the result of underreplication of the polytene tissues that are responsible for the majority of larval growth. To further examine the genetic nature of the polytene chromosome underreplication observed in Mcm10d08029 we examined polytene chromosomes in salivary glands from a variety of genotypes (supporting information, Figure S1). Underreplicated polytene chromosomes were observed in wandering third instar larvae with Mcm10Scim19/Mcm10d08029 and Mcm10d08029/Df(2L) genotypes, respectively. Interestingly underreplicated polytene chromosomes were observed in Mcm10d08029/+ larvae, indicating that the Mcm10 truncation allele is dominant with respect to polytene chromosome replication. Underreplication in these heterozygous larvae can be rescued by multiple doses of the Mcm10 transgene (Figure S1)
Figure 6.—
Effects of the two Mcm10 alleles on polytene chromosomes, DNA replication in larval brains, and mitotic index. (A) Confocal micrographs of polytene chromosome spreads from the genotypes indicated. Mcm10d08029 polytene chromosomes are underreplicated compared to wt control and Mcm10Scim19. (B) Fluorescent micrographs of wandering third instar larval brains showing DNA (blue) and EdU incorporation (green). wt brains are larger and show less EdU incorporation than either of the Mcm10 mutant alleles. (C) Graph of fraction of cells in mitosis for brain squashes of genotypes indicated.
DNA replication and mitotic indexes:
Given the widely reported role for mcm10 in DNA replication (Homesley et al. 2000; Gregan et al. 2003; Ricke and Bielinsky 2004; Chattopadhyay and Bielinsky 2007; Zhu et al. 2007), we sought to test the competency of the two Mcm10 alleles for DNA replication during the more canonical cell cycle in the brain tissues of the wandering third instar larvae. We utilized EdU incorporation assays. Incorporation of EdU was performed for 30 min on dissected third instar brains in growth media. Visualization of the brains from the various genotypes revealed, at first inspection, that the brains of wild-type larvae were slightly larger than those of either Mcm10Scim19 or Mcm10d08029 larvae, suggesting cell proliferation is slower in the mcm10 mutant alleles (Figure 6B). The second observation is that the number of cells that incorporated EdU in both Mcm10 mutant alleles was higher than that in wild type (Figure 6B). An increase in the cell number incorporating nucleotide analogs has also been observed in third instar brain tissue from mutants in the DNA replication genes mcm2 and mcm4 and is likely indicative of an S-phase delay (Feger et al. 1995; Treisman et al. 1995).
To test for cell-cycle delay, mitotic indexes were determined for the genotypes indicated in Figure 6C. Mitotic indexes for combinations of the Mcm10Scim19 allele with Mcm10+ showed a dosage-dependent trend with Mcm10Scim19/Df(2L), Mcm10Scim19/Mcm10Scim19, Mcm10Scim19/+, and Mcm10Scim19/Mcm10Scim19; p[Mcm10+]/p[Mcm10+] larvae having 6.0, 3.6, 2.6, and 1.1 times fewer nuclei, respectively, in mitosis than those of wild type. These observations suggest that Mcm10Scim19 represents a hypomorphic semidominant allele of Mcm10. Mitotic indexes for various combinations of the Mcm10d08029 allele with wild type and Df(2L) suggest that the truncation allele is dominant not only for endoreplication defects but also for cell-cycle delay in larval brain tissue (Figure 6C). Only the addition of three wild-type copies of Mcm10 was able to push the fraction of nuclei in mitosis close to that of the wild-type control. In addition, the observation that, unlike Mcm10Scim19, Mcm10d08029 homozygotes are not different from Mcm10d08029/Df(2L) with respect to mitotic index suggests that the defects observed in the Mcm10d08029 allele are not likely due to reductions in the level of protein.
When taken together, the decreased brain sizes, the increased number of cells in S phase, and the reduced mitotic indexes suggest that both Mcm10 mutants are delayed in S phase. This delay in S phase is consistent with observations made in human cell lines where depletion of Mcm10 by siRNA resulted in S-phase delay (Chattopadhyay and Bielinsky 2007; Park et al. 2008a,b).
Chromosome condensation:
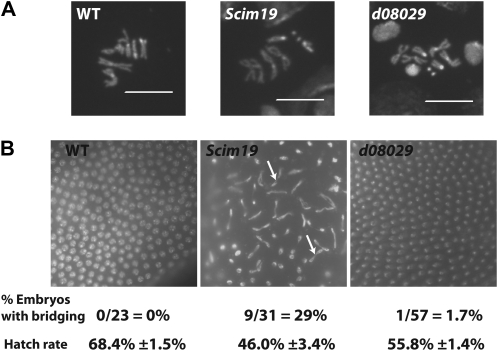
It has been previously reported that RNAi-mediated depletion of mcm10 in Drosophila KC cells results in metaphase chromosomes that are undercondensed (Christensen and Tye 2003). Unlike the tissue culture experiments, examination of metaphase chromosomes in both Mcm10Scim19 and Mcm10d08029 backgrounds did not reveal any analogous chromosome condensation defects (Figure 7A).
Figure 7.—
Chromosomal phenotypes of the two Mcm10 alleles in larval brains and early embryos. (A) Fluorescent micrographs of representative mitotic figures from brain squashes of the indicated genotype. No significant differences were observed. (B) Fluorescent micrographs of nuclei in early embryos from the homozygous females for the two Mcm10 alleles and the wild-type control as well as the percentage of embryos showing two or more anaphase bridges per field of view. Significant cell-cycle asynchrony is observed in the Mcm10Scim19 background as well as anaphase bridges (open arrows), while no anaphase bridges were observed in wt or Mcm10d08029.
Early embryo:
The early embryo cell cycles 10–12 differ from those in the larval brain in that they occur in a syncytium, lack gap phases, proceed in synchrony, and occur rapidly over ∼9 min compared to the 8 hr in the larval brain tissue (O'Farrell et al. 2004). Examination of homozygous early embryos deposited by homozygous females at cell cycles 10–12 in the respective Mcm10 mutant alleles reveals that nuclear divisions occurred in normal synchrony in Mcm10d08029 but are asynchronous in the Mcm10Scim19 background (Figure 7B). Moreover, anaphase bridges are observed in 29% of the Mcm10Scim19 embryos in cell cycles 10–12. This asynchrony and bridging in the Mcm10Scim19 homozygous embryos likely has negative consequences for embryo viability as hatch rates are only 46% in the Mcm10Scim19 background. Hatch rates are also lower in the Mcm10d08029 background but this is not attributable to any observed defects in synchrony or bridging. The observed asynchrony and anaphase bridges in Mcm10Scim19 may be a consequence of entry into mitosis prior to the completion of DNA replication. This hypothesis is consistent with the observed S-phase delay in the brain tissue of Mcm10Scim19 larvae. However, unlike in Mcm10Scim19, the S-phase delay in the Mcm10d08029 background did not translate into a similar asynchrony in the early embryo. The absence of a defect in the Mcm10d08029 mutant may be due to the fact that Mcm10 protein levels, albeit truncated, are sufficient in the early embryo for rapid DNA synthesis to occur. Alternatively, the cell-cycle asynchrony in the Mcm10Scim19 background may be indicative of defects in the condensation/decondensation of chromosomes that must occur very rapidly in the early embryonic nuclei.
Nurse cell nuclei condensation/decondensation:
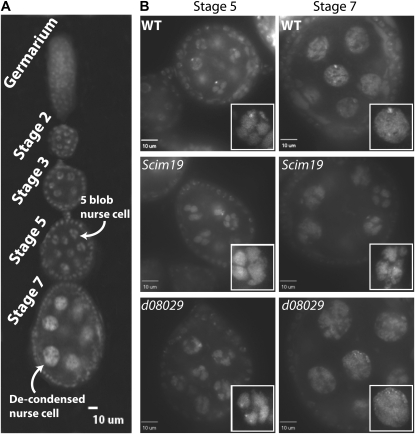
Ovaries in Drosophila are made up of multiple ovarioles, which, in turn, are made up of a string of egg chambers that become progressively more mature as they move away from the germaruim (Figure 8A). Each of these egg chambers contains 15 nurse cells and one oocyte. As the egg chambers mature, nurse cell nuclei are characterized by highly condensed “five-blob” polytene chromosome structures. This five-blob structure persists until stage 5 when the nurse cell chromosomes begin to decondense. Chromosome decondensation is then fully complete by stage 7 in all nuclei (Dej and Spradling 1999). This decondensation is linked to a mitosis-like phase during endocycle 5 that promotes the dissociation of sister chromatids (Reed and Orr-Weaver 1997; Royzman and Orr-Weaver 1998; Dej and Spradling 1999). Careful examination of nurse cell nuclei chromosome decondensation events revealed temporal defects in this process only in Mcm10Scim19 egg chambers (Figure 8B). Nurse cell nuclei in stage 7 Mcm10Scim19 egg chambers are heterogeneous with respect to chromosome decondensation with some nuclei fully decondensed and others persisting in the five-blob stage (Figure 8B). Persistence of the five-blob stage was observed in nearly 75% of the stage 7 egg chambers examined (n = 24).
Figure 8.—
Confocal micrographs of Drosophila egg chambers and nurse cell nuclei. (A) String of egg chambers with different stages labeled. Note five-blob nurse cell nuclei at stage 5. By stage 7 all nuclei appear decondensed (some egg chambers have been reoriented for clarity). (B) Egg chambers at stages 5 and 7 from the two Mcm10 alleles and wt. The five-blob stage is present in all genotypes at stage 5. However, at stage 7 some nuclei in the Mcm10Scim19 background have not decondensed and remain in the five-blob state (see inset).
Position-effect variegation:
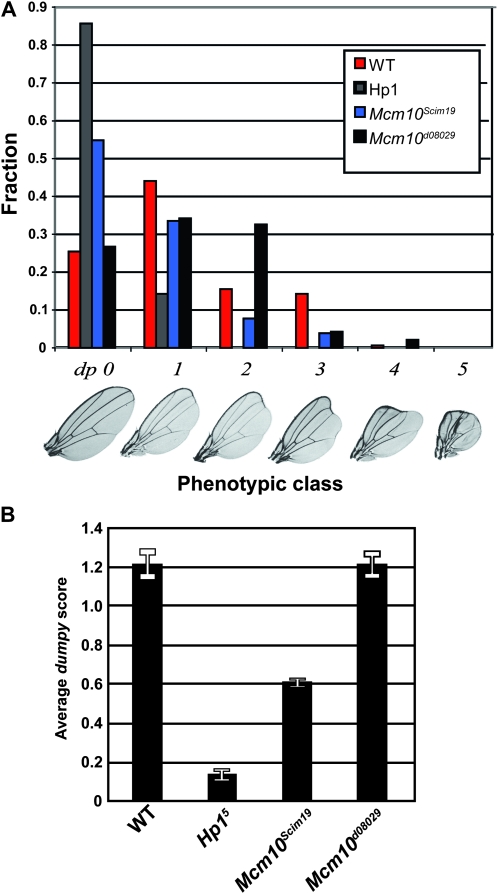
Given that Mcm10 interacts with Hp1 (Christensen and Tye 2003) (this study), we wanted to test whether Mcm10 has a role in heterochromatin formation. To address this we have taken advantage of variegating dumpy (dp) alleles that result in variable wing morphology due to the proximity of dp to centric heterochromatin (R. MacIntyre, personal communication). Wing phenotypes were categorized into increasingly severe dp phenotypes (Figure 9A). Utilizing this system we measured the impacts of the two Mcm10 alleles on position-effect variegation (PEV) and compared these to wild type and an Hp1 mutation (Figure 9, A and B). As expected, the Hp1 mutation strongly suppresses PEV of the dp wing phenotype compared to wild type (Figure 9A). Analysis of the two Mcm10 alleles revealed that only the hypomorphic allele dominantly suppresses PEV, while the truncation allele does not. These results suggest that levels of Mcm10 are important for the formation and/or maintenance of heterochromatin while the last 85 aa of Mcm10 are dispensable with respect to heterochromatin formation.
Figure 9.—
PEV analysis of Mcm10 alleles and Hp15 using a variegating dumpy reporter line. (A) Fraction of flies in each phenotypic class (0–5) for the genotypes indicated. Hp15 results in a dramatic shift toward wild-type distribution of dumpy phenotypes. (B) Average “dumpy” score for the different genotypes. Both Hp15 and Mcm10Scim19 show significant suppression of dumpy PEV whereas Mcm10d08029 shows no shift from wild type.
DISCUSSION
Though initially identified as a protein with a role in DNA replication, the function of Mcm10 in other cellular processes is coming to light. Indeed, the examination of the conserved nature of Mcm10 suggests multiple functions. The highly conserved core with its single atypical zinc-finger domain is present in all eukaryotes examined to date and likely represents the most ancient function of this protein. This core has been shown to interact with single-stranded DNA, DNA polymerase α, and PCNA (Ricke and Bielinsky 2006; Chattopadhyay and Bielinsky 2007; Robertson et al. 2008; Warren et al. 2008, 2009). Genetic studies also support the assertion that this central core supports the essential function(s) of Mcm10 as mutations in this region of Mcm10 affect viability of cells and affect DNA replication (Merchant et al. 1997; Homesley et al. 2000; Ricke and Bielinsky 2006). However, mutations in this region have also revealed a role for Mcm10 in heterochromatic silencing (Liachko and Tye 2005). This observation may hint that, in fact, the core region of Mcm10 may also have roles outside of DNA replication.
As well as the core, higher eukaryotes also have another conserved domain at the C terminus of the protein that contains two additional zinc-finger motifs (Robertson et al. 2008; Warren et al. 2008, 2009). The observation that this additional domain is present only in higher eukaryotes suggests that Mcm10 has taken on additional roles that are mediated through this C-terminal domain. The expansion and conservation of this C-terminal domain may be a consequence of evolutionary pressures associated with DNA replication through the more complex and varied genomic contexts that are present in the larger genomes and differentiated tissues of higher eukaryotes. Interestingly the expansion of this C-terminal domain may also represent an expansion in the role of Mcm10 with respect to linking DNA replication to heterochromatic silencing. A recent study in S. cerevisiae demonstrated that the C-terminal domain of Mcm10 is responsible for mediating the interaction between Sir2 and members of the Mcm2-7 complex (Liachko and Tye 2009). An expansion of this domain in higher eukaryotes may then reflect additional roles in heterochromatin formation. In higher eukaryotes, this C-terminal domain has been shown to bind double-stranded and single-stranded DNA (Robertson et al. 2008). However, the significance of this binding has not yet been uncovered. Through a fragment-based two-hybrid mapping we have shown that the C terminus of Mcm10 also interacts with Orc2, Mcm2, and Hp1.
The fortuitous identification of the Mcm10d08029 C-terminal truncation allele that removes one of the two conserved zinc-finger motifs has allowed us to begin to examine the possible roles of the extreme C terminus of Mcm10. Our yeast two-hybrid results demonstrated that, within the context of a larger fragment of Mcm10, the last 85 aa of Mcm10 are required for interaction with Mcm2. Despite the conservation of this region and a loss of Mcm2 interaction, this portion of the protein is dispensable for viability in Drosophila. Larvae homozygous for the Mcm10d08029 allele have provided some insight into the significance of this region of the protein. Both the variable larval size and the underreplication of the salivary gland polytene chromosome suggest that the last 85 aa of Mcm10 are required for efficient DNA replication in endoreplicating tissues. Moreover, this Mcm10 function may be modulated through an interaction with Mcm2. Curiously, defects in endoreplication were not observed in the polytene nurse cells of egg chambers. This difference may be due to penetrance of the phenotype being lower in nurse cells or to the fact that nurse cells' endocycles are different from those in salivary glands, in that they undergo a post-S endocycle phase (Dej and Spradling 1999).
As in the Mcm10d08029 allele, brain preparations of larvae homozygous for the Mcm10Scim19 hypomorphic allele demonstrated S-phase delay as evidenced by both excessive EdU incorporation and a lowered mitotic index. Unlike the truncation allele, the hypomorphic allele did not show any defects in endoreplication. This suggests that wild-type levels of Mcm10 are not required for endoreplication but are required for efficient passage through S phase in normal mitotic cells.
Whether the S-phase delays observed in larval brains in the two Mcm10 alleles are due to the same defect remains to be conclusively determined. The degree of S-phase delay in both alleles may simply be a function of the reduced protein levels observed in Mcm10Scim19 (severe, Figure 3B) and those observed in Mcm10d08029 (moderate, Figure 3B). However, if the role of Mcm10 in normal mitotic S phase is embodied only in the last 85 aa, then it would follow that the S-phase delay in the truncation allele would be more severe than for the hypomorphic allele. However, the mitotic index is not substantially different between the two alleles. At the very least this, combined with the endoreplication defects in Mcm10d08029, argues that Mcm10 has multiple roles related to the different types of S phases found in Drosophila.
Unlike in larval brain tissue, visualization of early embryo cell cycles does reveal a more significant difference between the two alleles. In these synchronous, very rapid cell cycles that lack gap phases, embryos homozygous for the Mcm10Scim19 allele show compelling defects whereas Mcm10d08029 embryos do not. These defects include cell-cycle asynchrony and anaphase bridges. Both of these phenotypes may simply be attributable to an S-phase delay that is nonuniform in the Mcm10Scim19 allele. This would directly result in asynchrony in cell cycles and if mitosis proceeded prior to the completion of DNA, replication anaphase bridges would be observed. Moreover, the possible reason similar defects are not observed in the Mcm10d08029 allele is due to the fact that the S-phase delay is not severe enough in the early embryo to manifest as a defect. However, given that Mcm10 has been shown to have multiple roles in S phase, then it is more likely that the differences in these early embryos are a reflection of these additional Mcm10 functions (Wohlschlegel et al. 2002; Gregan et al. 2003; Lee et al. 2003; Ricke and Bielinsky 2004, 2006; Sawyer et al. 2004; Douglas et al. 2005; Liachko and Tye 2005; Zhu et al. 2007; Park et al. 2008a,b; Liachko and Tye 2009; Xu et al. 2009). In light of previous studies that demonstrated a role for Mcm10 in chromosome condensation, interaction with Hp1, and defects in heterochromatin formation (Christensen and Tye 2003; Liachko and Tye 2005, 2009), it is compelling to speculate that the defects observed in early embryos may not be due only to a direct role in DNA replication for Mcm10 but may also reflect a role in chromosome condensation and heterochromatin formation during S phase. Indeed, strikingly similar asynchrony and anaphase bridges have been observed in early embryos that are heterozygous for mutations in Hp1 (Kellum and Alberts 1995). Like the asynchrony observed in the Hp1 mutant, the similar defect in the hypomorphic Mcm10 allele may be due to defects in the rapid chromosome condensation or decondensation processes required during these exceptionally rapid cell cycles.
In support of a role for Mcm10 in chromosome condensation/decondensation are our observations of defects in chromosome decondensation in egg chamber nurse cells in Mcm10Scim19 homozygotes. Mcm10Scim19 animals did not display apparent defects in DNA replication in endocycling cells but do show delays in the developmentally regulated nurse cell decondensation that typically occurs at the end of endocycle 5. Similar defects in nurse cell nuclei decondensation have not been reported in DNA replication mutants but have been reported in numerous female-sterile mutations such as ovarian tumor (otu), string of pearls (sop), squid (sqd), tulipano (tlp), morula (mr), and rhino (rhi) (Cramton and Laski 1994; Heino et al. 1995; Reed and Orr-Weaver 1997; Gigliotti et al. 1998; Volpe et al. 2001; Goodrich et al. 2004). Rhino is of particular interest; it is a female-specific HP1-like chromodomain protein that has been postulated to modulate chromosome structure at the end of endocycle 5 (Volpe et al. 2001). Moreover, rhino has been shown to be under positive selection and to specifically localize to regions of heterochromatin where it has been proposed to suppress germline transposition events (Vermaak et al. 2005).
PEV analysis has also differentiated between the two Mcm10 alleles, further supporting a possible role for Mcm10 in heterochromatin formation. A reduction of Mcm10 dosage dominantly suppressed PEV whereas removal of the C-terminal 85 aa did not suppress it. This observation supports the conclusion that the last 85 aa of Mcm10 do not function in heterochromatin formation. If the role of Mcm10 in heterochromatin formation is mediated through the interaction with Hp1, then this is not surprising given that the interaction with Hp1 does not appear to be affected by the 85-aa truncation of Mcm10. Furthermore, these results suggest that the interaction between Mcm10 and Mcm2 is not required for Mcm10 function in heterochromatin formation. This conclusion is supported by studies in S. cerevisiae that have shown that the temperature sensitivity of specific Mcm10 alleles is suppressed by mutations in mcm2. However, this suppression via mcm2 does not suppress the heterochromatin maintenance defect of these same Mcm10 alleles (Liachko and Tye 2005). By extension this same study suggests that, at least in part, the function of Mcm10 in DNA replication is mediated through an interaction with mcm2. Our results showing that loss of Mcm2 interaction results in an S-phase delay are consistent with this hypothesis. A follow-up study in yeast elegantly demonstrated that Mcm10 mediates the interactions of both Mcm3 and Mcm7, respectively, with the silencing machinery outside of DNA replication (Liachko and Tye 2009). This work suggests that further studies concerning the role of Mcm10 in heterochromatin formation should address possible interactions with mcm3 and mcm7.
One impetus for our examination of these Mcm10 alleles in Drosophila was concern that our previous findings using the embryo-derived Drosophila KC tissue culture model may not be directly relevant to Mcm10 function in the whole organism because these cells are often aneuploid and are operating outside of normal cell-cycle, checkpoint, and developmental controls found in Drosophila tissues. Indeed, the S-phase delay observed in the Mcm10Scim19 hypomorph is at odds with the observation that no such S-phase delay was observed when Mcm10 was depleted by RNAi in KC cells (Christensen and Tye 2003). Additional differences between the tissue culture system and the larval brain come to light when examining metaphase chromosome condensation. Whereas significant condensation defects were observed in tissue culture when Mcm10 was depleted (Christensen and Tye 2003), no significant chromosome condensation defects were observed in Mcm10Scim19 larval brains. The discrepancies we observed between the two systems highlight the fact that findings in tissue culture systems are not always directly transferable to whole organisms.
The examination of these two Mcm10 alleles has begun to shed additional light on the role of Mcm10 in the cell (Table 1). When taken all together, the results presented here, based on examination of two different Mcm10 alleles, support the conclusion that Drosophila Mcm10 has multiple roles in S phase as well as a role in heterochromatin formation. Moreover, the comparison of the two Mcm10 alleles provides evidence that the endoreplication S-phase function of the C-terminal 85 aa is separable from the S-phase function of the remainder of the protein. In addition, the heterochromatic function of Mcm10 does not require the terminal 85 aa nor does it likely require interaction with Mcm2.
TABLE 1.
Comparison of phenotypes observed in the two Mcm10 alleles used in this study
|
Mcm10 allele |
||
|---|---|---|
| Mcm10Scim19 | Mcm10d08029 | |
| Nature of mutation | Hypomorphic, semilethal | C-terminal truncation, semilethal |
| Protein level | 23% of wt | 80% of wta |
| Interacts with Mcm2 | Yes | No |
| Pupae size | Normal | Smaller |
| Polytene chromosomes | Normal | Underreplicated |
| Mitotic index | Low | Low |
| Early embryo nuclei | Asynchronous, anaphase bridges | Normal |
| Nurse cell nuclei | Defective decondensation | Normal |
| PEV | Suppresses | No effect |
Lower protein level may be due to removal of antigenic residues by truncation.
Acknowledgments
We thank and are grateful to Bik Tye for providing support for the initial work and Ross MacIntyre for advice and reagents, especially his contribution of expertise and flies lines for the PEV analysis. We thank Patrick Ferree, Bonnie Bolkan, Daniel Barbash, Janice Werner, Michael Goldberg, Byron Wilson, and members of the Cornell Superfly group for input and advice. We also thank Alexey A. Soshnev from Carver College of Medicine for useful advice concerning ovary dissection and nurse cell visualization. In addition, at East Carolina University Tom Fink of the Imaging Core supported our imaging efforts. Support for undergraduate research on this project was provided by the Hughes Undergraduate Research and Cornell Presidential Research Scholars programs. Support for the initial work was provided by the National Science Foundation (NSF 0453773). Finally we thank East Carolina University for start-up funds and the National Institutes of Health (grant 1R15GM093328-01 awarded to T.W.C.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117234/DC1.
References
- Arbeitman, M. N., E. E. Furlong, F. Imam, E. Johnson, B. H. Null et al., 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297(5590): 2270–2275. [DOI] [PubMed] [Google Scholar]
- Asano, M., 2009. Endoreplication: The advantage to initiating DNA replication without the ORC? Fly (Austin) 3(2): 173–175. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., and A. K. Bielinsky, 2007. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell 18(10): 4085–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, T. W., and B. K. Tye, 2003. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 14(6): 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton, S. E., and F. A. Laski, 1994. String of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 137 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo, S., R. M. Ricke and A. K. Bielinsky, 2006. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 26(13): 4806–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej, K. J., and A. C. Spradling, 1999. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development 126(2): 293–303. [DOI] [PubMed] [Google Scholar]
- Dobie, K. W., C. D. Kennedy, V. M. Velasco, T. L. McGrath, J. Weko et al., 2001. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics 157 1623–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, N. L., S. K. Dozier and J. J. Donato, 2005. Dual roles for Mcm10 in DNA replication initiation and silencing at the mating-type loci. Mol. Biol. Rep. 32(4): 197–204. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., and T. L. Orr-Weaver, 2001. Endoreplication cell cycles: more for less. Cell 105(3): 297–306. [DOI] [PubMed] [Google Scholar]
- Feger, G., H. Vaessin, T. T. Su, E. Wolff, L. Y. Jan et al., 1995. dpa, a member of the MCM family, is required for mitotic DNA replication but not endoreplication in Drosophila. EMBO J. 14(21): 5387–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, J., M. Rousseau, S. Shenker, M. A. Ferraiuolo, Y. Hayashizaki et al., 2009. Chromatin conformation signatures of cellular differentiation. Genome Biol. 10(4): R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauhar, Z., M. Ghanim, T. Herreman, J. D. Lambert, T. R. Li et al., 2008. Drosophila melanogaster life-cycle gene expression dataset and microarray normalisation protocols. FlyBase personal communication, FBrf0205914.
- Gene Disruption Project and Exelixis, 2005 Genomic mapping of Exelixis insertion collection. FlyBase Computer File, FBrf0184340.
- Gerbi, S. A., and A. K. Bielinsky, 2002. DNA replication and chromatin. Curr. Opin. Genet. Dev. 12(2): 243–248. [DOI] [PubMed] [Google Scholar]
- Gigliotti, S., G. Callaini, S. Andone, M. G. Riparbelli, R. Pernas-Alonso et al., 1998. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J. Cell Biol. 142(5): 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J. S., K. N. Clouse and T. Schüpbach, 2004. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development 131(9): 1949–1958. [DOI] [PubMed] [Google Scholar]
- Gregan, J., K. Lindner, L. Brimage, R. Franklin, M. Namdar et al., 2003. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell 14(9): 3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth, A., W. Rocha, A. Verreault and G. Almouzni, 2007. Chromatin challenges during DNA replication and repair. Cell 128(4): 721–733. [DOI] [PubMed] [Google Scholar]
- Heino, T. I., V. P. Lahti, M. Tirronen and C. Roos, 1995. Polytene chromosomes show normal gene activity but some mRNAs are abnormally accumulated in the pseudonurse cell nuclei of Drosophila melanogaster otu mutants. Chromosoma 104(1): 44–55. [DOI] [PubMed] [Google Scholar]
- Hiratani, I., and D. M. Gilbert, 2009. Replication timing as an epigenetic mark. Epigenetics 4(2): 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst, M., and M. A. Marra, 2009. Epigenetics and human disease. Int. J. Biochem. Cell Biol. 41(1): 136–146. [DOI] [PubMed] [Google Scholar]
- Homesley, L., M. Lei, Y. Kawasaki, S. Sawyer, T. Christensen et al., 2000. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 14(8): 913–926. [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W., L. Fanti, D. T. Pak, M. R. Botchan, S. Pimpinelli et al., 1998. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J. Cell Biol. 142(2): 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, M., K. Yanagi, T. Mizuno, M. Yokoi, Y. Kawasaki et al., 2000. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 28(23): 4769–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P. A., S. B. Baylin, 2007. The epigenomics of cancer. Cell 128(4): 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, Y., S. Hiraga and A. Sugino, 2000. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5(12): 975–989. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and B. M. Alberts, 1995. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108(4): 1419–1431. [DOI] [PubMed] [Google Scholar]
- Lee, J. K., Y. S. Seo and J. Hurwitz, 2003. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl. Acad. Sci. USA 100(5): 2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko, I., and B. K. Tye, 2005. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics 171 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko, I., and B. K. Tye, 2009. Mcm10 mediates the interaction between DNA replication and silencing machineries. Genetics 181 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, A. M., Y. Kawasaki, Y. Chen, M. Lei and B. K. Tye, 1997. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 17(6): 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard, M. E., A. K. Jain and M. C. Barton, 2009. Analysis of epigenetic alterations to chromatin during development. Genesis. 47(8): 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell, P. H., J. Stumpff and T. T. Su, 2004. Embryonic cleavage cycles: How is a mouse like a fly? Curr. Biol. 14(1): R35–R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, D. T., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum et al., 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91(3): 311–323. [DOI] [PubMed] [Google Scholar]
- Park, J. H., S. W. Bang, S. H. Kim and D. S. Hwang, 2008. a Knockdown of human MCM10 activates G2 checkpoint pathway. Biochem. Biophys. Res. Commun. 365(3): 490–495. [DOI] [PubMed] [Google Scholar]
- Park, J. H., S. W. Bang, Y. Jeon, S. Kang and D. S. Hwang, 2008. b Knockdown of human MCM10 exhibits delayed and incomplete chromosome replication. Biochem. Biophys. Res. Commun. 365(3): 575–582. [DOI] [PubMed] [Google Scholar]
- Park, S. Y., and M. Asano, 2008. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc. Natl. Acad. Sci. USA 105(34): 12343–12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, B. H., and T. L. Orr-Weaver, 1997. The Drosophila gene morula inhibits mitotic functions in the endo cell cycle and the mitotic cell cycle. Development 124(18): 3543–3553. [DOI] [PubMed] [Google Scholar]
- Ricke, R. M., and A. K. Bielinsky, 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 16(2): 173–185. [DOI] [PubMed] [Google Scholar]
- Ricke, R. M., and A. K. Bielinsky, 2006. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 281(27): 18414–18425. [DOI] [PubMed] [Google Scholar]
- Robertson, P. D., E. M. Warren, H. Zhang, D. B. Friedman, J. W. Lary et al., 2008. Domain architecture and biochemical characterization of vertebrate Mcm10. J. Biol. Chem. 283(6): 3338–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman, I., and T. L. Orr-Weaver, 1998. S phase and differential DNA replication during Drosophila oogenesis. Genes Cells 3(12): 767–776. [DOI] [PubMed] [Google Scholar]
- Rusché, L. N., A. L. Kirchmaier and J. Rine, 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13(7): 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S. L., I. H. Cheng, W. Chai and B. K. Tye, 2004. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J. Mol. Biol. 340(2): 195–202. [DOI] [PubMed] [Google Scholar]
- Schwed, G., N. May, Y. Pechersky and B. R. Calvi 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13(2): 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef, M. M., C. King, M. Damaj, R. Badagu, D. W. Huang et al., 2001. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol. Biol. Cell 12(6): 1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman, J. E., P. J. Follette, P. H. O'Farrell and G. M. Rubin, 1995. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 9(14): 1709–1715. [DOI] [PubMed] [Google Scholar]
- Tye, B. K., 1999. MCM proteins in DNA replication. Annu. Rev. Biochem. 68 649–686. [DOI] [PubMed] [Google Scholar]
- Vermaak, D., S. Henikoff and H. S. Malik 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1(1): 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, A. M., H. Horowitz, C. M. Grafer, S. M. Jackson and C. A. Berg, 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, E. M., S. Vaithiyalingam, J. Haworth, B. Greer, A. K. Bielinsky et al., 2008. Structural basis for DNA binding by replication initiator Mcm10. Structure 16(12): 1892–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, E. M., H. Huang, E. Fanning, W. J. Chazin and B. F. Eichman, 2009. Physical interactions between MCM10, DNA, and DNA polymerase α. J. Biol. Chem. 284(36): 24662–24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., S. K. Dhar, T. A. Prokhorova, A. Dutta, J. C. Walter et al., 2002. Xenopus Mcm10 binds to origins of DNA replication after Mcm2–7 and stimulates origin binding of Cdc45. Mol. Cell 9(2): 233–240. [DOI] [PubMed] [Google Scholar]
- Xu, X., P. J. Rochette, E. A. Feyissa, T. V. Su and Y. Liu, 2009. MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J. 28(19): 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., C. Ukomadu, S. Jha, T. Senga, S. K. Dhar et al., 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 21(18): 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]