Abstract
While mitochondria are renowned for their role in energy production, they also perform several other integral functions within the cell. Thus, it is not surprising that mitochondrial dysfunction can negatively impact cell viability. Although mitochondria have received an increasing amount of attention in recent years, there is still relatively little information about how proper maintenance of mitochondria and its genomes is achieved. The Neurospora crassa mus-10 mutant was first identified through its increased sensitivity to methyl methanesulfonate (MMS) and was thus believed to be defective in some aspect of DNA repair. Here, we report that mus-10 harbors fragmented mitochondria and that it accumulates deletions in its mitochondrial DNA (mtDNA), suggesting that the mus-10 gene product is involved in mitochondrial maintenance. Interestingly, mus-10 begins to senesce shortly after deletions are visualized in its mtDNA. To uncover the function of MUS-10, we used a gene rescue approach to clone the mus-10 gene and discovered that it encodes a novel F-box protein. We show that MUS-10 interacts with a core component of the Skp, Cullin, F-box containing (SCF) complex, SCON-3, and that its F-box domain is essential for its function in vivo. Thus, we provide evidence that MUS-10 is part of an E3 ubiquitin ligase complex involved in maintaining the integrity of mitochondria and may function to prevent cellular senescence.
THE mus-10 mutant was isolated from a screen aimed at identifying Neurospora crassa strains that were sensitive to MMS and therefore likely to lack proper DNA repair mechanisms (Kafer and Perlmutter 1980). Epistasis analyses involving mus-10 suggested that it belonged to the uvs-6 epistasis group, which functions in recombination repair (Kafer and Perlmutter 1980; Kafer 1983). However, mus-10 did not display several phenotypes common to other members of the uvs-6 epistasis group: chromosomal instability, a high sensitivity to histidine, and the inability to produce viable ascospores in homozygous crosses (Newmeyer et al. 1978; Newmeyer and Galeazzi 1978; Kafer and Perlmutter 1980; Kafer 1981; Schroeder 1986; Watanabe et al. 1997; Handa et al. 2000; Sakuraba et al. 2000). Furthermore, the frequencies of spontaneous and radiation-induced mutation observed in mus-10 were similar to those of a wild-type strain (Kafer 1981). Past efforts to uncover the nature of these discrepancies or the function of the mus-10 gene product have been uninformative.
The majority of cellular ATP is produced in mitochondria through aerobic respiration, which couples electron flow through respiratory complexes within the mitochondrial inner membrane with oxidative phosphorylation. Besides their role in ATP synthesis, mitochondria are also involved in many other cellular processes including beta-oxidation (Bartlett and Eaton 2004), calcium homeostasis (Gunter et al. 2004; Rimessi et al. 2008), production of iron-sulfur clusters (Zheng et al. 1998; Gerber and Lill 2002; Lill and Muhlenhoff 2005; Rouault and Tong 2005), and apoptosis (Green 2005; Antignani and Youle 2006; Xu and Shi 2007). Although virtually all mitochondrial proteins are encoded within the nucleus, a small number of proteins are encoded by mitochondrial DNA (mtDNA). The integrity of the mitochondrial genome may affect cell survival as mutations in mtDNA accumulate in patients suffering from severe neurological diseases including Alzheimer's, Huntington's and Parkinson's, as well as several types of cancer (Chatterjee et al. 2006; Higuchi 2007; Krishnan et al. 2007; Reeve et al. 2008). The number of mtDNA mutations also increases with age, suggesting a link between mitochondrial dysfunction and ageing (Cortopassi and Arnheim 1990; Corral-Debrinski et al. 1992; Cortopassi et al. 1992; Simonetti et al. 1992; Reeve et al. 2008). Contrary to the single genome in the nucleus, there are several copies of mtDNA in each mitochondrion. Thus, defects in a few mitochondrial genomes do not necessarily lead to mitochondrial dysfunction. Many patients suffering from mitochondrial diseases exhibit heteroplasmy, a phenomenon in which a mixture of wild-type and mutant mtDNAs exist in a single cell. The ratio of wild-type to mutant mtDNAs is critical in determining the penetrance of the genetic defect, where mutant loads >60% are required to cause respiratory chain dysfunction within an individual cell (Boulet et al. 1992; Chomyn et al. 1992; Sciacco et al. 1994).
Even though N. crassa strains are generally deemed immortal if they can be subcultured ∼50 times, a wild-type strain was recently reported to senesce after 12,000 hr of growth, implying that this fungus undergoes natural or programmed ageing (Maheshwari and Navaraj 2008; Kothe et al. 2010). However, replicative life span is also influenced by genetic background as certain mutations can cause progressive deterioration of growth, ultimately leading to death. One such example is the nuclear-encoded natural death (nd), which when mutant causes a senescence phenotype correlating with the accumulation of multiple mtDNA deletions (Sheng 1951; Seidel-Rogol et al. 1989). The deletions of mtDNA in nd occurred between two 70- to 701-bp direct repeats, suggesting that the nd gene product regulates recombination, repair, or replication of mtDNA (Bertrand et al. 1993). Another nuclear mutation, senescence (sen), was isolated from N. intermedia and introgressed into N. crassa (Navaraj et al. 2000). Deletions were also observed in the mtDNA of sen mutants, but unlike those occurring in nd were flanked by 6- to 10-bp repeats typically associated with GC-rich palindromic sequences (D'Souza et al. 2005). The nature of the sequences that flanked the mtDNA deletions in these two mutants supported the existence of two distinct systems of mtDNA recombination in N. crassa: a general system of homologous recombination (system I) and a site-specific mechanism (system II), mediated in part by nd and sen, respectively (Bertrand et al. 1993; D'Souza et al. 2005). The nd and sen mutations have been mapped to linkage groups I and V, respectively, but neither gene has been cloned and the precise function of their gene products remains unclear. Two ultraviolet (UV)-sensitive mutants, uvs-4 and uvs-5, are thought to undergo senescence, but unfortunately, these strains have not been studied in great detail (Schroeder 1970; Perkins et al. 1993; Hausner et al. 2006). Premature senescence has also been observed in cytoplasmic mutants of N. crassa including the E35 and ER-3 stopper mutants that harbor large mtDNA deletions, as well as strains that accumulate mitochondrial plasmids capable of inserting into mtDNA through homologous recombination (de Vries et al. 1986; Akins et al. 1989; Myers et al. 1989; Niagro and Mishra 1989; Court et al. 1991; Alves and Videira 1998).
While trying to establish the role of MUS-10 in DNA repair, we discovered that the mus-10 mutant exhibited a shortened life span, an abnormal mitochondrial morphology and mtDNA instability. We cloned the mus-10 gene through its ability to complement the MMS sensitivity of the mus-10 mutant and revealed that it encoded a novel F-box protein. This suggested that MUS-10 is part of an Skp, Cullin, F-box containing (SCF) E3 ubiquitin ligase complex that targets proteins for degradation by the 26S proteasome. The data we present in this article offer proof that an SCF complex can regulate both mitochondrial maintenance and cellular senescence.
MATERIALS AND METHODS
Neurospora strains and cosmid libraries:
The N. crassa strains used in this study are listed in Table 1. Growth and handling of N. crassa were performed as previously described (Davis and de Serres 1970). Some Neurospora strains, as well as the pMOcosX (Orbach 1994) and pLORIST (Kelkar et al. 2001) cosmid libraries were obtained from the Fungal Genetics Stock Center (FGSC; University of Missouri, Kansas City, MO). The original mus-10 mutant (FGSC 5148) was twice backcrossed to C1-T10-28a to produce two mus-10 isolates, KB27(10)-13A and KB27(10)-18a. Age-matched wild-type and mus-10 mutant strains, K-byWT and K-byM10A, respectively, were obtained from a cross between KB27(10)-18a and 74-OR31-16A. A mus-10 knockout strain, KTO-m10H2-1, was generated using a mus-52 mutant (FGSC 9719) and standard protocols (Ninomiya et al. 2004). Removal of mus-52∷Bar from KTO-m10H2-1 was achieved through a cross with C1-T10-34A producing KTO-10H-10A. To facilitate targeted integration at the his-3 locus, KTO-10H-10A was crossed to a his-3 mutant (FGSC 6103) to create the mus-10 his-3 double mutant KRA-m10his3-5.
TABLE 1.
N. crassa strains used in this study
| Strain | Genotype | Origin, source, or reference |
|---|---|---|
| C1-T10-28a | a | Tamaru and Inoue (1989) |
| C1-T10-34A | A | Tamaru and Inoue (1989) |
| C1-T10-37A | A | Tamaru and Inoue (1989) |
| 74-OR31-16A | A al-2 pan-2 cot-1 | de Serres (1980) |
| FGSC 5148 | A mus-10 | FGSCa |
| KB27(10)-13A | A mus-10 | This study |
| KB27(10)-18a | a mus-10 | This study |
| K-byWT | Undetermined | This study |
| K-byM10A | A mus-10 pan-2 | This study |
| FGSC 9719 | a mus52∷Bar | FGSC |
| KTO-m10H2-1 | a mus-10∷Hygr mus52∷Bar | This study |
| KTO-10H-10A | A mus-10∷Hygr | This study |
| FGSC 6103 | A his-3 | FGSC |
| KRA-m10his3-5 | mus-10∷Hygr his-3 | This study |
| KRA-m10M10F-2 | mus-10∷Hygr his-3+:mus-10-FLAG | This study |
| KRA-m10dFM10F-1 | mus-10∷Hygr his-3+:mus-10ΔF-box-FLAG | This study |
Fungal Genetics Stock Center, University of Missouri, Kansas City, MO, 64110.
Measurement of linear growth and life span:
Apical growth of hyphae was measured in race tubes that were ∼30 cm in length (Ryan et al. 1943). Race tubes containing Vogel's minimal agar medium with 0.5 or 1.2% sucrose were inoculated with conidia at one end of the tube and incubated at 25° with constant light. The position of the growth front was marked once or twice a day to facilitate measurement of the apical growth rate. When hyphae reached the opposite side of the tube, a small piece of mycelia-containing medium was transferred to a fresh tube. To measure apical growth rates over an extended period of time, the entire process was repeated several times. Strains that were unable to traverse the race tube after numerous transfers to new medium were deemed to have a shortened life span.
Spot test analysis:
The MMS sensitivity of various N. crassa strains was examined through spot tests. Briefly, conidia were harvested and washed twice with sterile water. The concentration of each conidial suspension was adjusted to 1 × 106 conidia/ml. These mixtures were then subjected to five 1:4 dilutions. A 10-μl aliquot of each suspension was spotted onto agar plates containing Vogel's minimal medium and sorbose. When required, MMS (0.015%) was added to the medium. Plates were incubated at 30° for 2 days and then photographed.
Isolation of mtDNA:
Sucrose gradient-purified mitochondria were isolated from mycelia using previously described methods (Lambowitz 1979; Seidel-Rogol et al. 1989; Rowley et al. 1994). TE200 (200 mm Tris-HCl pH 8.0, 1 mm EDTA) was mixed with the mitochondria and then centrifuged at 15,000 rpm for 15 min. The mitochondrial pellet was resuspended in 250 μl of TE200, 40 μl of 20% SDS, and 290 μl of phenol:chloroform (1:1). After vigorous mixing, the sample was centrifuged at 12,000 rpm for 10 min. The aqueous phase was extracted two more times, first with phenol:chloroform and then with chloroform alone. The mtDNA was precipitated from the aqueous phase with ethanol, dissolved in TE buffer containing RNase A and then stored at −25°.
Cloning and sequencing of mus-10 mtDNA:
ApaI digestion of mtDNA obtained from the fifth subculture of mus-10 generated a 6.6-kbp fragment that was extracted from an agarose gel and cloned into pBluescript SK+ (Stratagene) to produce pmtApaI. To identify the deletion breakpoints, smaller regions of the 6.6-kbp ApaI insert were sequentially subcloned into pBluescript using XbaI and then HindIII, generating pmtXbaI and pmtHindIII, respectively. The ends of all three inserts were sequenced using the universal T3 and T7 promoter primers and a BigDye sequencing kit (Applied Biosystems). Sequencing samples were run on an ABI PRISM 3100 genetic analyzer (Applied Biosystems). The sequences obtained in this manner were compared with mtDNA sequences from the Neurospora database (Assembly 3; Galagan et al. 2003).
Mitochondrial staining:
Plates containing Vogel's minimal medium, 1.2% sucrose and 2% agar were inoculated with conidia and incubated overnight at 30°. To observe mitochondria in live cells, mycelia were stained with MitoFluor Red (Molecular Probes) or MitoTracker Green FM (Invitrogen). After 20 min at room temperature, a piece of mycelia-containing medium was transferred to a glass slide and examined by fluorescence microscopy. Mitochondria stained with MitoFluor Red were observed using a BX60 microscope (Olympus) and images were captured with a black-and-white camera (C4742-95; Hamamatsu). When MitoTracker Green FM was used, mitochondria were visualized and photographed using a confocal laser-scanning microscope (FV1000-D; Olympus).
Transformation of Neurospora:
Neurospora transformations were performed as described with slight modifications (Ninomiya et al. 2004). Briefly, the conidial suspension was washed with 1 m sorbitol three times after which the concentration was adjusted to 2 × 109 conidia/ml. Linearized DNA (1–5 μg) was added to 100 μl of the conidial suspension and incubated on ice for 5 min. An aliquot of 40 μl was then transferred to a chilled electroporation cuvette (2 mm width). Electroporation was performed using a BTX Electro Cell Manipulation 600 (Genetronics) set at 1.5 kV and 186 ohm. After electroporation, the suspension was quickly removed from the cuvette and mixed with 960 μl of Vogel's minimal medium containing 1.2% sucrose. The conidia were incubated at 30° for 3 hr, mixed with molten top agar, and then spread over a selection medium. For the transformations that facilitated cloning of the mus-10 gene, hygromycin B was used at a concentration of 500 μg/ml.
Cloning of mus-10:
A 1.4-kbp SalI fragment of pCB1003 (Carroll et al. 1994) containing the Escherichia coli hph gene controlled by the Aspergillus nidulans trpC gene promoter was subcloned into pBluescript SK+ to produce pHS. NotI digestion of a pLORIST cosmid, H013 B4, generated an 8.5-kbp fragment that was cloned into the corresponding site of pHS to produce the plasmid pH 13-N8. A portion of pH 13-N8 was removed by SacII digestion and subsequent recircularization with T4 DNA ligase to create the plasmid pH 13-SS. This plasmid included 3.1 kbp of sequence from H013 B4 and contained a single open reading frame, NCU02379.3, which was later confirmed as the mus-10 gene.
MUS-10 antibody production:
The 1.9-kb mus-10 open reading frame (ORF) was amplified through PCR using a cDNA template and the primers MUS10-Ab-F (5′-GTACCATATGACGTCCTCCTCCTCACTGGA-3′) and MUS10-Ab-R (5′-TACGAAGCTTGTCGTCGGGGTACGATTCCT-3′). This fragment was cloned into pET-21a (Novagen) using NdeI and HindIII restriction sites added by the primers used for PCR amplification. The resulting plasmid, pmus10-Ab-4, was transformed into Rosetta 2(DE3)pLysS E.coli competent cells (Novagen) to produce RS2m4-1. Expression of full-length MUS-10 protein in E. coli was performed as per the manufacturer's instructions. The cells were harvested by centrifugation and stored at −20° until processed.
To facilitate purification of MUS-10 inclusion bodies, frozen cell pellets were thawed on ice prior to addition of 3 ml of resuspension buffer [50 mm Tris-HCl pH 8.5, 5 mm EDTA, 1 mm phenylmethanesulfonyl fluoride (PMSF)] per gram of cells. The cells were lysed by sonication (8 × 15 sec bursts; UR-200P, Tomy Seiko, Tokyo, Japan). The insoluble fraction was pelleted through centrifugation and resuspended in 1 ml of wash buffer (50 mm Tris-HCl pH 8.5, 5 mm EDTA, 1% sodium deoxycholate). This mixture was subjected to a second round of sonication and the insoluble matter was again collected by centrifugation. The pellet was resuspended in 1 ml deoxycholate (1%) and incubated at 37° overnight. Following a final round of sonication and centrifugation, solubilization buffer (3 m urea, 10 mm Tris-HCl pH 8.0) was used to solubilize the purified inclusion bodies to a final concentration of 0.4 mg/ml. This mixture was sent to the antibody manufacturer Japan Lamb (Hiroshima, Japan) where it was injected into rabbits to facilitate production of polyclonal antibodies.
Protein isolation and Western blot analysis:
Liquid media were inoculated with various N. crassa strains to a final concentration of 1 × 106 conidia/ml and grown at 30° for 18 hr with vigorous shaking. When indicated, MMS (0.05%) was added to the culture after 16 hr of growth and harvested with the untreated cultures 2 hr later. Previously published protocols were used for the preparation of cytosolic and crude mitochondrial protein fractions (Chae and Nargang 2009), purification of mitochondria through sucrose gradients (Lambowitz 1979; Rowley et al. 1994), SDS–PAGE (Laemmli 1970), and Western blotting (Good and Crosby 1989). Western blot analysis was performed using the anti-MUS-10 antibody described above (1/400), as well as three commercially available mouse monoclonal antibodies: ANTI-FLAG M2 antibody (1/10,000; Sigma, F3165), anti-α-tubulin (1/200,000; Sigma, T6074), and anti-COX3 (1/30,000; Molecular Probes, A-6408). Goat anti-mouse and goat anti-rabbit IgG, HRP conjugated secondary antibodies (Promega) were used at concentrations of 1/10,000 and 1/3000, respectively.
Yeast two-hybrid analysis:
Yeast two-hybrid experiments were performed according to the manufacturer's instructions (Matchmaker Two-hybrid System 2 and 3, Clontech). Briefly, the primers m10-1 (5′-CCATGGATATGACGTCCTCCTCCTCA-3′) and m10-2 (5′-GGATCCCTAGTCGTCGGGGTACGA-3′) were employed in RT–PCR to amplify a full-length mus-10 cDNA. The primers used in PCR introduced NcoI and BamHI restriction sites used to clone the mus-10 cDNA into pACT2 and pGBKT7, producing the plasmids pACT2-mus-10 and pGBKT7-mus-10, respectively. The GAL4 activation domain is encoded in pACT2 while pGKBT7 contains the GAL4 DNA binding domain. The plasmids pACT2-scon-3 and pAS2-scon-3 were generated in a similar manner, but using the primers scon3U (5′-CCATGGAGATGGCGGAGAACGACG-3′) and scon3L (5′-GGATCCCTAACGGTCTTCCGCCCA-3′). For the latter construct, scon-3 was fused to the GAL4 DNA binding domain of pAS2-1 rather than that of pGBKT7. Plasmids carrying the GAL4 activation domain were cotransformed into yeast (Y187) with one of several constructs encoding the GAL4 DNA binding domain. Transformants were spread over plates containing medium lacking tryptophan and leucine, which selected for plasmids derived from pGBKT7 (or pAS2-1) and pACT2, respectively. Colonies resulting after 3 days at 30° were subjected to a filter assay for detection of β-galactosidase activity using the protocol described by Clontech.
Generation of FLAG-tagged wild-type and F-box deficient MUS-10:
Two primers, S40-LnFG5 (5′-ccctcgaggatccggtagtatggactacaaagaccatgacggtgattataaagatcatgacatt-3′) and S41-LnFG3 (5′-ttgggcccttacttgtcatcgtcatccttgtaatccttgtaatcaatgtcatgatctttataatca-3′), which contain 22-bp complementary sequences at their termini, were used in a PCR reaction in the absence of template DNA to produce a 3xFLAG tag. This fragment was digested with XhoI and ApaI and cloned into the corresponding sites of pMF272, thereby removing the GFP gene contained in this plasmid (Freitag et al. 2004). This plasmid was named pFLAGC. m10Fc-5 (5′-GCTCTAGAGATGACGTCCTCCTCCTCACT-3′) and m10-272Fc-3 (5′-GGATCCGTCGTCGGGGTACGATTCCTTA-3′) were used to amplify the full-length mus-10 ORF (637 codons) from mus-10 cDNA. This fragment was cloned into pFLAGC using XbaI and BamHI restriction sites introduced by the primers. This cloning procedure placed the mus-10 ORF under the control of the ccg-1 gene promoter and placed a 3xFLAG tag on the C terminus of the MUS-10 protein. A similar procedure was used to insert a truncated form of the mus-10 ORF (codons 54–637) into pFLAGC, but in this case, the cloning was achieved using a different forward primer, dF-m10Fc-5 (5′-TCTAGAACATGTCGTTTACTTTCTGGGAGCCTG-3′). Both constructs were transformed into a mus-10 his-3 double mutant, KRA-m10his3-5, using electroporation. Desired transformants were selected by their ability to grow on media lacking histidine as elements from pMF272 promote targeted integration and reversion at the his-3 locus (Freitag et al. 2004). In this manner, two N. crassa transformants were recovered, KRA-m10M10F-2 and KRA-m10dFM10F-1, which produced a full-length and F-box-deficient MUS-10 protein, respectively.
RESULTS
The mus-10 mutant has a shortened life span:
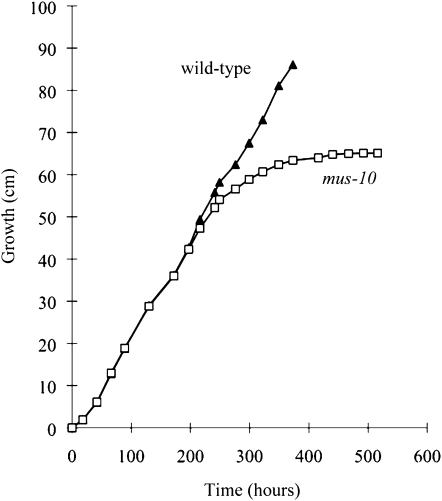
During our analysis of mus-10, we noticed that the viability of its conidia decreased through successive subculturing. To confirm our suspicions, we compared the apical growth rate of a wild-type strain and a mus-10 mutant using race tubes. To ensure the inocula in these experiments were of similar age, mus-10 was backcrossed to wild type and the resulting ascospores were randomly isolated and cultured. These age-matched progeny were characterized as carrying wild-type or mutant mus-10 alleles on the basis of their sensitivity to MMS. Conidia from these strains were then used to inoculate race tubes. The apical growth rate of the wild-type strain remained constant over all of the time points examined (Figure 1). Conversely, growth of mus-10 began to deteriorate after ∼200 hr and completely stopped after ∼380 hr (Figure 1). These data verified that mus-10 exhibited a senescence phenotype.
Figure 1.—
Apical growth rates of wild-type and mus-10 mutant strains. Conidia from age-matched wild-type (K-byWT) and mus-10 (K-byM10A) were used to inoculate race tubes containing Vogel's minimal agar medium supplemented with 0.5% sucrose. Hyphal growth was recorded once or twice a day. Once the mycelia had traversed the tube, a piece of the growth front was transferred to a race tube containing fresh medium and the entire process was repeated. All race tubes were incubated at 25° under constant light.
mtDNA deletions in the mus-10 mutant:
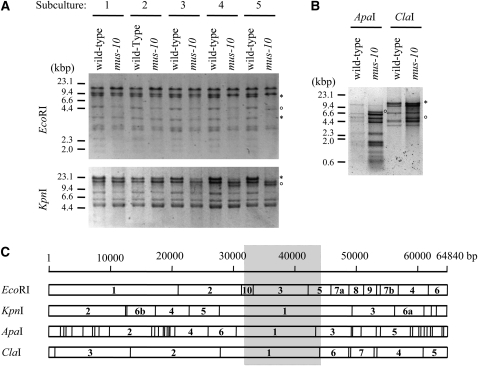
The nd and sen mutants of N. crassa suffer from a shortened life span that correlates with the accumulation of mtDNA rearrangements (Sheng 1951; Seidel-Rogol et al. 1989; Bertrand et al. 1993; Navaraj et al. 2000; D'Souza et al. 2005). To investigate whether mtDNA rearrangements became more prevalent in mus-10 as it aged, we isolated mtDNAs from five sequential subcultures of age-matched wild type and mus-10. Samples of mtDNA were digested with EcoRI or KpnI and the resulting fragments were resolved using agarose gel electrophoresis. The mtDNAs obtained from the first three subcultures of wild type and mus-10 displayed virtually identical restriction digest patterns (Figure 2A). However, mtDNAs from the fourth and fifth subcultures of mus-10 produced an altered banding pattern characterized by the disappearance of the 8.8-kbp and 3.7-kbp EcoRI fragments (designated as EcoRI-3 and EcoRI-5, respectively; Bertrand et al. 1993) and the emergence of a 4.4-kbp EcoRI band (Figure 2A). Similarly, the 21.6-kbp KpnI fragment seemed to be replaced by a novel band of ∼10 kbp (Figure 2A). No such changes were observed in age-matched wild-type strains. Unfortunately, the three smallest EcoRI fragments (EcoRI-8, -9 and -10) could not be clearly observed in our experiments, and thus we could not determine whether these bands were modified in any way. mtDNAs from older wild type and mus-10 (fifth subculture) also produced differing restriction digest patterns when digested with ApaI and ClaI (Figure 2B). On the basis of these results, we estimated that the mtDNA of mus-10 carried a 10- to12-kbp deletion (Figure 2C).
Figure 2.—
Restriction analysis of mtDNA. (A) mtDNAs were isolated from five sequential subcultures (labeled 1–5) of wild-type and mus-10 mutant strains. These mtDNAs were digested with EcoRI or KpnI, and subjected to agarose gel electrophoresis. Bands emerging or disappearing in the fourth and fifth subcultures of mus-10 are indicated with an open circle (°) and asterisk (*), respectively. (B) mtDNAs from the fifth subculture of wild-type and mus-10 mutant strains were digested with ApaI or ClaI. Restriction digest fragments that differed between the two strains are indicated as described in A. (C) Restriction maps of N. crassa mtDNA digested with ApaI, ClaI, EcoRI, or KpnI. Position 1 in this figure corresponds to the nucleotide at the 5′ end of the largest EcoRI fragment. Numbers located within the mtDNA fragments indicate their size relative to the other fragments generated from digestion with a given restriction enzyme. The number “1” is used to describe the largest fragment, while “a” and “b” are employed when two fragments of similar size are produced, where “a” specifies the larger fragment. The shaded area shows the predicted location of a large mtDNA deletion (∼10–12 kbp) observed in latter cultures of mus-10.
Cloning and analysis of mus-10 mtDNA:
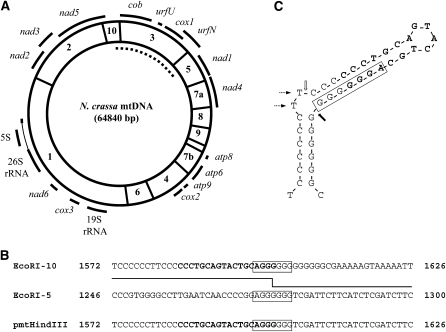
To uncover the region of mtDNA deleted in mus-10, we first needed to determine the sequence of the flanking regions. We reasoned that the 6.6-kbp ApaI restriction fragment observed in mtDNA from the fifth subculture of mus-10, but absent in that of wild type, likely resulted from a large deletion involving the 12.9-kbp and possibly the 5.8-kbp ApaI fragments (Figure 2C). This novel ApaI band was cloned into the corresponding site of pBluescript. The ends of the 6.6-kbp insert were sequenced with the universal T7 and T3 promoter primers but unfortunately, no mtDNA deletions were detected in the sequenced region when compared with mtDNA sequences from the Neurospora database (assembly 3; Galagan et al. 2003). This implied that the deletion breakpoint occurred further into the cloned mtDNA fragment. Consequently, a 2.2-kbp region of mtDNA from the cloned ApaI restriction fragment was subcloned into pBluescript using XbaI. Sequence analysis was performed as described above, but the deletion breakpoints could not be identified. Subsequent sequencing of pmtHindIII, which contained 1.2 kbp of mtDNA obtained from digestion of the 6.6-kbp ApaI restriction fragment with HindIII, finally revealed a 10,488-bp deletion in the mtDNA of mus-10. This deletion completely removed EcoRI-3 as well as parts of EcoRI-5 and EcoRI-10, thereby eliminating two unidentified reading frames (urfU and urfN), cox1 (cytochrome c oxidase subunit 1), cob (apocytochrome b), and part of nad1 (NADH dehydrogenase subunit 1) (Figure 3A).
Figure 3.—
Characterization of the mtDNA deletion observed in mus-10. (A) A circular EcoRI restriction map of N. crassa mtDNA. The sizes of the EcoRI fragments are indicated as described in Figure 2C. The location of genes and rRNAs are indicated with solid black lines. The dotted line denotes the mtDNA deleted in mus-10. (B) Location of the mtDNA deletion breakpoints of mus-10. The 10,488-bp deletion detected in mus-10 was flanked by 7-bp direct repeats (indicated with boxes) present in EcoRI-10 and EcoRI-5. Numbers beside the mtDNA sequences indicate the position of nucleotides relative to the closest 5′ EcoRI restriction site. The recombining sequences that gave rise to the mtDNA observed in pmtHindIII are indicated with a solid line. The 18-bp PstI palindrome associated with deletions of mtDNA in mus-10 and sen is shown in boldface type. (C) Predicted stem-loop structure of the mtDNA sequence (nucleotides 1572–1610 of EcoRI-10) surrounding the 5′ deletion junction. The nucleotides that comprise the PstI palindrome are shown in boldface type. The 7-bp direct repeat of EcoRI-10 is indicated with a box. The two unpaired thymine residues that are thought to trigger BER or MMR pathways are shown with dotted arrows. The positions where single strand breaks likely occurred to facilitate deletions of mtDNA in mus-10 and sen are indicated with solid and open block arrows, respectively.
Further examination revealed that the deleted region was flanked by 7-bp direct repeats (AGGGGGG; Figure 3B), resembling sequences associated with system II, the site-specific mtDNA recombination pathway. Interestingly, the first four nucleotides of the 5′ repeat overlapped an 18-bp PstI palindrome (5′-CCCTGCAGTACTGCAGGG-3′) that occurs in mtDNA of N. crassa 67 times (Cahan and Kennell 2005) and is thought to form a stem-loop structure (Figure 3C; Yin et al. 1981; Nargang et al. 1983; de Vries et al. 1986).
Mitochondrial morphology of the mus-10 mutant:
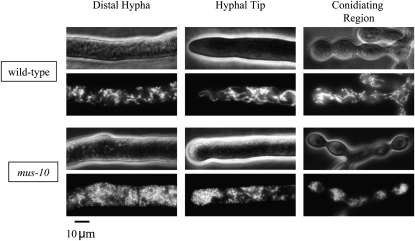
Our restriction digest analysis of mtDNA suggested that loss of the mus-10 gene had an adverse effect on mitochondria. To explore this hypothesis, we used fluorescence microscopy to examine the morphology of mitochondria within mus-10. Growing hyphae were stained with a mitochondria-specific fluorescent dye and then visualized under a microscope. In wild-type mycelia, long tubular mitochondria were observed in both distal and conidiating hyphae (Figure 4). Conversely, mus-10 had abnormal mitochondria that exhibited a spherical morphology (Figure 4). Surprisingly, the occurrence of this phenotype was not age related, as spherical mitochondria were observed in earlier subcultures during which hyphae were growing normally (Figure 4). This implies that changes to mitochondrial structure occur prior to the formation of abnormal mtDNAs.
Figure 4.—
The morphology of mitochondria in wild-type and mus-10 mutant strains. Growing hyphae from the first subculture of wild-type (K-byWT) and mus-10 (K-byM10A) strains were stained with MitoFluor Red and then visualized using fluorescence microscopy (lower). These hyphae were also observed using phase-contrast microscopy (upper).
Identification of the mus-10 gene:
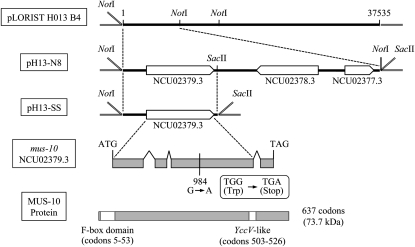
The mus-10 gene was previously mapped to a site on LG VII near met-7 (7%; Kafer and Perlmutter 1980). To identify the mus-10 gene, we used a gene-rescue approach in which mus-10 conidia were transformed with digested cosmids that spanned a 27.7-kbp region defined by our linkage and RFLP analyses (data not shown). In this manner, we discovered an 8.5-kbp NotI fragment derived from the pLORIST cosmid H013 B4 that could rescue the MMS sensitivity of mus-10. This fragment was cloned into pHS to produce pH 13-N8 (Figure 5). The sequence in this fragment contained three annotated ORFs: NCU02379.3, NCU02378.3, and NCU02377.3 (Figure 5). Removal of the latter two ORFs from pH 13-N8 was achieved through SacII digestion and subsequent recircularization of the desired fragment to generate the plasmid pH 13-SS. The 3.1-kbp insert in this plasmid contained a single ORF (NCU02379.3) that conferred wild-type MMS resistance to mus-10. To confirm the identity of the mus-10 gene, we isolated genomic DNA from the original mus-10 mutant and employed PCR to amplify the candidate gene. Sequence analysis of this PCR product revealed that mus-10 carries a single G-to-A transition at position 984 of the predicted ORF sequence, producing a nonsense mutation (Figure 5).
Figure 5.—
Identification of the mus-10 gene. NotI digestion of pLORIST H013 B4 produced an 8.5-kbp fragment that could restore growth of mus-10 on medium containing MMS. This fragment was cloned into pHS to produce pH 13-N8. Sequence analysis of pH 13-N8 revealed three predicted ORFs, NCU02379.3, NCU02378.3, and NCU02377.3. To identify the mus-10 gene, a portion of pH 13-N8 was removed with SacII producing pH 13-SS. This plasmid contained a single ORF, NCU02379.3, and could complement the mus-10 mutant phenotype. Sequencing revealed that the mus-10 mutant carried a G-to-A transition at position 984 of this ORF, which generated a nonsense mutation. ORF NCU02379.3 is predicted to encode a 637-amino acid polypeptide with a molecular weight of 73.7 kDa. Analysis of the amino acid sequence revealed an F-box domain (codons 5–53) and a YccV-like domain (codons 503–526).
The mus-10 gene was predicted to encode a 637 residue polypeptide with a molecular weight of 73.7 kDa. The ORF sequence and intron–exon boundaries were confirmed through sequence analysis of cDNA generated through RT–PCR. An F-box was identified at the N terminus of the MUS-10 protein (Figure 5), suggesting that it belongs to the F-box protein family whose members are normally components of SCF E3 ubiquitin ligase complexes that direct proteins to the 26S proteasome, ultimately leading to their degradation. These findings are concurrent with the hypothesis that SCF complexes can regulate mitochondrial function (Cohen et al. 2008; Deng et al. 2008). A YccV-like domain, which has been shown to bind hemimethylated DNA (D'Alencon et al. 2003), was also observed in MUS-10, suggesting that it may be capable of interacting with DNA (Figure 5).
Localization of MUS-10:
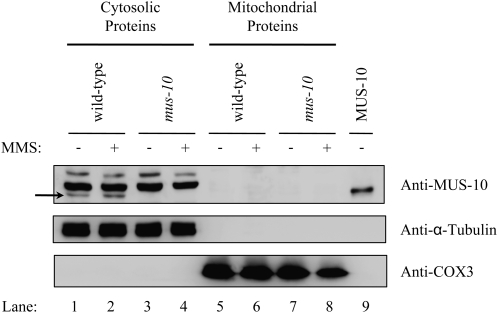
To help elucidate the function of MUS-10, we wanted to establish its subcellular location. Western blot analysis was performed on cytosolic and purified mitochondrial proteins isolated from a wild-type strain (C1-T10-37A) and a mus-10 knockout (KTO-10H-10A). To ensure the absence of cytosolic proteins in the mitochondrial fraction and vice versa, blots were also examined using antibodies against α-tubulin and COX3, which are found in cytosol and mitochondria, respectively (Figure 6). Use of an anti-MUS-10 antibody revealed a band in the cytosolic fraction of wild type that was absent in mus-10 (Figure 6, lanes 1 and 3). Conversely, MUS-10 was not observed in mitochondria from either strain (Figure 6, lanes 5 and 7).
Figure 6.—
MUS-10 localization. Western blot analysis was performed using cytosolic proteins (200 μg) and sucrose gradient-purified mitochondria (50 μg) isolated from a wild-type (C1-T10-37A) and mus-10 mutant strain (KTO-10H-10A) grown in the presence (+) or absence (−) of MMS. Anti-α-tubulin and anti-COX3 were used as controls to identify cytosolic and mitochondrial marker proteins, respectively. A total of 250 ng of recombinant MUS-10 (MUS-10) was loaded in the gel to verify functionality of the MUS-10 antibody. Since use of the anti-MUS-10 antibody produced several nonspecific bands, the location of MUS-10 is indicated with a solid arrow.
Since MUS-10 appears to influence mitochondrial morphology and also protects cells against the effects of MMS, we reasoned that exposure to MMS may induce movement of MUS-10 from the cytosol to mitochondria. However, MUS-10 localization was not altered in cells grown in the presence of MMS (Figure 6). Taken together, these results suggest that MUS-10 may function in the cytosol.
MUS-10 is part of an SCF complex:
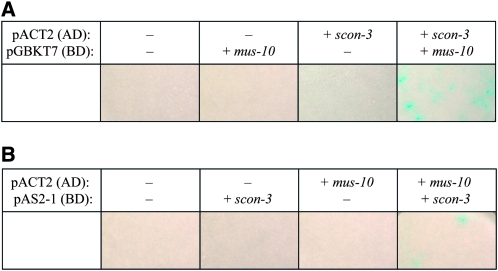
Proteins with an F-box motif (F-box proteins) generally function in SCF E3 ubiquitin ligase complexes along with two additional core components, Cullin and Skp1, although they are thought to physically interact only with the latter (Bai et al. 1996; Jackson and Eldridge 2002; Willems et al. 2004; Ho et al. 2008). To investigate whether MUS-10 physically interacts with SCON-3 (the Neurospora Skp1 homolog; Sizemore and Paietta 2002) a yeast two-hybrid assay was performed. cDNAs of mus-10 and scon-3 were cloned into yeast expression vectors containing the Gal4p activation or DNA binding domains. A filter assay showed that β-galactosidase was produced when yeast cells were cotransformed with constructs carrying mus-10 and scon-3 fused to the DNA binding and activation domains of Gal4p, respectively, while no β-galactosidase activity was observed in any of the control experiments (Figure 7A). Although fewer yeast colonies were generated in reciprocal experiments, β-galactosidase activity could still be detected (Figure 7B). These data are consistent with a physical interaction between MUS-10 and SCON-3.
Figure 7.—
A physical interaction between MUS-10 and SCON-3. Yeast two-hybrid analysis was performed to determine whether the MUS-10 protein interacted with a core component of N. crassa SCF complexes, SCON-3. (A) Yeast cells (Y187) were cotransformed with two plasmids, one of which contained the GAL4 activation domain (AD) of pACT2 while the other included a GAL4 DNA binding domain (BD) from pGBKT7. While empty vectors (−) were used in some of these experiments, transformations were also performed with derivatives of pACT2 and pGBKT7 that contained in-frame scon-3 (+ scon-3) and mus-10 (+ mus-10) cDNAs, respectively. Selection of cotransformants was achieved using nutritional markers present within pACT2 and pGBKT7, which confer the ability to grow on medium lacking leucine and tryptophan, respectively. Colonies emerging after a 3-day incubation at 30° were subjected to a filter assay capable of detecting β-galactosidase activity, which would only be observed if the two proteins being examined could physically interact. (B) Reciprocal experiments performed as described in A. In these experiments, pAS2-1, which contains the GAL4 DNA binding domain and the TRP1 gene, was used in place of pGBKT7.
The F-box motif of MUS-10 is required for its function:
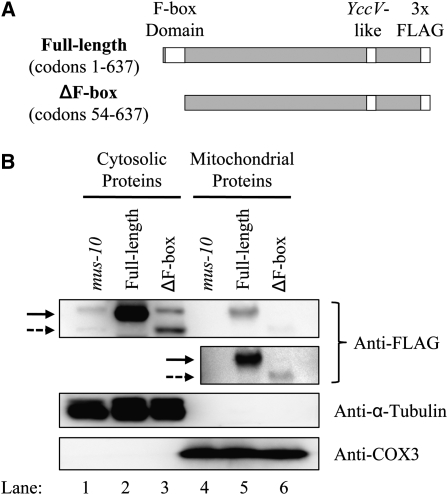
The F-box motif normally interacts with Skp1 and is thought to be essential for F-box protein function (Bai et al. 1996). To confirm the importance of the MUS-10 F-box motif, we examined whether a MUS-10 protein lacking the F-box motif could rescue the phenotypes of the mus-10 mutant. Full-length and F-box-deficient versions of the mus-10 gene were inserted into pFLAGC, which placed them under the control of the constitutive ccg-1 gene promoter (Freitag et al. 2004) and added a 3xFLAG-tag to the C terminus of each protein (Figure 8A). Transformation of these constructs into a mus-10 knockout produced strains that were revealed through Western blot analysis to contain relatively high levels of FLAG-tagged full-length or truncated MUS-10 protein within the cytosol (Figure 8B, lanes 2 and 3). Unfortunately, the anti-FLAG antibody also recognized a nonspecific protein similar in size to full-length MUS-10 (lanes 1 and 3). However, given the intensity of the band observed in mus-10 transformed with the full-length construct (lane 2), it is clear that FLAG-tagged MUS-10 is being produced in this strain. Surprisingly, a small amount of full-length and ΔF-box-deficient MUS-10 were detected in mitochondria with the relative levels of the two proteins being similar to that observed in the cytoplasm (Figure 8B, lanes 2 and 3 vs. lanes 5 and 6). This may suggest that overexpression of the two MUS-10 forms leads to or increases their association with mitochondria. While this result will be addressed further in discussion, it is important to note that the ΔF-box MUS-10 was also observed in mitochondria and thus, removal of the first 53 amino acids did not remove an N-terminal mitochondrial signal sequence.
Figure 8.—
FLAG-tagged forms of MUS-10. (A) Diagram of full-length (codons 1–637) and truncated (codons 54–637) MUS-10 used in our experiments. Insertion of cDNAs encoding the two forms of MUS-10 into pFLAGC placed their expression under the control of the constitutive ccg-1 gene promoter and added a 3xFLAG tag on the C terminus of each protein. Since pFLAGC is derived from pMF272, it contains a portion of the N. crassa his-3 gene, which can be used for targeted integration and reversion at the his-3 locus. (B) Western blot analysis was performed as described in Figure 6. Plasmids encoding the two different FLAG-tagged versions of MUS-10 (shown in A) were used to transform mus-10 (KRA-m10his3-5) conidia. Two resulting his+ transformants, KRA-m10M10F-2 and KRA-m10dFM10F-1, expressed full-length and ΔF-box MUS-10, respectively. Background levels were determined using untransformed mus-10. Full-length MUS-10 protein is indicated with a solid arrow, while the ΔF-box form is shown with a dotted arrow. To facilitate viewing of MUS-10 in the mitochondrial fraction, a second, longer exposure was also included (Anti-FLAG, lower).
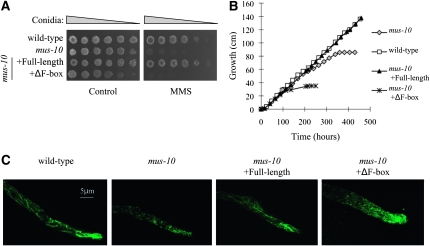
After we confirmed expression of both FLAG-tagged MUS-10 proteins, we performed spot test analysis to determine whether these proteins could rescue the MMS sensitivity of mus-10. As anticipated, strains expressing full-length MUS-10 protein displayed wild-type growth on MMS plates while those carrying the truncated form of MUS-10 lacking the F-box motif remained susceptible to MMS (Figure 9A).
Figure 9.—
The F-box of MUS-10 is essential for its function. (A) Spot test analysis was performed using conidia from C1-T10-37A (wild type), KTO-10H-10A (mus-10), KRA-m10M10F-2 (mus-10 + full-length) and KRA-m10dFM10F-1 (mus-10 + ΔF-box). Conidial suspensions were adjusted to a concentration of 1 × 106 conidia/ml and subjected to five 1:4 serial dilutions. A 10-μl aliquot of each mixture was spotted onto agar plates containing Vogels's minimal sorbose medium lacking (control) or supplemented with 0.015% MMS (MMS). Plates were photographed after 2 days at 30°. (B) Measurements of apical growth were performed as described in Figure 1. In these experiments, race tubes contained 1.2% sucrose instead of 0.5%. (C) Mitochondrial morphology. Live hyphae were stained with MitoTracker Green and visualized using a confocal laser-scanning microscope.
To examine whether these transformants exhibited a shortened life span, their growth rates were monitored in race tubes. Transformants that expressed full-length MUS-10 displayed wild-type growth, while those harboring the F-box deficient form senescence prematurely (Figure 9B). Although the strain expressing the truncated form of MUS-10 appeared to undergo senescence earlier than the original mus-10 mutant (Figure 9B), it should be noted that deficiencies leading to senescence likely continued to accumulate during the transformation procedure, and thus conidia from this transformant are likely “older” than those of the original mus-10 mutant. Fluorescence microscopy revealed that tubular mitochondria could be restored in mus-10 through transformation with a construct encoding wild-type, but not truncated, MUS-10 protein (Figure 9C). Taken together, these data confirm that the F-box domain of MUS-10 is essential for its function in vivo.
DISCUSSION
Although sensitivity of mus-10 to MMS and UV light implied a deficiency in one or more DNA repair pathways, the precise function of the mus-10 gene was unclear. In this present work we report that MUS-10 belongs to the F-box protein family and is thus likely involved in proteasome-mediated protein turnover. This finding suggests that even though mus-10 is more susceptible to MMS than wild type, the mus-10 gene product may not have a direct role in DNA repair. Indeed, other instances of this phenomenon have been reported in other organisms. In S. cerevisiae, an increased sensitivity to MMS was observed in strains deficient for Tim13p, whose role in mitochondrial protein import is well established (Hanway et al. 2002). It was hypothesized that the heightened susceptibility of the tim13Δ strain to MMS resulted from the reduced import of mtDNA repair proteins. Conversely, overexpression of a nuclear BER protein in the mitochondria of human cells was thought to cause an imbalance of mitochondrial DNA repair proteins making them more vulnerable to MMS (Fishel et al. 2003). Since MUS-10 is likely to function in ubiquitin-mediated proteolysis, it is conceivable that MUS-10 deficiency promotes accumulation of one or more mitochondrial proteins that affect mtDNA repair, replication, or recombination, leading to an elevated sensitivity to MMS.
Similar to nd and sen, strains deficient for mus-10 displayed a shortened life span and accumulated mtDNA deletions (Seidel-Rogol et al. 1989; Bertrand et al. 1993; Navaraj et al. 2000; D'Souza et al. 2005). Analysis of the mtDNA deletion observed in latter subcultures of mus-10 revealed the absence of EcoRI-3 and parts of EcoRI-5 and EcoRI-10. This deletion removed at least three components of the electron transport chain, cob, cox1, and nad1, which likely impaired respiration leading to senescence and subsequent death of mus-10. Similarly, the stop–start growth phenotype of the E35 stopper mutant was attributed to loss of nad2 and nad3 (de Vries et al. 1986; Alves and Videira 1998), while the ER-3 stopper mutant was shown to harbor a ∼25-kbp deletion of mtDNA that removed several genes including cob and cox1 (Niagro and Mishra 1989; Niagro and Mishra 1990). Senescence in these and other N. crassa strains, including nd, sen, and mus-10, correlated with the accumulation of mtDNAs that harbor large deletions, which may incur a replicative advantage over wild-type molecules due to their smaller size. In a recent report, quantitative real-time PCR was used to demonstrate that in mice, mitochondrial genomes that contain large deletions (∼10 kb) accumulated faster than those carrying small deletions (∼3.8 kb) (Fukui and Moraes 2009). While these data support the notion that smaller mtDNAs have a replicative advantage over larger ones, it does not exclude the possibility that the number and/or nature of the genes present in a mitochondrial genome can influence copy number.
The PstI palindrome observed at the 5′ flank of the mus-10 mtDNA deletion is capable of forming a GC-rich imperfect stem loop, a structure which has been hypothesized to stall DNA replication and/or act as substrates for mismatch repair (MMR) or base excision repair (BER) systems (D'Souza et al. 2005; Hausner et al. 2006). Either scenario could lead to single and/or double strand breaks that are thought to promote mtDNA recombination. Indeed, the stem-loop structure formed by the PstI palindrome flanking the mus-10 mtDNA deletion contains two unpaired nucleotides that could potentially trigger BER or MMR pathways resulting in endonucleolytic cleavage of the phosphodiester bond following the AGGGGGG repeat (Figure 3C). Interestingly, the same PstI palindrome and unpaired nucleotides were implicated in the generation of mtDNA deletions in sen (recombination junction J2; D'Souza et al. 2005), but in this case, the cleavage event occurred on the opposite strand (Figure 3C), implying that mtDNA repair pathways can target either strand of the mismatched and/or unpaired sequence.
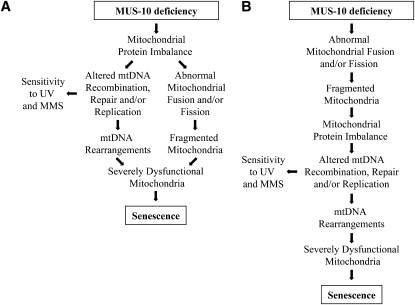
Our examination of mitochondrial morphology and mtDNA rearrangements in mus-10 suggests that fragmented mitochondria arise prior to modification in the mtDNA restriction digest profile. While we cannot exclude the possibility that mtDNA rearrangements arise in mus-10 much earlier than they can be visualized through restriction digest and agarose gel electrophoresis, our observations do raise the question of whether mitochondrial fragmentation leads to altered mtDNA or if these events occur through independent mechanisms. We propose two models that address the relationship between mitochondrial morphology and mtDNA integrity, both of which rely on MUS-10 functioning as an E3 ubiquitin ligase. In our first model (Figure 10A), MUS-10 deficiency is proposed to promote the accumulation of a wide variety of mitochondrial proteins including, but not limited to, mediators of fission and/or fusion, components of import machinery, and proteases. Accumulation of mitochondrial proteases would subsequently lead to lower levels of their substrates. The resulting mitochondrial protein imbalance has two concurrent effects: (i) deficiencies in mtDNA replication, recombination, and/or repair, leading to mtDNA rearrangements and sensitivity to MMS and UV, and (ii) disruption of the fission/fusion equilibrium, resulting in mitochondrial fragmentation. The persistence of such problems promotes further mitochondrial defects, which initiates a vicious cycle leading to the accumulation of dysfunctional mitochondria that ultimately cause senescence. Our second model (Figure 10B) proposes that loss of MUS-10 leads to the production of fragmented mitochondria through inhibited mitochondrial fusion and/or accelerated division. This model predicts that smaller mitochondria are more likely to harbor an imbalanced protein complement and are thus prone to defective maintenance of mtDNA. This compromises respiration, which in turn leads to senescence.
Figure 10.—
Models to explain the relationship between mus-10 and senescence. (A) In this model, loss of MUS-10 prevents operation of a mitochondrial E3 ubiquitin ligase leading to accumulation and/or deficiency of numerous mitochondrial proteins. The resulting protein imbalance leads to defects in both mtDNA maintenance and mitochondrial morphology through independent mechanisms. Defective mtDNA recombination, repair, and/or replication leads to increased MMS/UV-light sensitivity and mtDNA rearrangements. Mitochondrial fragmentation and altered mtDNA promote further mitochondrial dysfunction, which ultimately results in impaired respiration and eventual senescence. (B) This model is similar to the one proposed in A except that in this case, dysfunction of mtDNA recombination, repair, and/or replication results from a mitochondrial protein imbalance caused by mitochondrial fragmentation and is thus not a direct result of MUS-10 deficiency.
Regardless of whether there is a causal relationship between defective mtDNA and mitochondrial morphology in mus-10, the fragmented mitochondria observed in this strain likely arise through abnormally high amounts of fission or by inhibited fusion. Mitochondrial fusion and fission are complicated processes involving many proteins and are thought to enable mixing of metabolites and mtDNA thereby allowing optimal mitochondrial function (Cerveny et al. 2007; Hoppins et al. 2007; Knott and Bossy-Wetzel 2008; Hoppins and Nunnari 2009). Therefore circumstances that disrupt the balance between these opposing forces can be detrimental to the cell. Given that the mus-10 gene product is a mediator of proteolysis, the altered mitochondrial morphology may result from the accumulation of one or more proteins that promote division or hinder fusion. Indeed, links between E3 ubiquitin ligase complexes and mitochondrial structure have been reported. Mutations in Parkin, an E3 ubiquitin ligase, have been associated with defective mtDNA repair and progression of Parkinson's disease in humans (Dawson 2006; Dodson and Guo 2007). Recent evidence suggests that Parkin functions to promote mitochondrial fission and/or inhibit fusion (Deng et al. 2008; Poole et al. 2008). In Saccharomyces cerevisiae, the F-box protein Mdm30p is involved in ubiquitylation and subsequent degradation of the mitochondrial fusion mediator Fzo1p (Fritz et al. 2003; Escobar-Henriques et al. 2006; Cohen et al. 2008). The mdm30 mutant was shown to accumulate Fzo1p and exhibit aggregated mitochondria. A physical interaction between Mdm30p and Fzo1p has also been observed (Escobar-Henriques et al. 2006). Interestingly, Mdm30p of yeast has a weak homology to N. crassa MUS-10, and thus it is tempting to speculate that MUS-10 can interact with FZO-1 and/or other mitochondrial outer membrane proteins. Such an interaction might explain why we observed full-length and truncated forms of MUS-10 in mitochondria upon their overexpression as elevated levels of these proteins may promote binding of MUS-10 to its outer membrane targets. However, we cannot exclude the possibility that MUS-10 is imported into mitochondria and that our detection of the FLAG-tagged MUS-10 proteins in mitochondria stems from increased import arising from high levels of expression. In this case, mitochondrial import would have to rely on internal mitochondrial signal sequences as the N-terminal truncated form of MUS-10 also associated with mitochondria.
There are many unanswered questions regarding the function of MUS-10 and its relationship with mitochondria. However, the N. crassa mus-10 mutant could provide great insight into the role of ubiquitin-mediated proteolysis in the maintenance of mitochondrial morphology and mtDNA and thus may be used as a model for studying ageing and mitochondrial diseases in humans.
Acknowledgments
We thank Niji Ohta for her help with our sequencing experiments. We also express our gratitude to Yosuke Morishima and Shigeyuki Kawano who helped photograph mitochondria. This work was funded by grants-in-aid for scientific research 11640619, 08F08756, and 20570001. The Rational Evolutionary Design of Advanced Biomolecules, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, Japan Science and Technology Agency also supported this work. M.C. was given a grant-in-aid from the Japan Society for the Promotion of Science.
Sequence data from this article have been deposited with the DDBJ database under accession no. AB495263.
References
- Akins, R. A., R. L. Kelley and A. M. Lambowitz, 1989. Characterization of mutant mitochondrial plasmids of Neurospora spp. that have incorporated tRNAs by reverse transcription. Mol. Cell. Biol. 9 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, P. C., and A. Videira, 1998. The membrane domain of complex I is not assembled in the stopper mutant E35 of Neurospora. Biochem. Cell. Biol. 76 139–143. [PubMed] [Google Scholar]
- Antignani, A., and R. J. Youle, 2006. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr. Opin. Cell Biol. 18 685–689. [DOI] [PubMed] [Google Scholar]
- Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl et al., 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86 263–274. [DOI] [PubMed] [Google Scholar]
- Bartlett, K., and S. Eaton, 2004. Mitochondrial beta-oxidation. Eur. J. Biochem. 271 462–469. [DOI] [PubMed] [Google Scholar]
- Bertrand, H., Q. Wu and B. L. Seidel-Rogol, 1993. Hyperactive recombination in the mitochondrial DNA of the natural death nuclear mutant of Neurospora crassa. Mol. Cell. Biol. 13 6778–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet, L., G. Karpati and E. A. Shoubridge, 1992. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 51 1187–1200. [PMC free article] [PubMed] [Google Scholar]
- Cahan, P., and J. C. Kennell, 2005. Identification and distribution of sequences having similarity to mitochondrial plasmids in mitochondrial genomes of filamentous fungi. Mol. Genet. Genomics 273 462–473. [DOI] [PubMed] [Google Scholar]
- Carroll, A. M., J. A. Sweigard and B. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41 22. [Google Scholar]
- Cerveny, K. L., Y. Tamura, Z. Zhang, R. E. Jensen and H. Sesaki, 2007. Regulation of mitochondrial fusion and division. Trends Cell Biol. 17 563–569. [DOI] [PubMed] [Google Scholar]
- Chae, M. S., and F. E. Nargang, 2009. Investigation of regulatory factors required for alternative oxidase production in Neurospora crassa. Physiol. Plant 137 407–418. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A., E. Mambo and D. Sidransky, 2006. Mitochondrial DNA mutations in human cancer. Oncogene 25 4663–4674. [DOI] [PubMed] [Google Scholar]
- Chomyn, A., A. Martinuzzi, M. Yoneda, A. Daga, O. Hurko et al., 1992. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl. Acad. Sci. USA 89 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, M. M., G. P. Leboucher, N. Livnat-Levanon, M. H. Glickman and A. M. Weissman, 2008. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 19 2457–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski, M., T. Horton, M. T. Lott, J. M. Shoffner, M. F. Beal et al., 1992. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 2 324–329. [DOI] [PubMed] [Google Scholar]
- Cortopassi, G. A., and N. Arnheim, 1990. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 18 6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi, G. A., D. Shibata, N. W. Soong and N. Arnheim, 1992. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl. Acad. Sci. USA 89 7370–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court, D. A., A. J. Griffiths, S. R. Kraus, P. J. Russell and H. Bertrand, 1991. A new senescence-inducing mitochondrial linear plasmid in field-isolated Neurospora crassa strains from India. Curr. Genet. 19 129–137. [DOI] [PubMed] [Google Scholar]
- d'Alencon, E., A. Taghbalout, C. Bristow, R. Kern, R. Aflalo et al., 2003. Isolation of a new hemimethylated DNA binding protein which regulates dnaA gene expression. J. Bacteriol. 185 2967–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, A. D., H. Bertrand and R. Maheshwari, 2005. Intramolecular recombination and deletions in mitochondrial DNA of senescent, a nuclear-gene mutant of Neurospora crassa exhibiting “death” phenotype. Fungal Genet. Biol. 42 178–190. [DOI] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. De Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Meth. Enzymol. 17A 79–143. [Google Scholar]
- Dawson, T. M., 2006. Parkin and defective ubiquitination in Parkinson's disease. J. Neural Transm. 70(Suppl.): 209–213. [DOI] [PubMed] [Google Scholar]
- de Serres, F. J., 1980. Mutagenesis at the ad-3A and ad-3B loci in haploid UV-sensitive strains of Neurospora crassa. II. Comparison of dose-response curves for inactivation and mutation induced by UV. Mutat. Res. 71 181–191. [DOI] [PubMed] [Google Scholar]
- de Vries, H., B. Alzner-DeWeerd, C. A. Breitenberger, D. D. Chang, J. C. de Jonge et al., 1986. The E35 stopper mutant of Neurospora crassa: precise localization of deletion endpoints in mitochondrial DNA and evidence that the deleted DNA codes for a subunit of NADH dehydrogenase. EMBO J. 5 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, H., M. W. Dodson, H. Huang and M. Guo, 2008. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. USA 105 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson, M. W., and M. Guo, 2007. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr. Opin. Neurobiol. 17 331–337. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques, M., B. Westermann and T. Langer, 2006. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 173 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel, M. L., Y. R. Seo, M. L. Smith and M. R. Kelley, 2003. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 63 608–615. [PubMed] [Google Scholar]
- Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker and N. D. Read, 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. [DOI] [PubMed] [Google Scholar]
- Fritz, S., N. Weinbach and B. Westermann, 2003. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell. 14 2303–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, H., and C. T. Moraes, 2009. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18 1028–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Gerber, J., and R. Lill, 2002. Biogenesis of iron-sulfur proteins in eukaryotes: components, mechanism and pathology. Mitochondrion 2 71–86. [DOI] [PubMed] [Google Scholar]
- Good, A. G., and W. L. Crosby, 1989. Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 90 1305–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D. R., 2005. Apoptotic pathways: ten minutes to dead. Cell 121 671–674. [DOI] [PubMed] [Google Scholar]
- Gunter, T. E., D. I. Yule, K. K. Gunter, R. A. Eliseev and J. D. Salter, 2004. Calcium and mitochondria. FEBS Lett. 567 96–102. [DOI] [PubMed] [Google Scholar]
- Handa, N., Y. Noguchi, Y. Sakuraba, P. Ballario, G. Macino et al., 2000. Characterization of the Neurospora crassa mus-25 mutant: the gene encodes a protein which is homologous to the Saccharomyces cerevisiae Rad54 protein. Mol. Gen. Genet. 264 154–163. [DOI] [PubMed] [Google Scholar]
- Hanway, D., J. K. Chin, G. Xia, G. Oshiro, E. A. Winzeler et al., 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA 99 10605–10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner, G., K. A. Nummy, S. Stoltzner, S. K. Hubert and H. Bertrand, 2006. Biogenesis and replication of small plasmid-like derivatives of the mitochondrial DNA in Neurospora crassa. Fungal Genet. Biol. 43 75–89. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., 2007. Regulation of mitochondrial DNA content and cancer. Mitochondrion 7 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, M. S., C. Ou, Y. R. Chan, C. T. Chien and H. Pi, 2008. The utility F-box for protein destruction. Cell. Mol. Life Sci. 65 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins, S., and J. Nunnari, 2009. The molecular mechanism of mitochondrial fusion. Biochim. Biophys. Acta 1793 20–26. [DOI] [PubMed] [Google Scholar]
- Hoppins, S., L. Lackner and J. Nunnari, 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76 751–780. [DOI] [PubMed] [Google Scholar]
- Jackson, P. K., and A. G. Eldridge, 2002. The SCF ubiquitin ligase: an extended look. Mol. Cell 9 923–925. [DOI] [PubMed] [Google Scholar]
- Kafer, E., 1981. Mutagen sensitivities and mutator effects of MMS-sensitive mutants in Neurospora. Mutat. Res. 80 43–64. [DOI] [PubMed] [Google Scholar]
- Kafer, E., 1983. Epistatic grouping of repair-deficient mutants in Neurospora: comparative analysis of two uvs-3 alleles, uvs-6 and their mus double mutant strains. Genetics 105 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer, E., and E. Perlmutter, 1980. Isolation and genetic analysis of MMS-sensitive mus mutants of neurospora. Can. J. Genet. Cytol. 22 535–552. [DOI] [PubMed] [Google Scholar]
- Kelkar, H. S., J. Griffith, M. E. Case, S. F. Covert, R. D. Hall et al., 2001. The Neurospora crassa genome: cosmid libraries sorted by chromosome. Genetics 157 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott, A. B., and E. Bossy-Wetzel, 2008. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann. N Y Acad. Sci. 1147 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe, G. O., M. Kitamura, M. Masutani, E. U. Selker and H. Inoue, 2010. PARP is involved in replicative aging in Neurospora crassa. Fungal Genet. Biol. 47 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, K. J., A. K. Reeve and D. M. Turnbull, 2007. Do mitochondrial DNA mutations have a role in neurodegenerative disease? Biochem. Soc. Trans. 35 1232–1235. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A. M., 1979. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 59 421–433. [DOI] [PubMed] [Google Scholar]
- Lill, R., and U. Muhlenhoff, 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30 133–141. [DOI] [PubMed] [Google Scholar]
- Maheshwari, R., and A. Navaraj, 2008. Senescence in fungi: the view from Neurospora. FEMS Microbiol. Lett. 280 135–143. [DOI] [PubMed] [Google Scholar]
- Myers, C. J., A. J. Griffiths and H. Bertrand, 1989. Linear kalilo DNA is a Neurospora mitochondrial plasmid that integrates into the mitochondrial DNA. Mol. Gen. Genet. 220 113–120. [DOI] [PubMed] [Google Scholar]
- Nargang, F. E., J. B. Bell, L. L. Stohl and A. M. Lambowitz, 1983. A family of repetitive palindromic sequences found in Neurospora mitochondrial DNA is also found in a mitochondrial plasmid DNA. J. Biol. Chem. 258 4257–4260. [PubMed] [Google Scholar]
- Navaraj, A., A. Pandit and R. Maheshwari, 2000. Senescent: a new Neurospora crassa nuclear gene mutant derived from nature exhibits mitochondrial abnormalities and a “death” phenotype. Fungal Genet. Biol. 29 165–173. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D., and D. R. Galeazzi, 1978. A meiotic UV-sensitive mutant that causes deletion of duplications in Neurospora. Genetics 89 245–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer, D., A. L. Schroeder and D. R. Galeazzi, 1978. An apparent connection between histidine, recombination, and repair in Neurospora. Genetics 89 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niagro, F. D., and N. C. Mishra, 1989. An ethidium bromide induced mutant of Neurospora crassa defective in mitochondrial DNA. Curr. Genet. 16 303–305. [DOI] [PubMed] [Google Scholar]
- Niagro, F. D., and N. C. Mishra, 1990. Biochemical, genetic and ultrastructural defects in a mitochondrial mutant (ER-3) of Neurospora crassa with senescence phenotype. Mech. Ageing Dev. 55 15–37. [DOI] [PubMed] [Google Scholar]
- Ninomiya, Y., K. Suzuki, C. Ishii and H. Inoue, 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach, M. J., 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150 159–162. [DOI] [PubMed] [Google Scholar]
- Perkins, D. D., J. A. Kinsey, D. K. Asch and G. D. Frederick, 1993. Chromosome rearrangements recovered following transformation of Neurospora crassa. Genetics 134 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, A. C., R. E. Thomas, L. A. Andrews, H. M. McBride, A. J. Whitworth et al., 2008. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. USA 105 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, A. K., K. J. Krishnan and D. Turnbull, 2008. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann. N Y Acad. Sci. 1147 21–29. [DOI] [PubMed] [Google Scholar]
- Rimessi, A., C. Giorgi, P. Pinton and R. Rizzuto, 2008. The versatility of mitochondrial calcium signals: from stimulation of cell metabolism to induction of cell death. Biochim. Biophys. Acta 1777 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, T. A., and W. H. Tong, 2005. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell. Biol. 6 345–351. [DOI] [PubMed] [Google Scholar]
- Rowley, N., C. Prip-Buus, B. Westermann, C. Brown, E. Schwarz et al., 1994. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77 249–259. [DOI] [PubMed] [Google Scholar]
- Ryan, F. J., G. W. Beadle and E. L. Tatum, 1943. The tube method of measuring the growth rate of Neurospora. Am. J. Bot. 30 784–799. [Google Scholar]
- Sakuraba, Y., A. L. Schroeder, C. Ishii and H. Inoue, 2000. A Neurospora double-strand-break repair gene, mus-11, encodes a RAD52 homologue and is inducible by mutagens. Mol. Gen. Genet. 264 392–401. [DOI] [PubMed] [Google Scholar]
- Schroeder, A. L., 1970. Ultraviolet-sensitive mutants of Neurospora. I. Genetic basis and effect on recombination. Mol. Gen. Genet. 107 291–304. [DOI] [PubMed] [Google Scholar]
- Schroeder, A. L., 1986. Chromosome instability in mutagen sensitive mutants of Neurospora. Curr. Genet. 10 381–387. [DOI] [PubMed] [Google Scholar]
- Sciacco, M., E. Bonilla, E. A. Schon, S. DiMauro and C. T. Moraes, 1994. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum. Mol. Genet. 3 13–19. [DOI] [PubMed] [Google Scholar]
- Seidel-Rogol, B. L., J. King and H. Bertrand, 1989. Unstable mitochondrial DNA in natural-death nuclear mutants of Neurospora crassa. Mol. Cell. Biol. 9 4259–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, T. C., 1951. A gene that causes natural death in Neurospora crassa. Genetics 36 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, S., X. Chen, S. DiMauro and E. A. Schon, 1992. Accumulation of deletions in human mitochondrial DNA during normal aging: analysis by quantitative PCR. Biochim. Biophys. Acta 1180 113–122. [DOI] [PubMed] [Google Scholar]
- Sizemore, S. T., and J. V. Paietta, 2002. Cloning and characterization of scon-3+, a new member of the Neurospora crassa sulfur regulatory system. Eukaryot. Cell 1 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru, H., and H. Inoue, 1989. Isolation and characterization of a laccase-derepressed mutant of Neurospora crassa. J. Bacteriol. 171 6288–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Y. Sakuraba and H. Inoue, 1997. Genetic and molecular characterization of Neurospora crassa mus-23: a gene involved in recombinational repair. Mol. Gen. Genet. 256 436–445. [DOI] [PubMed] [Google Scholar]
- Willems, A. R., M. Schwab and M. Tyers, 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695 133–170. [DOI] [PubMed] [Google Scholar]
- Xu, G., and Y. Shi, 2007. Apoptosis signaling pathways and lymphocyte homeostasis. Cell Res. 17 759–771. [DOI] [PubMed] [Google Scholar]
- Yin, S., J. Heckman and U. L. RajBhandary, 1981. Highly conserved GC-rich palindromic DNA sequences flank tRNA genes in Neurospora crassa mitochondria. Cell 26 325–332. [DOI] [PubMed] [Google Scholar]
- Zheng, L., V. L. Cash, D. H. Flint and D. R. Dean, 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273 13264–13272. [DOI] [PubMed] [Google Scholar]