Abstract
Transposons of the Mutator (Mu) superfamily have been shown to play a critical role in the evolution of plant genomes. However, the identification of Mutator transposons in other eukaryotes has been quite limited. Here we describe a previously uncharacterized group of DNA transposons designated Phantom identified in the genomes of a wide range of eukaryotic taxa, including many animals, and provide evidence for its inclusion within the Mutator superfamily. Interestingly three Phantom proteins were also identified in two insect viruses and phylogenetic analysis suggests horizontal movement from insect to virus, providing a new line of evidence for the role of viruses in the horizontal transfer of DNA transposons in animals. Many of the Phantom transposases are predicted to harbor a FLYWCH domain in the amino terminus, which displays a WRKY–GCM1 fold characteristic of the DNA binding domain (DBD) of Mutator transposases and of several transcription factors. While some Phantom elements have terminal inverted repeats similar in length and structure to Mutator elements, some display subterminal inverted repeats (sub-TIRs) and others have more complex termini reminiscent of so-called Foldback (FB) transposons. The structural plasticity of Phantom and the distant relationship of its encoded protein to known transposases may have impeded the discovery of this group of transposons and it suggests that structure in itself is not a reliable character for transposon classification.
TRANSPOSABLE elements (TEs) are mobile pieces of parasitic DNA that can replicate and move around in the host genome and are classified on the basis of their transposition intermediate (Craig et al. 2002). Class 1 transposable elements are mobilized via an RNA intermediate while class 2, or DNA transposons, mobilize via a DNA intermediate. TEs can be found in bacteria, archaea, and eukaryotes. Indeed, members of some superfamilies of cut and paste DNA transposons are common to all three domains of life, suggesting either their existence prior to the diversification of the three domains from a common ancestor or frequent interdomain horizontal transfer (HT) (Feschotte and Pritham 2007). Cut and paste transposons display a relatively simple structure where autonomous copies carry a single transposase gene flanked on either side by transposase binding sites (often the binding sites are embedded in terminal inverted repeats, TIRs) (Craig et al. 2002; Feschotte and Pritham 2007). Nonautonomous copies typically do not carry any transposase gene but instead carry just the binding sites and thus have the ability to move in trans utilizing the transposase encoded by an autonomous copy located elsewhere in the genome (Craig et al. 2002; Feschotte and Pritham 2007).
One class 2 TE superfamily, Mutator (Mu)/IS256, is common in bacteria, archaea, and plants but has been described in few other eukaryotes (Talbert and Chandler 1988; Lisch 2002; Chalvet et al. 2003; Xu et al. 2004; Pritham et al. 2005; Hua-Van and Capy 2008). The Mu system was first described by Donald Robertson in a line of Zea mays that exhibited increased mutation rates (50- to 100-fold) as compared to wild-type stocks (Walbot and Rudenko 2002). It was later discovered that these mutant stocks contained a 1.5-kb DNA insertion in the first intron of the Adh-1 gene, causing changes in gene expression (Strommer 1982; Bennetzen et al. 1984). This insertion was sequenced and identified as a cut and paste DNA transposon called, Mu1, with TIRs and 9-bp target site duplications (TSDs) (Barker et al. 1984). Mu elements identified in maize have conserved TIRs ∼220 bp in length, induce an 8- to 9-bp TSD, and have variable internal sequences (Lisch 2002). Mutator TEs have been described as the most mutagenic plant transposon (Lisch 2002). Fairly early on, a relationship was noted between the maize Mutator transposase with the transposase encoded by IS256 prokaryotic mobile elements (Byrne et al. 1989, 1990; Eisen et al. 1994). In addition, both Mutator TEs and IS256 insertions were shown to be flanked by 8- to 10-bp TSDs, which likely reflected a conserved feature of transposase function and further supported a common origin (Eisen et al. 1994).
Most of the work on Mutator has been done in plants where they have been shown to have a variety of impacts on the evolution of the genomes they invade. They not only cause an increase in mutation rates and changes in gene expression (Lisch 2002; Walbot and Rudenko 2002), but they have also been known to shuffle genomic sequences, including genes, in rice, Arabidopsis thaliana, and Lotus japonicus (Jiang et al. 2004; Hoen et al. 2006; Holligan et al. 2006; van Leeuwen et al. 2007). The transposons that have picked up gene fragments are called Pack-MULEs and typically do not carry a transposase (Jiang et al. 2004). Mutator-like (MULE) transposases have also been noted for their propensity to become domesticated by the genome and to have given rise to several key genes involved in light sensing in plants (Hudson et al. 2003; Cowan et al. 2005; Babu et al. 2006; Lin et al. 2007; Saccaro et al. 2007). In addition, Mutator elements have been reported to move via horizontal transfer between grass species (Diao et al. 2006).
MULEs have been described in various grasses (Yoshida et al. 1998; Mao et al. 2000; Lisch et al. 2001; Saccaro et al. 2007), in A. thaliana (Yu et al. 2000; Singer et al. 2001), and other eudicot plants (Holligan et al. 2006; van Leeuwen et al. 2007), and in two fungi (Fusarium oxysporum (Chalvet et al. 2003) and Yarrowia lipolytica (Neuveglise et al. 2005), the parabasalid, Trichomonas vaginalis (Lopes et al. 2009), and Entamoeba (Pritham et al. 2005). In addition to classic Mutator TEs, sequences distantly related to Mutators, but not complete transposable elements were described from Entamoeba invadens and E. moshkovskii (Pritham et al. 2005). The putative transposases from Entamoeba display only weak sequence identity to the pfam00872 Mutator transposase and were called Phantom (Pritham et al. 2005).
Here we present a comprehensive computational analysis of Phantom sequences and their distribution across the eukaryotic tree of life. Detailed structural analysis of these sequences reveals that they are bonafide class 2 TEs, which share structural and coding characteristics with Mutator TEs. The taxonomic distribution of Phantom illustrates that these elements are widespread in animals, found in a few distantly related protists and two insect viruses.
MATERIALS AND METHODS
Data mining and identification of elements:
Candidate Phantom elements were identified using the E. invadens Phantom translated ORF [accession no. AANW02000107.1, coordinates 4912–7645 (complete element) and 5257–7012 (ORFs)] as a query in TblastN searches at National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov), beginning January 2007 until the final preparation and submission of this article. Additional blast searches were conducted (primarily BlastN and TBlastN) using default parameters and without filtering for simple and complex repeats to identify copies of Phantom in different taxa. Searches were conducted against various GenBank databases including whole genome shotgun reads (WGS), nucleotide collection (NR), high throughput genomic sequences (HTGS), genome survey sequences (GSS), and expressed sequence tags (EST) databases. Accession numbers, reading frame, and nucleotide coordinates of all significant hits were annotated for further evaluation. A hit was considered significant when the e-value was <10−4. TIRs were identified by pairwise comparisons taking 3000 bp upstream and downstream of each contig using Blast. TSDs were identified by aligning 100 bp upstream and downstream from the TIRs of the elements. To maximize the probability of identifying all probable Phantom elements, newly identified elements and putative proteins were used as queries using Blast against the WGS and NR database. Autonomous Phantom elements were used to identify related nonautonomous elements. The nonautonomous elements share the same TIRs but do not contain a transposase gene. Majority rule consensus sequences for Phantom were generated by constructing majority rule alignments using Clustal (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and MacVector 7.2.2. TE copy numbers for each genome were estimated on the basis of the results of TBlastN and BlastN using consensus sequences to search against the WGS and NR databases in GenBank. Hits with e-values lower than 10−4 were considered significant. Consensus sequences for all multi-element families can be found in supporting information, File S1.
Identification of open reading frames and conserved domains:
Both the Translate (www.expasy.org/tools/dna.html) and ORF Finder tools (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) were utilized to identify open reading frames (ORFs) within Phantom elements through conceptual translations. When necessary, frameshifts were judiciously introduced according to nucleotide alignments of closely related sequences. The function of hypothetical Phantom proteins was predicted by homology to proteins of known function and by the presence of conserved domains identified through a conserved domain database (CDD) search (Marchler-Bauer et al. 2009).
Identification of paralogous “empty” sites:
To illustrate the mobility of Phantom elements paralogous sites (empty sites) not containing a Phantom insertion were analyzed. Empty sites were identified by homology searches utilizing BlastN (word size 7, expectancy 1000) with a query constructed from the sequences directly flanking the insertion site containing the unduplicated target site. The chimeric query sequence (∼100 bp in length) was created by extracting the flanking sequence (∼50 bp) upstream from the element insertion containing the target site duplication and extracting the flanking sequence (∼50 bp) downstream from the element insertion (lacking the element and target site duplication). Paralogous empty sites are defined as duplicated regions in the host genome homologous to the region where a Phantom insertion is found but that lacks the Phantom insertion.
Alignments and phylogenetic analysis:
Alignments of Phantom, Jittery, Hop, MuDR, and IS256/6120 putative catalytic core domains (∼200 amino acids) were constructed using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and MUSCLE (http://www.ebi.ac.uk/Tools/muscle/) using default parameters and visually refined using GeneDoc v3. Phylogenetic trees were created with Mr. Bayes 3.14 (Ronquist and Huelsenbeck 2003) using the amino acid model with a discrete γ-distribution with four rate categories and random starting trees. Two independent runs with four Markov chains each operating for one million generations with a sampling frequency set to 100 were utilized. Convergence was considered for the two runs when the standard deviation split frequencies was <0.001. The temperature difference between the “cold” chain and the “heated” chain was set to default parameters (temp = 0.2) to improve the chain swap. The first 200 trees recovered in these searches were discarded as burn, on the basis of stabilization of likelihood scores.
RESULTS
Phantom proteins from Entamoeba are encoded by bonafide transposable elements:
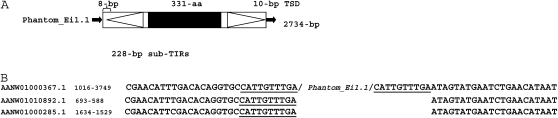
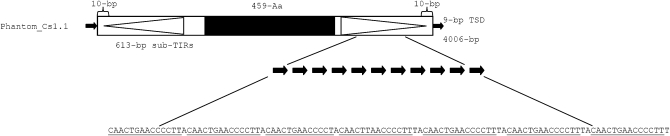
Sequences encoding putative proteins with weak similarity to Mutator transposase and designated Phantom have been previously identified in the genomes of E. invadens and E. moshkovskii (Pritham et al. 2005). However, whether these proteins were part of bonafide TEs and represent a new lineage of the Mutator TEs had not been further investigated. To characterize these putative TEs, the Entamoeba transposases were used as queries in TBlastN searches against the E. invadens genome shotgun sequence. This search yielded 49 significant hits. These contigs were subjected to pairwise alignment to reveal the boundaries of sequence identity and evaluated for the presence of structural features typical of DNA transposons, such as potential TIRs and TSD. Discrete units of ∼2730 bp in length containing a single uninterrupted ORF (330–400 AA), bracketed by long (194–228 bp) and imperfect (82–100%) subterminal inverted repeats (sub-TIRs) flanked by a 7- to 12-bp TSD (Figure 1A) were identified. The proper boundaries of the TE were confirmed by the identification of paralogous (empty) sites containing the unduplicated target site (Figure 1B). These findings allowed us to show that Entamoeba Phantom proteins were carried by bonafide TEs, which display the structural features typical of DNA transposons (Figure 1). Together these data reveal that Entamoeba species harbor a novel lineage of DNA transposons distantly related to previously described TEs of the Mutator superfamily, in addition to an entirely distinct grouping of Mutators TEs previously described and called EMULEs (Pritham et al. 2005).
Figure 1.—
Phantom transposable elements in Entamoeba invadens. (A) Schematic representation of a complete Phantom element in E. invadens. Small solid arrows represent target site duplications, subterminal inverted repeats are represented by the large open arrows, solid box represents the open reading frame (ORF). (B) Target site duplications (TSD) created upon Phantom_Ei insertions. TSDs are underlined.
Identification of Phantom in other taxa:
To identify other related TEs, queries representing the E. invadens Phantom-translated ORF were used in TBlastN searches against all the species with sequences deposited in the GenBank databases. Related ORFs were identified in 31 different species from various eukaryotic taxa including the planarian Schmidtea mediterranea, the annelid Helobdella robusta, a wide variety of nematodes and insects, the sea urchin Strongylocentrotus purpuratus, the ascidians Ciona intestinalis and C. savigny, several mammals (but not other vertebrates), one species of Candida yeast, three species of Phytophthora (oomycetes), and the Trichomonad T. vaginalis (Table 1). Much to our surprise Phantom-like transposases were also identified in two insect viruses, Chelonus bracovirus and Glypta fumiferanae ichnovirus.
TABLE 1.
Distribution of the Phantom clade of the Mutator superfamily
| Taxa | Abb. | Accession | DB | CD | CC | FLCC |
|---|---|---|---|---|---|---|
| Archamoebae | ||||||
| Entamoeba dispara | Ed | AANV01001934.1 | WGS | MULE | ||
| E. invadensb | Ei | AANW01000293.1 | WGS | 200 | 2 | |
| Cnidarians (Hydrozoa) | ||||||
| Hydra magnipapillatac | Hm | XM002166453 | NR | 60 | ||
| Planarians | ||||||
| Schmidtea mediterraneab | Sm | AAWT01029681.1 | WGS | FLYWCH | 600 | 8 |
| Annelids | ||||||
| Helobdella robustab | Hr | AC171129.2 | NR | FLYWCH | 1 | 1 |
| Mollusca | ||||||
| Aplysia californicac | Ac | AASC01110148.1 | WGS | MULE | 30 | 1 |
| Nematodes | ||||||
| Caenorhabditis briggsaec | Cb | XM_001664648.1 | NR | 20 | ||
| C. elegansc | Ce | U37429.1 | NR | MULE | 30 | 7 |
| Heterodera glycinesa | Hg | CB934986.1 | EST | |||
| Meloidogyne chitwoodia | Mc | CB830714.1 | EST | |||
| M. haplac | Mh | ABLG01001649.1 | WGS | 17 | ||
| M. incognitac | Mi | CZ172697.1 | GSS | 20 | ||
| Trichinella spiralisb | Ts | AC188123.1 | NR | MULE | 700 | 2 |
| Insects | ||||||
| Acyrthosiphon pisumc | Ap | AC202214.3 | NR | MULE | 900 | |
| Aedes aegypti strainb | Aa | AAGE02008886.1 | WGS | >1000 | 3 | |
| Apis melliferaa | Am | AADG05002861.1 | WGS | |||
| Chironomus tentansa | Ct | CAC37683.1 | NR | MULE | ||
| C. pallidivittatusa | Chp | CAC37681.1 | NR | MULE | ||
| Culex pipiens quinquefasciatusb | Cp | AAWU01011212.1 | WGS | MULE | >1000 | 1 |
| Drosophila ananassaeb | Da | AAPP01015916.1 | WGS | MULE | >1000 | 1 |
| D. yakubac | Dy | AAEU02002585.1 | WGS | MULE | 7 | |
| Ixodes scapularisb | Is | ABJB010984717.1 | WGS | 300 | 1 | |
| Nasonia vitripennisb | Nv | AAZX01008412.1 | WGS | FLYWCH | 17 | 1 |
| Tribolium castaneumb | Tc | AAJJ01003811.1 | WGS | FLYWCH | >1000 | 3 |
| Echinoida | ||||||
| Strongylocentrotus purpuratusb | Sp | AAGJ02149146.1 | WGS | 40 | 1 | |
| Ascidians (Urochordata) | ||||||
| Ciona intestinalisb | Ci | AABS01001273 | NR | FLYWCH | 7 | 2 |
| Ciona savignyib | Cs | AACT01008187.1 | NR | FLYWCH | >1000 | 1 |
| Leptocardii (Cephalochordata) | ||||||
| Branchiostoma floridaec | Bf | DE195457.1 | GSS | MULE | 44 | |
| Vertebrata (Chordata) | ||||||
| Homo sapiensb | Hs | AADC01162133.1 | WGS | 1 | ||
| Pan troglodytesc | Pt | AC200913.3 | NR | >1000 | ||
| Canis familiarisa | Cf | AACN010093066 | NR | |||
| Equus caballusb | Ec | AAWR01022474 | WGS | FLYWCH | 300 | 1 |
| Monodelphis domesticaa | Md | AAFR03063600.1 | WGS | |||
| Rattus norvegicusa | Rn | AAHX01085823.1 | WGS | |||
| Fungi (Ascomycetes) | ||||||
| Candida glabrataa | Cg | CR380951.1 | NR | |||
| Stramenopiles (Oomycetes) | ||||||
| Phytophthora infestansb | Pi | AATU01012134.1 | WGS | MULE | 240 | 8 |
| P. sojaeb | Ps | AAQY01002515.1 | WGS | MULE | 100 | 13 |
| P. ramorumb | Pr | AAQX01003219.1 | WGS | MULE | 30 | 3 |
| Parabasalids (Trichomonads) | ||||||
| Trichomonas vaginalisb | Tv | NW_001580983.1 | WGS | MULE | 780 | 9 |
| Viruses (dsDNA) | ||||||
| Chelonus inanitus bracovirusc | Cib | CAC82100.3 | NR | MULE | ||
| Glypta fumiferanae ichnovirusb | Gfi | AB289994.1 | NR | MULE | 1 | 1 |
| Yeasts | ||||||
| Kluyveromyces lactisa | Kl | CR382123.1 | NR | |||
| Yarrowia lipolyticaa | Yl | CR382128.1 | NR | |||
Abb., species abbreviation; accession, accession number for one representative hit; DB, database where the hit is deposited (GEN, genomic sequences; EST, expressed sequence tag; WGS, whole-genome sequencing; GSS, genomic survey sequence; NR, nucleotide sequences); CD, conserved domain; CC, copy number (tBlastn and Blastn > e−04); FLCC, full-length copy number.
Significant hit to query (e = 10−4).
Full-length copy number of Phantom.
Protein only.
Determining the coding potential and structural features of these novel TEs:
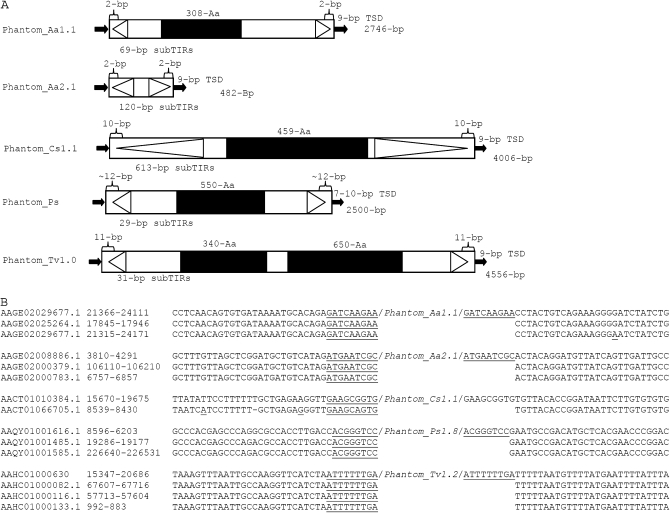
To determine the structural characteristics and identify TIRs and TSD common to Phantom TEs pairwise comparisons of upstream and downstream flanking sequences (up to 3000 bp if available) were carried out (Figure 2A). To ensure that complete Phantom elements were properly delineated, searches for paralogous (empty) sites in the genome were performed (Figure 2B). Majority rule consensus sequences were constructed for each family of elements identified. These analyses allowed the identification of 77 complete Phantom TEs in 21 different species from various eukaryotic taxa representing 38 families (Table 1). The full-length copy numbers of Phantom in these taxa are generally low (one to three full-length copies) except for Phantom elements in S. mediterranea (eight full-length copies, 600 total copies), T. vaginalis (nine full-length copies, 780 total copies) and 2 Phytophthora species (approximately eight full-length copies, >300 total copies). Interestingly, some of the elements identified in P. infestans and P. sojae share high sequence identity (>99%) as compared to the consensus sequence, suggesting that these TEs have been recently active. Together these analyses expand the distribution of TEs related to Phantom to three of the five eukaryotic supergroups (as described by Keeling et al. 2005) including the Excavates and Chromalveolates, as well as the Unikonts.
Figure 2.—
Schematic representation of Phantom elements. TSDs are represented by solid arrows, open arrows represent sub-TIRs, and ORFs are represented by solid boxes. (B) TSDs created upon Pantom_Aa, Phantom_Cs, Phantom_Ps, and Phantom_Tv insertions. TSDs are underlined. Aa, Aedes aegypti; Cs, Ciona savigny; Ps, Phytophthora sojae; and Tv, Trichomonas vaginalis.
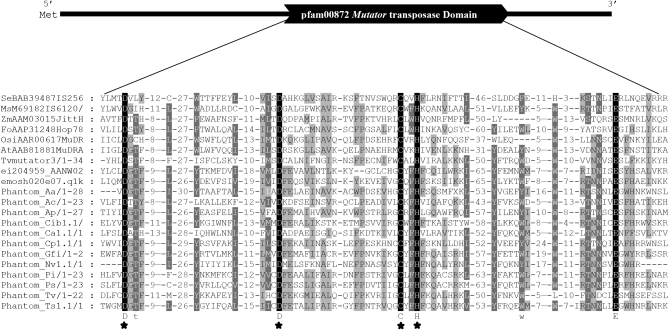
Conceptual translations were used to identify ORFs that were annotated on the basis of homology to known proteins and domains present in the NCBI protein and CDD. These analyses indicate that Phantom TEs generally encode a single putative protein (300–700 AA) with multiple conserved domains (Marchler-Bauer et al. 2009). A pfam00872 MULE transposase domain (e-values range 10−8–10−1) was readily identifiable in several of the translated Phantom ORFs (Table 1, Figure 3) therefore we refer to this putative protein as the transposase.
Figure 3.—
MUSCLE alignment of the conserved domain found in transposases from the IS256/Mutator superfamily corresponding to the pfam00872. Solid residues have related physical or chemical properties and occurred in every sequence. Shaded residues occur in some of the sequences. The star symbol below the residues D, D, C, H refers to conserved residues previously described by Hua-Van and Capy (2008) and Lisch (2002). Aa, Aedes aegypti; Ac, Aplysia californica; Ap, Acyrothosiphon pisum; At, Arabidopsis thaliana; Ce, Caenorhabditis briggsae; Cib, Chelonus inanitus bracovirus; Ca, Candida albicans; Cp, Culex pipiens quinquefasciatus; Ei, Entamoeba invadens; Em, Entamoeba moshkovskii; Gfi, Glypta funiferanae ichnovirus; Ms, Mycobacterium smegmatis; Nv, Nasonia vitripennis; Os, Oryza sativa; Pi, Phytophthora infestans; Ps, Phytophthora sojae; Se, Staphylococcus epidermis; Tv, Trichomonas vaginalis; Ts, Trichinella spiralis; Zm, Zea mays; Se, IS256; and Ms, Pfam0082.
Further analyses using multiple sequence alignments of Phantom proteins and other previously identified Mutators and bacterial and archaeal IS sequences revealed a region of ∼200 aa in Phantom that is homologous to the previously identified Mutators including MULEs, Jittery, Hop, and some bacterial and archaeal IS sequences (Robertson 1978; Barker et al. 1984; Yu et al. 2000; Chalvet et al. 2003; Xu et al. 2004; Pritham et al. 2005), presumably the catalytic core of the transposase. Multiple sequence alignments containing Phantom, Jittery, Hop, MuDR, and related IS256 sequences were constructed using MUSCLE. The alignments were edited by removing regions of low sequence conservation, which resulted in an ∼200-aa conserved region located in the C terminus of the proteins. This region is marked by the DDCHE motif (Figure 4). This region was previously identified in Mutator elements including IS256, the MURA protein in MuDRs and TvMULEs (Lisch 2002; Hua-Van and Capy 2008; Lopes et al. 2009).
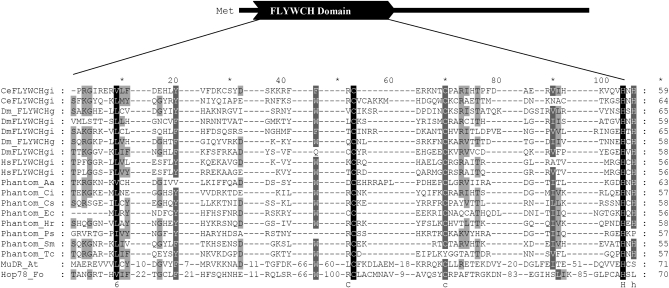
Figure 4.—
MUSCLE alignment of the conserved FLYWCH domain found in Phantoms, MuDR, and Hop78. Solid residues have related physical or chemical properties and occurred in every sequence. Shaded residues occur in some of the sequences. Aa, Aedes aegypti; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Cs, Ciona savigny; Dm, Drosophila melanogaster; Ec, Equus caballus; Fo, Fusarium oxysporum; Hr, Helobdella robusta; Hs, Homo sapiens; Ps, Phytophthora sojae; Sm, Schmidtea mediterranea; and Tc, Triboleum castaneum.
Mutator transposases frequently harbor an N-terminal DNA binding domain (DBD) and a C-terminal catalytic domain. To determine what kind of domains are detected in Phantom transposases, the proteins were used to query the CDD at NCBI (Marchler-Bauer et al. 2009). A Mutator TPASE domain (pfam00872) and/or a FLYWCH domain (pfam04500) was detected in many of the transposase proteins (Figure 4). Cellular proteins that harbor the FLYWCH domain are all involved in transcriptional regulation and have been identified in the genome of Drosophila melanogaster, Homo sapiens, and Caenorhabditis elegans (Babu et al. 2006). The presence of a FLYWCH domain in a Mutator transposase has not to our knowledge been previously reported. However, the FLYWCH domain bears a WRKY–GCM1 fold also found in the DNA binding domain of other MULE transposases (Babu et al. 2006). This suggests that DNA binding domains displaying a WRKY–GCM1 fold are an ancient component of all Mutator transposases and therefore, the presence in Phantom transposases supports Phantom as a bonafide member of the Mutator superfamily.
Phantom elements belong to three different structural variant groups:
Phantom elements range in size from 2 to 5 kb and with few exceptions belong to three different structural variant groups. The elements in the first group are characterized by TIRs that are between 200 and 800 bp in length and are reminiscent of those typically associated with the MuDR elements of the Mutator superfamily (Lisch 2002). The second and most prevalent group of elements has sub-TIRs, characterized by inverted repeats between 10 and 880 bp in length and located 2–15 bp downstream from the termini (Figure 2A). Sub-TIRs are not specific to Phantoms and have been previously described for Jittery (Xu et al. 2004) elements of the Mutator superfamily, as well as other elements belonging to different superfamilies, some examples including Microuli (Tu and Orphanidis 2001) and Microns (Akagi et al. 2001). The third and final group includes those elements with TIRs of variable length (60–624 bp) characterized by repeated internal units (9–16 bp in length) reminiscent of the TIRs that characterize Foldback-like (FB-like) elements (Bingham and Zachar 1989). Phantom elements with Foldback-like TIRs are found in Aedes aegypti, Ciona intestinalis, C. savigny, and Culex pipiens (Figure 5). Foldback-like TIRs have not been previously reported in other Mutator elements.
Figure 5.—
Schematic representation of Foldback-like TIRs occurring in some Phantom elements. TSDs are represented by solid arrows, open arrows represent sub-TIRs, and ORFs are represented by solid boxes. Tandemly repeated sequences within the sub-TIRs are represented by small solid arrows. Repeated sequence motifs are underlined.
Nonautonomous Phantoms were identified in the mosquitoes, A. aegypti and C. pipiens and D. ananassae and Tribolium castaneum. The nonautonomous elements identified in A. aegypti can be classified as MITEs as they have reached high copy numbers and are fairly homogenous in size. The elements identified in D. ananassae and T. castaneum have not reached high copy numbers and are likely old as they have incurred other insertions. These elements are flanked by 8- to 9-bp TSDs and their TIRs share a strong similarity to the autonomous Phantoms identified, indicating that these elements are likely moved in trans by the transposases encoded by autonomous Phantoms in these species.
Phantoms form a well-supported clade with Mutator/IS256 elements:
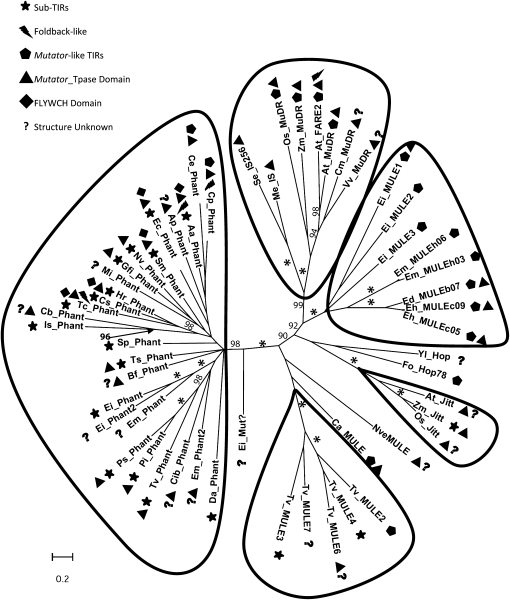
Mutator elements were previously described in plants, fungi, Entamoeba, and T. vaginalis (Taylor and Walbot 1987; Mao et al. 2000; Lisch et al. 2001; Lisch 2002; Chalvet et al. 2003; Xu et al. 2004; Pritham et al. 2005; Hua-Van and Capy 2008; Lopes et al. 2009). A phylogenetic analysis was generated from the (200 aa) catalytic domain alignment using a Bayesian method (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). The Bayesian tree revealed five distinct well-supported groups including Phantom, MuDR/IS256, Jittery, TvMULES, and EMULEs (Figure 6). The branching pattern supports Phantom in forming a unique group affiliated to the Mutator superfamily.
Figure 6.—
Comparative phylogenetic analysis of Phantom proteins and the conserved domains of TvMULEs, CaMULEs, MuDRs, Jittery, Hop, EMULEs, FARE2, and IS256 transposases. Bayesian tree constructed using Mr. Bayes v3.14 from an alignment portion of the Phantom transposase corresponding to the conserved DDCHE motif from representative species and rooted with IS256 proteins from bacteria and archaea. GenBank accession numbers for previously identified sequences in parenthesis. Aa, Aedes aegypti; Ap, Acyrthosiphon pisum; At, Arabidopsis thaliana; Bf, Branchiostoma floridae; Ca, Candida albicans; Cb, Caenorhabditis briggsae; Ce, Caenorhabditis elegans; Cib, Chelonus inanitus bracorvirus; Cm, Cucumis melo; Cp, Culex pipiens; Cs, Ciona savigny; Da, Drosophila ananassae; Ec, Equus caballus; Ed, Entamoeba dispar; Eh, Entamoeba histolytica; Ei, Entamoeba invadens; Emosh, Entamoeba moshkovskii; Fo, Fusarium oxysporum; Gfi, Glypta fumiferanae ichnovirus; Hr, Helobdella robusta; Is, Ixodes scapularis; Mi, Meloidogyne incognita; mosh, Entamoeba moshkovskii; Ms, Mycobacterium smegmatis; Nv, Nassonia vitripennis; Nve, Nematostella vectensis; Os, Oryza sativa; Pi, Phytophthora infestans; Ps, Phytopthora sojae; Se, Staphylococcus epidermidis; Sm, Schmidtea mediterranea; Sp, Strongylocentrotus purpuratus; Tc, Tribolium castaneum; Ts, Trichinella spiralis; Tv, Trichomonas vaginalis; Vv, Vitis vinifera; Yl, Yarrowia lipolytica; and Zm, Zea mays. TvMULEs used are the majority rule consensus sequences from the T. vaginalis genome. The corresponding accession numbers for MULEs, MuDRs, Jittery, Hop, FARE2, and IS transposases are as follows: Se_IS256: BAB394871; Me_IS: NP634554; Os_MuDR: NP918808; Zm_MuDR: M76978.1; At_FARE2: AAD15518; At_MuDR: NP178710; Cm_MuDR: RepBase:AY582736; Vv_MuDR: CAAP02002575.1; Ei_MULE1: 205732; Ei_MULE2: 206048; Em_MULE3: 206038; Em_MULEh06: 131h06; Em_MULEh03: 010h03; Ed_MULEb07: 98816116b07; Eh_MULEc09: 2841599c09; Yl_Hop: XP504344; Fo_HOP78: AAP31248; At_Jitt: AAD51282; Zm_Jitt: AF247646.1; Os_Jitt: XP475123; NveMULE: MuDR-2_Nv; Ca_MULE: AACQ01000039.1; and Ei_Mut: AANW02000228.1.
DISCUSSION
A novel group of DNA transposons found in many eukaryotic genomes:
A lineage of TEs identified in diverse eukaryotes with genome sequences present in the database were described and are called Phantom. Most families of Phantom elements display common features including a single putative transposase gene flanked by terminal or subterminal inverted repeats and a TSD variable in sequence usually 9 bp in length, but ranges from 7 to 12 bp, which is consistent with previously identified Mutators including Pack MULEs, CUMULEs, MULEs in A. thaliana, Hop, and IS256 sequences (Eisen et al. 1994; Yu et al. 2000; Chalvet et al. 2003; Jiang et al. 2004; van Leeuwen et al. 2007). The size, structure, and organization of Phantom elements and the TSD are consistent with the Mutator superfamily (Feschotte and Pritham 2007).
Structural features of Phantom elements:
Three structural variants (TIR, sub-TIR, and FB) typify Phantom elements. The first category (TIR) encompasses the elements, which display a structure typical of MuDR elements including inverted repeats (200–800 bp in length) located precisely on either flank of the element. The second category (sub-TIR) are quite untraditional in their structure in that the termini of the element are not part of the inverted repeat; instead, they are characterized by a nonrepetitive region of 2–15 bp flanking the inverted repeat. The noncanonical sub-TIR structure made it difficult to properly demarcate these elements. This structure was validated by the identification of paralagous empty sites, which illustrate mobility in the past. Elements in the third structural category (FB) display long complicated TIRs characterized by highly repeated subunits reminiscent of TEs previously classified as Foldback elements (Figure 5). Our phylogenetic analysis also revealed that FARE2, a TE from A. thaliana described as a Foldback element, forms a clade (pp = 98) with the transposases encoded by MuDR TEs from plants (Figure 6). Therefore, FARE2 can be considered as a Mutator and not a member of a distinct Foldback superfamily of TEs. No relationship between the structure of the TE and the domains present in the transposase could be detected.
The labiality in structural morphologies is not unique to Phantom elements; recent studies have shown that the FB element, Galileo, is a member of the P-element superfamily where the canonical families display short TIRs (Marzo et al. 2008). Since the TIRs typically contain the transposase binding sites required for the cleavage and integration of DNA transposons (Craig et al. 2002), we propose that structural variation might evolve to avoid recognition by the host. It is possible that the repetitive structure inherent to the TIRs may be detected by the host genome and become the target of silencing. For example, it has been shown that the TIRs of Mu elements in maize are methylated, resulting in transcriptional silencing (Lisch 2002). A TE without TIRs, might avoid transcriptional silencing and might successfully outcompete TEs with TIRs. Another strategy to outcompete TEs with a simple TIR structure might be to increase the number of transposase binding sites within the TIR. It has been proposed that the tandemly repeated sequences in FB TIRs increase the chances of transposition by increasing DNA binding sites for the transposases (Potter 1982). The co-occurrence of structural variability within the P-element superfamily suggests that the structure of DNA transposons and in particular the transposase binding sites may be under selective pressure to be flexible, perhaps in response to host genome defense and should not be relied upon for classification. Therefore, FB structure does not signify an alliance to a FB superfamily as has been previously reported.
Most of the Phantom elements have coding capacity for a protein that ranges in size between 350 and 700 aa. Many of the proteins contain a conserved DDCHE motif (Figure 3) and a pfam00872–MULE transposase domain, further suggesting an allegiance to the Mutator superfamily. Phylogenetic analysis based on an alignment of the transposase domain with Phantom and selected Mutator transposases reveals that Phantom forms a well-supported group separate from previously described Mutators. In addition, Phantom elements identified in A. aegypti, C. intestinalis, C. savigny, E. caballus, H. robusta, P. sojae, S. mediterranea, and T. castaneum contain a conserved FLYWCH DNA binding domain (Figure 4). FLYWCH is a DNA binding domain classified under the WRKY–GCM1 superfamily of DBDs (Babu et al. 2006). The WRKY–GCM1 DBDs are a common feature of some MULE and plant MuDR transposases (Babu et al. 2006).
Our results reveal that Phantom transposases harbor a FLYWCH domain that is a member of the WRKY–GCM1 superfamily. The cellular proteins that harbor the FLYWCH domain are limited to animals, while Phantom transposases have a broader phylogenetic distribution, suggesting that the transposases were the source of these DNA binding domains. This pattern is consistent with the hypothesis that MuDR and MULE transposases are the progenitor of the DNA binding domain, found in all WRKY–GCM1 transcription factor proteins (Babu et al. 2006).
Distribution of Phantom elements in eukaryotes:
TEs of the Mutator superfamily are widespread in plants (supergroup Plantae) but previously were reported in few other eukaryotes including Entamoeba and various fungi (supergroup Unikont) (Talbert and Chandler 1988; Lisch 2002; Chalvet et al. 2003; Xu et al. 2004; Pritham et al. 2005) and in the genome of T. vaginalis (supergroup Excavate) (Hua-Van and Capy 2008; Lopes et al. 2009). This study broadens the distribution of the Mutator superfamily by revealing the widespread occurrence in animal genomes including human, as well in the genomes of three Phytophthora species, which are part of the Chromalveolate supergroup. In addition, related transposases were also identified in two insect viruses, C. bracovirus and Gf. ichnovirus. It is interesting to point out that Phantom elements have a broader phylogenetic distribution than other lineages of the Mutator superfamily. This observation suggests that the Phantom lineage is the most ancient clade of the Mutator superfamily and that Hop, Jittery, and MuDR may be viewed as more derived clades or alternatively that Phantom elements may be subject to horizontal transfer more readily than other Mutators.
Horizontal transfer of Phantom:
Numerous examples of related TEs occurring in distantly related animal genomes have been documented and can only be explained by invoking horizontal transfer, although the mechanism remains a mystery (Daniels et al. 1990; Robertson 2002; Pace et al. 2008). It has been hypothesized that viruses may make good vectors as they are known to frequently pickup host genes and are infectious. Indeed, TEs have been previously identified in viral genomes. For example, piggybac and TED were identified when they passed from the Lepidopteran host to the infectious baculovirus (Lerch and Friesen 1992; Wang and Fraser 1993). These HT events were caught in the act during experiments in the laboratory. More recently, a reptilian SINE was identified bioinformatically in the genome of the taterapox virus that infects mammals, revealing that HT of TEs to viruses occurs in nature (Wang and Fraser 1993; Ozers and Friesen 1996; Piskurek and Okada 2007). Interestingly, we have identified Phantom transposases and/or complete Phantom elements in two double-stranded polydsDNA viruses, Gf. ichnovirus (Phantom_Gfi) and C. bracovirus (Phantom_Cib) that are known to infect wasps (Table 1). The host species of these viruses has not been sequenced; however, phylogenetic analysis based on the putative transposases encoded by these elements reveals a monophyletic clade between Phantom_Gfi and the Phantom transposase from the wasp Nasonia vitripennis (pp = 69). This clade is nestled within a well-supported clade (pp = 97) of other insect and invertebrate transposases, which lends support to the HT occurring from the insect to the virus rather than vice versa. Phantom elements found in dsDNA viruses adds to the growing body of evidence (Fraser et al. 1983; Friesen and Nissen 1990; Jehle et al. 1998; Lerch and Friesen 1992; Piskurek and Okada 2007; Xu et al. 2006) that dsDNA viruses may act as vectors for horizontal movement of TEs between eukaryotes.
Acknowledgments
The authors thank Assiatu Barrie, Cedric Feschotte, and Jainy Thomas for their critical review of this manuscript. The authors also thank Robert Makowsky and Jeff Streicher for their assistance with the Bayesian analyses. This work was funded by start-up funds from University of Texas at Arlington and C.P.M. was supported by the Society for the Advancement of Chicanos and Native Americans in Science with a Genome Scholars fellowship.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.116673/DC1.
References
- Akagi, H., Y. Yokozeki, A. Inagaki, K. Mori and T. Fujimura, 2001. Micron, a microsatellite-targeting transposable element in the rice genome. Mol. Genet. Genomics 266 471–480. [DOI] [PubMed] [Google Scholar]
- Babu, M. M., L. M. Iyer, S. Balaji and L. Aravind, 2006. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34 6505–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, R. F., D. V. Thompson, D. R. Talbot, J. Swanson and J. L. Bennetzen, 1984. Nucleotide sequence of the maize transposable element Mul. Nucleic Acids Res. 12 5955–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., J. Swanson, W. C. Taylor and M. Freeling, 1984. DNA insertion in the first intron of maize Adh1 affects message levels: cloning of progenitor and mutant Adh1 alleles. Proc. Natl. Acad. Sci. USA 81 4125–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham, P. E., and Z. Zachar, 1989. Retrotransposons and the FB transposon from Drosophila melanogaster, pp. 485–502 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, DC.
- Byrne, M. E., D. A. Rouch and R. A. Skurray, 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81 361–367. [DOI] [PubMed] [Google Scholar]
- Byrne, M. E., M. T. Gillespie and R. A. Skurray, 1990. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob. Agents Chemother. 34 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvet, F., C. Grimaldi, F. Kaper, T. Langin and M. J. Daboussi, 2003. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20 1362–1375. [DOI] [PubMed] [Google Scholar]
- Cowan, R. K., D. R. Hoen, D. J. Schoen and T. E. Bureau, 2005. MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol. Biol. Evol. 22 2084–2089. [DOI] [PubMed] [Google Scholar]
- Craig, N. L., R. Craigie, M. Gellert and A. M. Lambowitz (Editors), 2002. Mobile DNA II. ASM, Washington, DC.
- Daniels, S. B., K. R. Peterson, L. D. Strausbaugh, M. G. Kidwell and A. Chovnick, 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao, X., M. Freeling and D. Lisch, 2006. Horizontal transfer of a plant transposon. PLoS Biol. 4 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J. A., M. I. Benito and V. Walbot, 1994. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22 2634–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., and E. J. Pritham, 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen, D. R., K. C. Park, N. Elrouby, Z. Yu, N. Mohabir et al., 2006. Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol. Biol. Evol. 23 1254–1268. [DOI] [PubMed] [Google Scholar]
- Holligan, D., X. Zhang, N. Jiang, E. J. Pritham and S. R. Wessler, 2006. The transposable element landscape of the model legume Lotus japonicus. Genetics 174 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van, A., and P. Capy, 2008. Analysis of the DDE motif in the Mutator superfamily. J. Mol. Evol. 67 670–681. [DOI] [PubMed] [Google Scholar]
- Hudson, M. E., D. R. Lisch and P. H. Quail, 2003. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34 453–471. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, X. Zhang, S. R. Eddy and S. R. Wessler, 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573. [DOI] [PubMed] [Google Scholar]
- Keeling, P. J., G. Burger, D. G. Durnford, B. F. Lang, R. W. Lee et al., 2005. The tree of eukaryotes. Trends Ecol. Evol. 20 670–676. [DOI] [PubMed] [Google Scholar]
- Lerch, R. A., and P. D. Friesen, 1992. The baculovirus-integrated retrotransposon TED encodes gag and pol proteins that assemble into viruslike particles with reverse transcriptase. J. Virol. 66 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., L. Ding, C. Casola, D. R. Ripoll, C. Feschotte et al., 2007. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., 2002. Mutator transposons. Trends Plant Sci. 7 498–504. [DOI] [PubMed] [Google Scholar]
- Lisch, D. R., M. Freeling, R. J. Langham and M. Y. Choy, 2001. Mutator transposase is widespread in the grasses. Plant Physiol. 125 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, F. R., J. C. Silva, M. Benchimol, G. G. Costa, G. A. Pereira et al., 2009. The protist Trichomonas vaginalis harbors multiple lineages of transcriptionally active Mutator-like elements. BMC Genomics 10 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., T. C. Wood, Y. Yu, M. A. Budiman, J. Tomkins et al., 2000. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res. 10 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott et al., 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37 D205–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo, M., M. Puig and A. Ruiz, 2008. The foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus. Proc. Natl. Acad. Sci. USA 105 2957–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuveglise, C., F. Chalvet, P. Wincker, C. Gaillardin and S. Casaregola, 2005. Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot. Cell 4 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozers, M. S., and P. D. Friesen, 1996. The Env-like open reading frame of the baculovirus-integrated retrotransposon TED encodes a retrovirus-like envelope protein. Virology 226 252–259. [DOI] [PubMed] [Google Scholar]
- Pace, J. K., 2nd, C. Gilbert, M. S. Clark and C. Feschotte, 2008. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. USA 105 17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek, O., and N. Okada, 2007. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. USA 104 12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, S. S., 1982. DNA sequence of a foldback transposable element in Drosophila. Nature 297 201–204. [DOI] [PubMed] [Google Scholar]
- Pritham, E. J., C. Feschotte and S. R. Wessler, 2005. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol. Biol. Evol. 22 1751–1763. [DOI] [PubMed] [Google Scholar]
- Robertson, D. S., 1978. Characterization of a mutator system in maize. Mutat. Res. 51 21–28. [Google Scholar]
- Robertson, H. M., 2002. Evolution of DNA transposons in eukaryotes, pp. 1093–1110 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM, Washington, DC.
- Ronquist, F., and J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saccaro, Jr., N. L., M. A. Van Sluys, A. de Mello Varani and M. Rossi, 2007. MudrA-like sequences from rice and sugarcane cluster as two bona fide transposon clades and two domesticated transposases. Gene 392 117–125. [DOI] [PubMed] [Google Scholar]
- Singer, T., C. Yordan and R. A. Martienssen, 2001. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommer, J. N., 1982. Regulatory mutants of the maize Adh1 gene caused by DNA insertions. Nature 300 542–544. [Google Scholar]
- Talbert, L. E., and V. L. Chandler, 1988. Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5 519–529. [DOI] [PubMed] [Google Scholar]
- Taylor, L. P., and V. Walbot, 1987. Isolation and characterization of a 1.7-kb transposable element from a mutator line of maize. Genetics 117 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z., and S. P. Orphanidis, 2001. Microuli, a family of miniature subterminal inverted-repeat transposable elements (MSITEs): transposition without terminal inverted repeats. Mol. Biol. Evol. 18 893–895. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, H., A. Monfort and P. Puigdomenech, 2007. Mutator-like elements identified in melon, Arabidopsis and rice contain ULP1 protease domains. Mol. Genet. Genomics 277 357–364. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and G. Rudenko, 2002. MuDR/Mu transposable elements of maize, pp. 533–564 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM, Washington, DC.
- Wang, H. G., and M. J. Fraser, 1993. TTAA serves as the target site for TFP3 lepidopteran transposon insertions in both nuclear polyhedrosis virus and Trichoplusia ni genomes. Insect Mol. Biol. 1 109–116. [DOI] [PubMed] [Google Scholar]
- Xu, H. F., Q. Y. Xia, C. Liu, T. C. Cheng, P. Zhao et al., 2006. Identification and characterization of piggyBac-like elements in the genome of domesticated silkworm, Bombyx mori. Mol. Genet. Genomics 276 31–40. [DOI] [PubMed] [Google Scholar]
- Xu, Z., X. Yan, S. Maurais, H. Fu, D. G. O'Brien et al., 2004. Jittery, a Mutator distant relative with a paradoxical mobile behavior: excision without reinsertion. Plant Cell 16 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., K. Tamaki, K. Watanabe, M. Fujino and C. Nakamura, 1998. A maize MuDR-like element expressed in rice callus subcultured with proline. Hereditas 129 95–99. [DOI] [PubMed] [Google Scholar]
- Yu, Z., S. I. Wright and T. E. Bureau, 2000. Mutator-like elements in Arabidopsis thaliana: structure, diversity and evolution. Genetics 156 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]