Abstract
Several studies have proposed that brain glutamate signaling abnormalities and glial pathology play a role in the etiology of major depressive disorder (MDD). These conclusions were primarily drawn from postmortem studies in which forebrain brain regions were examined. The locus coeruleus is the primary source of extensive noradrenergic innervation of the forebrain and as such exerts a powerful regulatory role over cognitive and affective functions, which are dysregulated in MDD. Furthermore, altered noradrenergic neurotransmission is associated with depressive symptoms and is thought to play a role in the pathophysiology of MDD. In the present study we used laser-capture microdissection to selectively harvest locus coeruleus (LC) tissue from postmortem brains of MDD patients, patients with bipolar disorder (BPD), and from psychiatrically-normal subjects. Using microarray technology we examined global patterns of gene expression. Differential mRNA expression of select candidate genes was then interrogated using quantitative real-time PCR and in situ hybridization. Our findings reveal multiple signaling pathway alterations in the LC of MDD, but not BPD subjects. These include glutamate signaling genes SLC1A2, SLC1A3, GLUL, growth factor genes FGFR3 and TrkB, and several genes exclusively expressed in astroglia. Our data extend previous findings of altered glutamate, astroglial and growth factor functions in MDD for the first time to the brainstem. These findings indicate that such alterations: 1) are unique to MDD and distinguishable from BPD, and 2) affect multiple brain regions, suggesting a whole-brain dysregulation of such functions.
Keywords: laser-capture microdissection, human, monoamine, norepinephrine, postmortem, microarray
Introduction
Major depression (MDD) is a chronic and debilitating mood disorder with a 16% lifetime prevalence 1 and is associated with excess mortality, especially from cardiovascular disease and through suicide 2. Mood disorders, including MDD and bipolar disorder (BPD) are characterized by dysfunction of noradrenergic neurotransmission. Drugs that increase brain norepinephrine (NE)/serotonin (5-HT) levels act as antidepressants 3, 4, whereas drugs that deplete NE/5-HT stores induce a depressive state 5, 6. Currently, most antidepressant drugs directly or indirectly target 5-HT and/or NE systems of the brain, as these neurotransmitters have been suggested to be mechanistically relevant to the etiology of mood disorders 7. However, drugs such as selective 5-HT reuptake inhibitors only produce remission in 37% of all patients, and if they show therapeutic responses, then only after 2-5 weeks 8. These observations suggest that brain monoamine deficiency may be part of a much more complex pathogenesis of major depression that requires further investigation.
The locus coeruleus (LC), which is located in the rostral pontine tegmentum 9, 10, possesses the largest number of NE-producing neurons in the mammalian brain. These neurons contain the pigment neuromelanin that gives the nucleus its characteristic dark color. LC neurons project widely throughout the entire central nervous system, including the cerebral cortex, thalamus, septum, hippocampus, hypothalamus, cerebellum, and spinal chord 11-13. In addition there are intense reciprocal connections between LC and the 5-HT producing dorsal raphe neurons, which provide the basis for neurochemical communication between both monoamine systems 14. With their wide-ranging and overlapping projections arising from the brainstem, both monoamine systems act in concert to modulate vigilance, sleep-wake cycle, memory, adaptive response to stress, and pain modulation 15. These behaviors are disturbed in patients with MDD, and as a consequence their LC displays molecular abnormalities. Therefore, the current study focused on the locus coeruleus (LC) which synthesizes the majority of NE released in the forebrain 16 The study is an attempt to more clearly understand the biology of cells in the in the LC (neurons and glia) in depression.
Previous postmortem studies focused on the LC have shown increased protein levels of presynaptic alpha 2 adrenoreceptors in MDD, which inhibits the firing of LC neurons and subsequent norepinephrine secretion 17. Protein levels of tyrosine hydroxylase, the rate limiting enzyme in the synthesis of norepinephrine, are likewise elevated in the LC of MDD patients 18, whereas binding to norepinephrine transporter is reduced 19. These results are consistent with the interpretation that: 1) noradrenergic neurotransmission is dysregulated in MDD, and 2) the LC represents a key region in the etiology of this disorder. The majority of post mortem studies involving the analysis of LC in MDD have focused on analyses of proteins involved in noradrenergic neurotransmission. Little is known, however, about mRNA expression, especially of non-adrenergic genes, in the LC of MDD patients.
Therefore, in the present study we used gene expression microarray technology in combination with laser-capture microdissection (LCM) to profile gene expression in the LC from post mortem brains of patients with MDD. These profiles were contrasted to those obtained from the LC of BPD patients and psychiatrically-normal subjects. Our approach yielded a number of candidate genes that may play a key role in the LC dysfunction in depression. Potentially altered gene transcripts were examined using quantitative real-time PCR (qPCR) and/or in situ hybridization (ISH). The results implicate MDD-specific dysfunctions in glutamate and growth factor signaling with special emphasis on altered astroglial transcripts, which is in strong agreement with previous studies from dorsolateral prefrontal cortex (DLPFC) and anterior cingulate cortex (AnCg) of MDD patients 20, 21.
Methods
Human Brain Tissue Acquisition and Preparation
All brains used in this study were collected by the Brain Donor Program at the University of California, Irvine with the consent of the decendents' relatives. Information regarding physical health, medication use, psychopathology, substance use, and details about the final hours of the decendents were obtained from medical records, coroner's investigation, medical examiner's conclusions, and interviews with relatives. Table 1 lists subjects in the current study and includes their age, gender, agonal state, postmortem interval, and brain pH. No significant differences in age, brain pH, or postmortem interval among the three diagnostic groups were detected (p > 0.05, one-way ANOVA). Previous post mortem studies concluded that brain tissue pH and a patient's agonal state (brief vs. protracted deaths) are among the most important factors in determining global gene expression patterns 22-24. Therefore, the present study only used brain samples with a pH > 6.7, and an agonal factor of zero, indicating a brief death.
Table 1.
List of demographical and clinical characteristics of included subjects. Averages ± S.E.M. are: age – Control (50.7±5.6), MDD (49.6±4.2), BPD (43.2±7.4); PMI – Control (20.0±1.3), MDD (25.5±1.4), BPD (22.7±4.4); pH – Control (6.94±0.04), MDD (7.03±0.04), BPD (6.95±0.04). Abbreviations: F - female, M - male, PMI - post-mortem interval, med.cond. - medical condition, SSRI – selective serotonin reuptake inhibitor
| diagnosis | subject | race | age | sex | PMI (hours) |
agonal factor |
brain pH |
cause of death |
medication |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Control | 2169 | Caucasian | 18 | M | 22 | 0 | 6.97 | accident | No |
| Control | 2292 | Caucasian | 55 | M | 15 | 0 | 6.89 | sudden med. cond. | No |
| Control | 2805 | Caucasian | 45 | M | 21 | 0 | 6.86 | sudden med. cond. | No |
| Control | 3228 | Pacific Islander | 39 | M | 18.15 | 0 | 6.81 | sudden med. cond. | No |
| Control | 3516 | Caucasian | 41 | M | 22.5 | 0 | 7.01 | sudden med. cond. | No |
| Control | 3519 | African American | 65 | M | 13.5 | 0 | 6.88 | sudden med. cond. | No |
| Control | 3520 | Caucasian | 74 | F | 18.5 | 0 | 7.21 | sudden med. cond. | No |
| Control | 3588 | Caucasian | 56 | M | 24.5 | 0 | 6.98 | sudden med. cond. | No |
| Control | 3706 | Caucasian | 63 | M | 24.5 | 0 | 6.88 | sudden med. cond. | No |
|
| |||||||||
| MDD | 2208 | Caucasian | 72 | F | 21 | 0 | 7.13 | suicide | Non-SSRI |
| MDD | 2267 | Caucasian | 19 | M | 18 | 0 | 7.11 | suicide | No |
| MDD | 3064 | Caucasian | 46 | M | 27 | 0 | 6.91 | sudden med. cond. | Non-SSRI |
| MDD | 3169 | Caucasian | 35 | M | 24.75 | 0 | 7.04 | accident | Non-SSRI |
| MDD | 3365 | Caucasian | 47 | M | 29 | 0 | 7.25 | suicide | No |
| MDD | 3426 | Caucasian | 63 | M | 28.5 | 0 | 7.17 | sudden med. cond. | No |
| MDD | 3031 | Caucasian | 49 | M | 27 | 0 | 7.19 | suicide | No |
| MDD | 2315 | Caucasian | 58 | M | 24 | 0 | 6.93 | suicide | SSRI |
| MDD | 2944 | Caucasian | 52 | M | 16 | 0 | 6.82 | sudden med. cond. | SSRI |
| MDD | 3071 | Caucasian | 49 | M | 31 | 0 | 7 | unknown | SSRI |
| MDD | 3168 | Caucasian | 39 | M | 27.5 | 0 | 6.79 | suicide | SSRI |
| MDD | 3481 | Caucasian | 66 | M | 32 | 0 | 7.05 | sudden med. cond. | SSRI |
|
| |||||||||
| BPD | 2311 | Caucasian | 23 | M | 9 | 0 | 7.12 | suicide | Lithium |
| BPD | 2466 | Caucasian | 26 | M | 19 | 0 | 6.92 | suicide | Lithium |
| BPD | 3085 | Caucasian | 63 | M | 40 | 0 | 6.87 | accident | No |
| BPD | 3241 | Caucasian | 59 | M | 15.5 | 0 | 6.99 | sudden med. cond. | Lithium |
| BPD | 2566 | Caucasian | 56 | F | 29 | 0 | 6.83 | suicide | No |
| BPD | 3079 | Caucasian | 32 | M | 23.75 | 0 | 6.94 | suicide | SSRI |
Brains were removed at autopsy, quickly chilled to approximately 4°C and then cut into series of 0.75 cm thick coronal slices that were quickly frozen and stored at -80°C as previously described25. Human brain slabs were then placed on dry ice blocks and dissected using fine-toothed saw to generate tissue blocks for subsequent cryostat sectioning. Such tissue blocks were approximately 4 × 3 cm in size and were stored at −80°C.
Tissue blocks containing LC were cryostat-cut into 10 μm coronal sections at −20°C, immediately thaw-mounted onto glass slides (1 section/slide), and then stored at −80° C. Every fiftieth section from each subject's slide set was processed for the detection of norepinephrine transporter (NET) mRNA via radiolabeled ISH as previously described 26. NET mRNA is highly enriched within the boundaries of the LC and can be reliably used to map its location 27. Following ISH, slides were stained with luxol fast blue combined with cresyl violet to visualize brainstem anatomy. Using these landmarks in combination with shape of the LC (as determined by NET ISH) and NET signal intensity and distribution, sections from all subjections were aligned to match along the anterior-posterior brain axis.
We used the ISH signal obtained from neighboring sections as guide to microdissect the LC by the means of laser capture (ISH-guided LCM). For each subject we collected a total of 4 slides 500 μm apart from within a 2 mm common region of the mid-rostral portion of the nucleus (~ +25 to +27 mm from obex 28), which resulted in processing of 8 bilateral LCs. Selected slides were removed from −80° C, thawed for 30 sec at room temperature, then dehydrated and defatted as previously described 29. ISH-guided LCM procedure was used to microdissect individual LC nuclei which is described in detail elsewhere 26.
RNA isolation and amplification
RNA isolation, including DNAse treatment, was performed using the PicoPure RNA Isolation kit (Molecular Devices, Sunnyvale, CA) according to the manufacturer's protocol. The final RNA elution volume was 15 μl. To assess RNA quality and concentration, 1 μl of each subject's isolated RNA sample was evaluated with on a 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA) and resulting electropherograms were quantified according to the method of Schoor et al. 30. This method has been applied to human brain tissue obtained through ISH-guided LCM 26.
RNA samples were subject to two rounds of RNA amplification (RiboAmp OA RNA kit, Molecular Devices). After the first round of amplification a portion of amplified double-stranded cDNA was saved for qPCR. Following two amplification rounds, 15 μg of biotinylated amplified RNA from each subject was then hybridized to HG-U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA) per manufacturer's instructions.
Microarray data analysis
Robust Multi-Chip Average (RMA) and Affymetrix Microarray Suite 5 (MAS5) calls algorithms were employed for Affymetrix CEL files analysis. Affymetrix chip description files were replaced by a custom probe set mapping files (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp), which independently reassigned all Affymetrix probe sets to an updated UniGene cluster (Build No. 199). Log 2-transformed intensity values of all transcripts from RMA output were statistically analyzed using Microsoft Excel 2003 software (Microsoft, Redmond, WA). Disease specific gene expression differences from controls were evaluated using Student's t-tests. Genes were considered to be significantly altered if: (1) the p values were ≤ 0.05, (2) the mRNA was detected in ≥ 50% of arrays in C, BPD, or MDD groups according to MAS5CALLS algorithm, and (3) a gene's log2 intensity value was ≥4 according to RMA algorithm. Pairwise fold change differences were calculated from group means of microarray intensity values. Pearson correlation calculations (two-tailed p-values) between microarray and qPCR data were made with GraphPad Prism 4 software (Graphpad, La Jolla, CA).
Canonical Pathway Analysis
Canonical pathway analysis was generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, Redwood City, CA). Microarray data were filtered to only include genes that: a) had log2 intensity value of ≥4; b) were detected on at least 50% of microarrays in any of the diagnostic groups; and c) had Student t-test p-values of ≤0.1 for either MDD vs. Control or BPD vs. Control comparisons; and d) and were altered in their mRNA expression either ≤ −1.05 or ≥1.05- fold. These relaxed parameters were used to maximize the number of candidate genes for downstream analyses. To investigate potential involvement of different functional pathways selected candidate genes were evaluated using Canonical Pathway analysis tool in the Ingenuity Pathways Knowledge Base (Ingenuity Pathways Analysis software (IPA), Ingenuity® Systems, www.ingenuity.com) The significance of the association between these data sets and each canonical pathway was evaluated using: 1) a ratio of the number of genes in the data set that belonged to a particular pathway relative to the total number of genes in such canonical pathway and 2) a p-value (Fischer's exact test) to determine the probability that the association is due to chance.
Quantitative Real-Time PCR (qPCR)
QPCR confirmation of microarray results utilized SYBR Green chemistry. A Bio-Rad iCycler (BioRad, Hercules, CA) along with SYBR-488 detection protocol combined with a touchdown PCR approach 26, 29 was employed for performance of amplification reactions and fluorescence quantification. Reactions were performed in 96 well PCR plates (Bio-Rad) with each well containing 5 μl of amplified double-stranded cDNA (aDNA; 50 pg/μl) that was set aside following the first round of RNA amplification. The concentration of aDNA was quantified for each sample using Quant-iT PicoGreen dsDNA kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. To each well 5 μl of forward and reverse strand primers (final concentration: 500 nM) and 10 μl of iQ SYBR Green Supermix (Bio-Rad) were added. Amplifications of all samples were carried out in triplicate and the average cycle threshold (Ct) was then calculated for each sample. Replicates that were ≥1 Ct away from the mean Ct were excluded; the mean Ct was calculated from the remaining duplicates. Subjects with only one Ct value were excluded from further analysis. Because input amount of aDNA was equivalent across all samples, raw Ct values were inversely proportional to the intensity levels of gene expression. We chose this approach rather than the ΔΔCt method, which relies on normalization to housekeeping genes, because of the potential for differential mRNA expression of such reference transcripts 31, 32. A similar approach in which standardized DNA input amounts for qPCR were used has recently been validated 26, 33.
The following formula was used to calculate relative fold changes: 2−(Cta − Ctb) in which Cta is cycle threshold in experimental subjects (MDD or BPD) and Ctb is cycle threshold in control subjects. The specificity of each reaction was then confirmed by the presence of a single peak on the melting curve, plotted as the negative derivative of fluorescence during incremental increases in well temperature. No template controls, in which aDNA was replaced with distilled H2O, did not yield amplification products. Primer design details are described elsewhere 26. Sequences of PCR primer pairs used in the present study are listed in Table S1.
In Situ Hybridization (ISH)
Detailed ISH methods using postmortem human brain tissue have been described elsewhere 34. Briefly, slides adjacent to the ones used for LCM were removed from -80°C storage, fixed in 4% paraformaldehyde at room temperature, rinsed in standard saline citrate buffer, incubated in a solution containing acetic anhydride in triethanolamine, and dehydrated in aqueous solutions with increasing alcohol concentrations. Sections were then hybridized with 35S-UTP- and 35S-CTP-doublelabeled cRNA probes, produced using standard in vitro transcription methodology. Riboprobes for NET (NM_001043) pos.1-1974 and TH (NM_000360) pos. 336-734 were synthesized from human cDNA fragments cloned in house. Clones for probes of glutamate-ammonia ligase (glutamine synthetase; GLUL/GLNS), solute carrier family 1 (glial high affinity glutamate transporters) members 2 and 3 (SLC1A2, SLC1A3) were a kind gift from Dr. Choudary (University of California – Davis). Probe sequences and information regarding their specificities have been previously published 20.
Following overnight incubation at 55°C, slides were washed, rinsed, dehydrated, and exposed to Kodak Biomax MR film (Eastman Kodak, Rochester, NY, USA). Exposure time was empirically determined using test slides to optimize the specific signal for densitometric analysis. Specificity of the hybridization was confirmed in control experiments using sense riboprobes.
Quantification of the Radioactive Signal
ISH autoradiograms were digitized and captured using ScanMaker 1000XL Pro Flatbed Scanner (Microtek, Carson, CA) in combination with SilverFast Ai Imaging Software (LaserSoft Imaging, Inc., Sarasota, FL). Adobe Photoshop CS software (Adobe Systems, San Jose, CA) was used to import images of adjacent autoradiograms labeled for TH, SLC1A2, SLC1A3, and GLUL mRNAs into individual layers. These digitized images were overlaid in Photoshop and registered to each other using major anatomical landmarks in each image as guides. Afterwards, 4 individual layers consisting of aligned TH, SLC1A2, SLC1A3, and GLUL autoradiogram images were imported into ImageJ software, version 1.37 (rsbweb.nih.gov/ij/) 35. In the TH autoradiogram anatomical boundaries of LC were identified, outlined, and transferred to the exactly matching position in the SLC1A2, SLC1A3, and GLUL labeled images where optical density within the whole area of the LC (as defined by TH mRNA distribution) was measured. Gray scale values were converted into relative optical density (ROD) measurements using standards from an optical density step tablet. Background was measured and subtracted from each specific LC measurement. Mean ISH signals were statistically analyzed using one-tailed Student's t-tests in Prism 4.0 software (GraphPad). Pearson correlation coefficients (two-tailed p-values) of ISH signals and qPCR Ct values were also computed with Prism 4.0 software (GraphPad).
Results
RNA quality
RNA quality was assessed by evaluating electropherogram signals prior to the appearance of the 18S peak and the AUC of the 28S peak. RNA samples with less than 65% of their total signal in the pre-18S peak area of the electropherogram and more than 4% of total signal under the 28S peak have been demonstrated to produce accurate and reliable results in microarray- and qPCR-based gene expression experiments 26, 30. Such analysis of our samples showed that the pre-18S peak area contained: 47.9±1.7% (Control), 47.8±1.4% (MDD), and 48.9±2.7% (BPD) of total signal, while the 28S peak region contained: 11.8±1.0% (Control), 10.8±0.9% (MDD), and 12.0±1.7% (BPD) of total signal. For technical reasons we are unable to obtain these parameters in three (1 control, 2 MDD) of all 27 samples. However, downstream gene expression results were not significantly different in these three samples. Our results indicate that the RNA extracted from all samples was suitable for valid analysis of microarray- and qPCR based gene expression data 26, 30
Overall Microarray results
Averaged microarray array intensity values derived from log2 RMA data were as followed (mean±S.E.M.): 6.75± 0.02 (control); 6.76± 0.04 (MDD); 6.77± 0.02 (BPD). Raw detection call rates calculated from the MAS5CALLS output were (mean±S.E.M.): 46.1 ± 0.9% (control); 45.0 ± 1.2% (MDD); 46.6 ± 0.4% (BPD). There were no significant differences (p>0.05) among diagnosis groups for RMA or MAS5CALLS, indicating an overall expression homogeneity among groups.
Glutamate signaling gene transcripts exhibit altered mRNA expression in LC of MDD subjects
Comparing mood disorder groups, microarray data revealed that at p≤0.1 significance level 1094 and 695 genes transcripts were upregulated in MDD and BPD patients, respectively, whereas 474 transcripts in MDD and 454 transcripts in BPD were downregulated. We utilized Ingenuity pathway analysis (IPA) as a discovery science approach for examination of LC microarray and for the identification of significantly altered canonical pathways. The pathway that was most significantly (p=0.004) altered in MDD was the one for glutamate signaling (Figure 1). It is defined by 47 different genes, and we found significantly altered mRNA expression in 8 of such genes. Of these 8 genes three were downregulated, in their mRNA expression and consisted of transcripts that encode high affinity glutamate transporters SLC1A2 and SLC1A3, and GLUL, are all of which are specifically expressed in glial cells but not in neurons. In contrast, 4 of the post-synaptic glutamate receptor transcripts were upregulated – GRIA1, GRIK1, GRM1, GRM5, whereas the presynaptic vesicular glutamate transporter 2 (VGLUT2 or SLC17A6) was upregulated in its gene expression as well. Of the eight altered glutamate signaling genes in MDD, which are all individually altered at the alpha level of p<0.05, only GRM5 also exhibited significant transcript upregulation in BPD subjects.
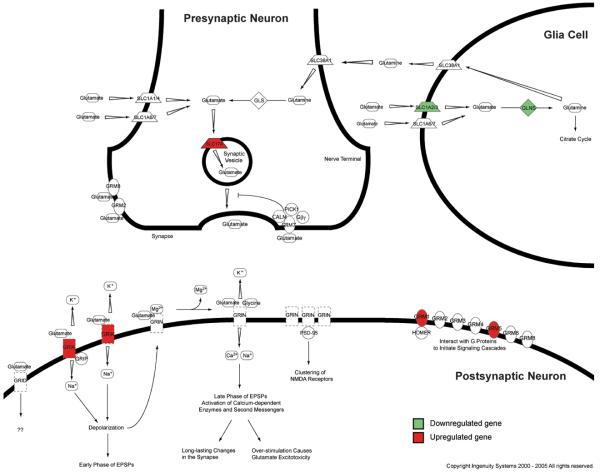
Figure 1.
Synaptic location of elements of canonical glutamate signaling pathway. Model of glutamate synapse consisting of pre- and postsynaptic neuron and glial cell depicting the cellular location of key glutamate signaling and interacting proteins according the IPA knowledge base. Marked in green are gene products that are downregulated, whereas highlighted in red are upregulated gene products in the LC of MDD patients.
Abbreviations: CALM – calmodulin; HOMER – homer homolog; Gβγ – beta/gamma subunit of G protein coupled receptor; GLNS – glutamine synthetase; GLS – glutaminase; GRIA – glutamate receptor, ionotropic AMPA; GRID – glutamate receptor, ionotropic, delta; GRIK – glutamate receptor, ionotropic, kainite; GRIN - glutamate receptor, ionotropic, NMDA; GRIP – glutamate receptor binding protein; GRM –glutamate receptor, metabotropic; PICK1 – protein interacting with C kinase 1; PSD-95 – postsynaptic density protein 95; SLC – member of solute carrier family
Subsequently, four glutamate signaling genes transcripts were probed using qPCR. This approach confirmed microarray results for the transcripts SLC1A2, SLC1A3, and GLUL whereas VGLUT2 transcript did not show altered mRNA expression (Table 2).
Table 2.
Results of microarray, qPCR, and ISH experiments. List of genes of interest (subdivided into 4 groups) and their expression changes in MDD and BPD patients evaluated by gene expression microarray, qRT-PCR, and/or ISH. Significant (p≤0.05) changes from Control determined by Student t-test are in bold. F-Change indicates the fold-change from control values.
| RefSeq ID | Gene Name | Symbol | Gene Expression Microarray Results | qRT-PCR results | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate signaling genes | log 2 mean intensity |
MD | BP | MD | BP | ||||||
| p value | F-Change | p value | F-Change | p value | F-Change | p value | F-Change | ||||
| NM_004171 | Glial high affinity glutamate transporter, member 2 | SLC1A2 | 6.1 | 0.036 | −1.26 | 0.370 | −1.05 | 0.044 | −1.72 | 0.470 | −1.02 |
| NM_004172 | Glial high affinity glutamate transporter, member 3 | SLC1A3 | 8.3 | 0.034 | −1.59 | 0.297 | −1.13 | 0.008 | −2.07 | 0.341 | −1.09 |
| NM_000828 | Glutamate receptor, ionotrophic, AMPA 3 | AMPA3 | 5.9 | 0.026 | 1.16 | 0.053 | 1.14 | ||||
| NM_175611 | Glutamate receptor, ionotropic, kainate 1 | GRIK1 | 6.1 | 0.024 | 1.21 | 0.332 | 1.04 | ||||
| NM_000838 | Glutamate receptor, metabotropic 1 | mGluR1 | 4.4 | 0.031 | 1.10 | 0.325 | 1.02 | ||||
| NM_000842 | Glutamate receptor, metabotropic 5 | mGluR5 | 5.5 | 0.010 | 1.23 | 0.038 | 1.15 | ||||
| NM_002065 | Glutamate-ammonia ligase (glutamine synthetase) | GLUL | 9.9 | 0.020 | −1.28 | 0.265 | −1.10 | 0.026 | −2.15 | 0.059 | −1.73 |
| NM_020346 | Solute carrier family 17, member 6, SLC17A6 | VGLUT2 | 10.1 | 0.018 | 1.28 | 0.138 | 1.17 | 0.446 | 1.04 | 0.383 | 1.10 |
| Glia associated genes | |||||||||||
| NM_001650 | Aquaporin 4 | AQP4 | 6.0 | 0.004 | −1.65 | 0.12 | −1.22 | 0.014 | −2.44 | 0.202 | −1.32 |
| NM_001428 | Enolase 1 (non-neuronal) | ENO1 | 6.2 | 0.324 | 1.02 | 0.44 | 1.01 | 0.095 | −1.31 | 0.075 | −1.36 |
| NM_000165 | Gap junction protein, alpha 1, 43kDa (connexin 43) | GJA1 | 10.1 | 0.038 | −1.86 | 0.44 | −1.05 | 0.015 | −2.10 | 0.489 | −1.02 |
| NM_006783 | Gap junction protein, beta 6 (connexin 30) | GJB6 | 7.0 | 0.024 | −1.93 | 0.24 | −1.26 | 0.025 | −1.95 | 0.361 | −1.19 |
| NM_002055 | Glial fibrillary acidic protein | GFAP | 10.0 | 0.054 | −1.32 | 0.40 | −1.05 | 0.034 | −1.83 | 0.289 | −1.20 |
| NM_006272 | S100 calcium binding protein, beta | SB100 | 10.3 | 0.020 | −1.21 | 0.06 | 1.11 | 0.014 | −1.62 | 0.306 | 1.07 |
| Growth factor genes | |||||||||||
| NM_002006 | Fibroblast growth factor 2 (basic) | FGF2 | 5.4 | 0.069 | −1.08 | 0.193 | −1.05 | 0.335 | −1.11 | 0.223 | 1.19 |
| NM_002010 | Fibroblast growth factor 9 (glia-activating factor) | FGF9 | 8.9 | 0.011 | 1.18 | 0.072 | 1.09 | 0.121 | 1.22 | 0.460 | −1.02 |
| NM_000142 | Fibroblast growth factor receptor 3 | FGFR3 | 5.9 | 0.032 | −1.17 | 0.983 | −1.02 | 0.010 | −2.19 | 0.292 | −1.18 |
| NM_000618 | Insuline-like growth factor 1 | IGF1 | 5.5 | 0.060 | 1.10 | 0.218 | 1.05 | ||||
| NM_001007097 | Neurotrophic tyrosine kinase, receptor, type 2 | TrkB | 6.6 | 0.027 | −1.45 | 0.375 | −1.07 | 0.009 | −1.66 | 0.462 | −1.01 |
| Neuronal markers genes | |||||||||||
| NM_001975 | Enolase 2 (gamma, neuronal) | ENO2 | 10.0 | 0.234 | 1.07 | 0.169 | 1.10 | 0.460 | 1.03 | 0.466 | 1.03 |
| NM_005382 | Neurofilament 3 (150kDa medium) | NEF3 | 12.6 | 0.460 | −1.01 | 0.406 | 1.02 | ||||
| NM_021076 | Neurofilament, heavy polypeptide 200kDa | NEFM | 12.2 | 0.398 | 1.02 | 0.422 | 1.02 | ||||
| NM_006158 | neurofilament, light polypeptide 68kDa | NEFL | 12.5 | 0.396 | −1.03 | 0.461 | −1.01 | ||||
Because of their profound impact on synaptic glutamate clearance SLC1A2, SLC1A3, and GLUL were selected for additional validation with ISH performed on sections adjacent to those used for LCM. Representative examples of LC ISH autoradiogram images are shown in Figure S1. In MDD subjects ISH optical density for SLC1A3 and GLUL was significantly decreased whereas the decrease in optical density for SLC1A2 did not reach statistical significance (Table 2). For BPD subjects no statistical significant ISH density changes were observed. Mean GLUL and SLC1A3 ISH ROD values for each subject are significantly correlated with their respective qPCR cycle threshold values (Table 3B). These results are consistent with the interpretation that the gene transcripts for SLC1A3 and GLUL are downregulated in MDD, confirming microarray and qPCR results.
Table 3.
Correlation between gene expression validation methods for differentially expressed genes. Table A) Pearson correlations between microarray and qPCR expression data with correlation-associated p-values for C, MDD, and BPD subjects. Table B) Pearson r correlations between qPCR expression and ISH signal intensity data with correlation-associated p-values for C, MDD, and BPD subjects.
| A | ||
|---|---|---|
| Gene | Pearson r | p value |
| AQP4 | 0.750 | <0.0001 |
| FGFR3 | 0.697 | <0.0001 |
| GFAP | 0.492 | 0.0091 |
| GJA1 | 0.906 | <0.0001 |
| GJB6 | 0.852 | <0.0001 |
| GLUL | 0.741 | <0.0001 |
| OXTR | 0.743 | <0.0001 |
| S100B | 0.748 | <0.0001 |
| SLC1A2 | 0.860 | <0.0001 |
| SLC1A3 | 0.766 | <0.0001 |
| TrkB | 0.725 | <0.0001 |
| B | ||
|---|---|---|
| Gene | Pearson r | p value |
| GLUL | 0.461 | 0.036 |
| SLC1A2 | 0.417 | 0.085 |
| SLC1A3 | 0.661 | 0.001 |
Glial deficits accompany glutamate signaling alterations in MDD
Since we detected that the strongest gene expression changes of glutamatergic transmission transcripts have glial origin, we hypothesized that there may be a general deficit of glial function. To test this notion we specifically examined mRNA expression of other genes known to be exclusively expressed by glia. This analysis revealed significant reductions in the mRNA expression of such transcripts, including GFAP, S100B, GJA1, GJB6, and AQP4 in MDD but not BPD subjects. QPCR experiments confirmed these gene expression changes (Table 2) and individual microarray and qPCR results for all glia-associated genes are strongly correlated (Table 3A, Figure S2). In contrast, mRNA expression of genes exclusively expressed in neurons, such as NEFL, NEFM, NEF3, and ENO2 were not altered in MDD or BPD, according to microarray results. The ENO2 transcript was then additional probed using qPCR, which confirmed microarray results. ENO1, a non-neuronal marker, displayed no microarray or qPCR gene expression differences in MDD and BPD. Taken together, these results suggest that there may be a selective insult to LC glial function in MDD.
Growth factor transcripts display altered gene expression in MDD
Finally we examined the mRNA expression of gene transcripts, which have been previously shown to be dysregulated in MDD specifically those that regulate FGF and BDNF functions as both have been implicated in the pathophysiology of MDD 21. Microarray gene expression of neurotrophic tyrosine receptor kinase receptor 2 (TrkB) and fibroblast growth factor receptor 3 (FGFR3) was significantly downregulated in the LC of MDD and were confirmed by qPCR. Additionally, the receptor ligands displayed altered microarray gene expression with FGF2 being downregulated whereas FGF9 as well as insulin-like growth factor 1 (IGF1) were upregulated. However, these microarray results were not confirmed by qPCR. All observed growth factor transcript alterations were specific to MDD subjects.
Discussion
The present study examined alterations in gene expression in the LC of patients with antemortem diagnosis of either MDD or BPD as compared to psychiatrically-normal subjects. We chose ISH-guided LCM to dissect tissue from individual LC nuclei, because our previous study has shown that this approach, compared to micropunches, provides increased anatomical resolution and more precise sampling which enriches specific mRNAs and thus improves sensitivity and the dynamic range of microarray-based gene expression profiling 26. Rather than individual LC cells, we laser-dissected the entire LC area to obtain transcriptional information from neurons and glia, as all cell types within a brain nucleus form a functional, and signaling network. While previous studies focused on neurons in the LC, recent studies have emphasized the importance and vulnerability of glia in mood disorders, such as major depression 20, 39, 40. Our initial studies were performed using high-density oligonucleotide microarrays in an unbiased discovery science approach to uncover potential alterations in the mRNA expression of functionally related genes. Changes in mRNA expression of selected candidate gene transcripts were then validated with qPCR and ISH. Based on microarray results, we thereby focused our efforts on genes that directly interface between astrocytes and components of glutamate signaling pathway. Our data indicate that several classes of genes were differentially expressed in MDD subjects, including those involved in: glutamate neurotransmission, glial function, and growth factor signaling. In contrast, none of these functional groups exhibited altered gene expression in the LC of BPD patients. These results suggest that dysregulation in these signaling systems in the LC may contribute to pathophysiological changes in MDD.
Limitations
The results presented here do not reflect gene expression alteration of the entire locus coeruleus. After alignment of LC-containing brain sections from all subjects, the mid-rostral portion of the LC was sampled. This region is known to send projections to many forebrain structures including hippocampus, cortex and hypothalamus 11-13; The functions of these regions are thought to be compromised by major depression. A MRI study has shown that neuromelanin signal intensity was selectively reduced in the medial and rostral part in locus coeruleus in depression in patients with major depression, implicating noradrenergic dysfunction 41. However, a previous postmortem study found no difference in neuromelanin-containing cells between control and major depressive subjects at any level of the LC 42.
Several postmortem studies have investigated LC nuclei of MDD patients for altered protein levels and found elevated immunoreactivity of TH 18, 42, nNOS 43, CRF 44 and reduced binding to NET 19. Our results, however, did not show mRNA expression changes of TH, CRF, and NET in the LC of MDD or BPD subjects. In fact, none of the transcripts of the norepinephrine biosynthesis enzymes exhibited gene expression alterations in MDD or BPD (data not shown). Translational regulatory mechanisms at the protein level may be responsible for these discrepancies.
We observed significant differences in the mRNA expression of glial and glutamatergic transcripts in MDD, but not BPD subjects. This observation may indicate that such changes are specific to MDD; however, it is limited by the small number of BPD subjects. However, our power analysis calculations indicate that given effects and associated variability significantly larger number of BPD subjects would be required to replicate observed gene expression alterations in MDD. For example, for SLC1A3, SLC1A2, and GLUL gene expression as measured by qPCR, power analysis calculations predict that 14, 15, and 12 MDD subjects respectively, would be required to observe differences vs. Controls at the p < 0.05 level. On the other, hand for BPD vs. Control comparisons 143, 159, and 90 BPD subjects would be required for each gene to observe the same effects. Similarly, we observed large differences (ranging from 2- to over 100- fold) in the power calculations for potential effects in MDD and BPD groups for all of the other transcripts we assayed by qPCR, including: FGF-2, FGF-9, FGF-R3, TrkB, SLC17A6, GFAP, S100B, GJA1, GJB6, AQP4, and ENO1. In our future studies we plan to increase the number of BPD subjects to confirm the specificity of these findings to MDD.
Most of the MDD subjects included in this study died from suicide, which was very likely related to their mood disorder. Therefore, the observed changes in LC gene expression described here may not apply to mildly to moderately depressed patients but may be limited to severely depressed, suicidal patients.
Gene expression alterations in glutamate signaling and glial genes in the LC of MDD may be functionally and anatomically related
Several human MRI studies have linked altered brain glutamate levels and major depression 45, 46 and have shown that therapeutic intervention can normalize brain glutamate concentrations 46, 47, which in turn lessens the severity of depressive symptoms. Glutamatergic neurons, mainly originating from the nucleus paragigantocellularis, provide a major excitatory input to the locus coeruleus 48. A second region, the medial prefrontal cortex, can also elicit glutamatergic responses in the LC 49. Glutamate exerts its postsynaptic activity through a variety of ionotropic and metabotropic glutamate receptors in the LC. Studies in rhesus monkey showed that the LC mainly expresses mRNA subunits for AMPA, GRM3. GRM1 and, GRM5 50. Local LC administration of AMPA elicits norepinephrine release from the LC 51 suggesting that glutamate in LC drives norepinephrine turnover. Excess levels of glutamate acting via glutamate receptors can cause neuronal damage (reviewed in 52) and is hypothesized to contribute to depression. Conversely, several antagonists of NMDA receptors have antidepressant properties (reviewed in 53). To prevent glutamate excitotoxicity, the glutamate transporters SLC1A2 and SLC1A3 take up excess glutamate into glia. Knockdown of glia glutamate transporters induces glutamate toxicity, whereas genetic deletion of neuronal glutamate transporters does not 54, 55. This emphasizes the physiological importance of glial glutamate transporters as the main source to prevent neuronal damage from excess glutamate.
The results presented here are in agreement with the existing hypothesis of altered glutamate neurotransmission in major depression. We found that the glutamate signaling pathway was the most significantly altered canonical pathway in the LC of MDD patients implying that altered mRNA expression of several glutamate signaling genes significantly impacts the signaling of the entire glutamate pathway. Expression of glutamate signaling genes was altered in different compartments of the LC synapse. The largest and qPCR-confirmed mRNA expression changes of glutamate signaling genes were limited to genes transcripts exclusively expressed in glia cells, including the two glia high affinity glutamate transporters SLC1A2 and SLC1A3. In situ hybridization also confirmed the downregulation of the SLC1A3 transcript. Both transporters play a crucial role in terminating glutamatergic neurotransmission by glial glutamate uptake and thereby maintaining synaptic glutamate concentrations. Glutamate transporters can concentrate glutamate more than 106-fold across cell membranes 56 to protect neurons from glutamate excitotoxicity 57.
Pharmacological enhancement of glial glutamate transporter function has antidepressant effects in humans 58 and in mouse models of depression 59. In addition to glutamate transporters, the gene expression of the enzyme that converts glutamate to glutamine, GLUL, also exhibited decreased mRNA expression in the LC of MDD patients. Gene expression of SLC1A2, SLC1A3, and GLUL have been shown to be significantly decreased in the anterior cingulated and dorsolateral prefrontal cortex of MDD patients 20. GLUL gene expression is also downregulated in the inferior frontal gyrus of depressed suicide victims 60. Downregulation of these three important glutamate signaling may not only perturb the balance between excitatory and inhibitory neurotransmitters but it can lead to cytotoxic glutamate concentrations affecting nearby neurons and glia 61. Our results show that several markers of glia, not of neurons, show strongly reduced mRNA expression in the LC of MDD patients (Table 2), suggesting glial dysfunction in the LC of MDD, but not BPD patients. Similar glia-linked glutamate signaling impairments were recently reported in a rat model of depression 62. Glia cells not only regulate and replenish the synaptic glutamate pool but glia also exchange signaling molecules with neurons and release neurotransmitters, including glutamate, to regulate the strength of synapses 63. Several postmortem studies have demonstrated reduced glial cell number and glial density in cortical and limbic brain regions of depressed subjects 39, 64-67. In our study, the non-neuronal marker ENO1 shows a trend (p=0.1, FC=−1.3) towards reduced gene expression in qPCR experiments, whereas mRNA expression of the astrocyte marker GFAP is significantly reduced in microarray and qPCR gene expression experiments in MDD subjects only (Table 2). This finding suggests that the observed gene expression changes of glial marker genes may reflect transcript expression alterations in astrocytes specifically. This notion is supported by the fact that nearly all glia transcripts in our study with significantly decreased mRNA expression in MDD subjects are mainly expressed in astrocytes 68-70. These include several signaling members that are crucial for astroglia function, such as GJA1, GJB6, S100B, and AQP4 all of which show strongly reduced mRNA expression in MDD subjects. GJA1 and GJB6 form astrocytic gap junctions, which form large molecule-permeable hemichannels providing high-conductance communication pathways between adjacent glia cells that are necessary for the propagation of ATP-induced Ca2+ waves among neighboring astrocytes 71. S100B is a peptide produced by CNS astroglia acting as trophic factor to promote axonal growth and synaptic remodeling 72. S100B for some time has been implicated in major depression 73 and its transcript has been recently shown to be downregulated in the orbitofrontal cortex of suicide victims with a history of major depression 60.
Recent reports of differential astrocytic distribution of glutamate transporters and receptors suggest that astrocytes form different subpopulations. In mouse hippocampus it has been shown that of these subgroups only astrocytes that express glutamate transporters participate in a gap junction network 74. Gap junction uncoupling of cultured cortical astrocytes is associated with a substantial loss in SLC1A2 gene expression and reduced glutamate uptake 75. In addition, the water transport regulating channel AQP4 is not only co-localized with SLC1A2 but loss of AQP4 function downregulates glutamate uptake and SLC1A2 gene expression in cultured astrocytes 76. Thus, the strong concomitant decreases in gene expression of glutamate transporters and their co-localized astrocytic constituents in MDD subjects in the present study suggest a structural and functional linkage between glutamate transporters, gap junctions, and aquaporin-4 channels and point to an overall glial deficit in the LC of MDD patients. Whether the reduced mRNA expression of these astroglia genes is accompanied by glia-related histological or morphological alterations in the LC needs to be investigated.
Neurotrophic receptor gene expression deficits in MDD as observed in the cortex are also present in the LC
Neurotrophic growth factors, such as BDNF and FGF-2, regulating neuronal plasticity, survival, and axonal growth, have been implicated in the pathophysiology of major depression 77. Centrally administered BDNF produced antidepressant effects in animal models of depression 78. Antidepressants have previously been shown to increase BDNF 79, 80 and FGF-2 81 levels and both growth factors promote and mediate antidepressant and anxiolytic effects in rodents 82-84. Conversely, gene expression of some FGF-ligands and -receptors 21 and the BDNF receptor TrkB is downregulated in the AnCg, DLPFC, and hippocampus of depressed and suicidal subjects 21, 85. Our results are in strong agreement with these findings. LC microarray gene expression of growth factors FGF-R3 and TrkB receptor was decreased, whereas the FGF receptor ligand FGF-9 was upregulated (Table 2). This replicates the gene expression pattern observed in AnCg and DLPFC of MDD subjects, suggesting that similar growth factor dysregulation mechanisms may exist in multiple brain regions of the depressed patients. In addition our study revealed that the growth factor transcript IGF-1 was upregulated in LC of MDD subjects. This is consistent with the findings that IGF-1 serum levels are increased in MDD patients, and that IGF-1 concentrations only decrease in responders to fluoxetine treatment 86.
Conclusion
In summary, the present study is the first to use laser-capture microdissected LC from mood disorder patients for gene expression profiling. The results show that several gene families, such as glutamate signaling genes, growth factor genes, and astrocytic genes, which have been previously shown to display altered mRNA expression in forebrain of MDD patients or suicide victims 20, 21, 60, are altered as well in LC of depressed subjects. However, also novel candidates such as gap junction genes exhibit a strongly reduced gene expression in MDD only. The results suggest that glutamate and astrocyte gene expression abnormalities in MDD are functionally linked and may contribute to the perturbation of the LC noradrenergic function. Whether the gene expression changes of glutamate and glia signaling genes reflect underlying pathophysiological changes or simply a diminished presence of certain glia constituents needs to be determined. Recent studies support the concept of altered glutamate transporter function in MDD 58.
Future experiments will need to stereologically quantify glia and neurons in the LC of MDD subjects to determine whether the ratio of glial cells to neurons is altered in patients with MDD. Equally important are studies designed to test whether the transcriptional gene expression chances in the LC of MDD patients are also found in corresponding translated proteins. LCM of single neurons and glia cells of the LC might be valuable tool to gain deeper insight in the signaling abnormalities in MDD in order to improve treatment options for mood disorders.
Supplementary Material
Acknowledgements
We thank S. Burke, J. Fitzpatrick, and M. Hoversten for expert technical assistance and F. Meng for his expertise in statistical analyses. The authors are members of a research consortium supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. An agreement exists between the fund and the University of Michigan, Stanford University, the Weill Medical College of Cornell University, the Universities of California at Davis, and at Irvine, to encourage the development of appropriate findings for research and clinical applications. This study has been supported by the NIMH Conte Center grant #L99MH60398. IAK is supported by the Young Investigator Award from NARSAD and NIH grant #1K99MH081927-01A1.
List of abbreviations
- 5-HT
Serotonin
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- AnCg
Anterior cingulate
- AQP4
Aquaporin 4
- BDNF
Brain-derived neurotrophic factor
- BPD
Bipolar disorder
- CRF
Corticotropin-releasing factor
- DLPFC
Dorsolateral prefrontal cortex
- ENO
Enolase
- FGF
Fibroblast growth factor
- GFAP
Glial fibrillary acidic protein
- GJA1
Gap junction protein, alpha 1
- GJB6
Gap junction protein, beta 6
- GLNS
Glutamine synthetase
- GLUL
Glutamine-ammonia ligase
- GRIA
Glutamate receptor, ionotropic AMPA
- GRIK
Glutamate receptor, ionotropic kainate
- GRM
Glutamate receptor, metabotropic
- IPA
Ingenuity Pathway Analysis
- ISH
In situ hybridization
- LC
Locus coeruleus
- LCM
Laser capture microdissection
- MAS5
Affymetrix Microarray Suite 5
- MDD
Major depressive disorder
- NE
Norepinephrine
- NEF
Neurofilament
- NEFL
Neurofilament, light polypeptide
- NEFM
Neurofilament, heavy polypeptide
- NET
Norepinephrine transporter
- NMDA
N-methyl-D-aspartic acid
- nNOS
Neuronal nitric oxide synthase
- qPCR
Quantitative real-time polymerase chain reaction
- RMA
Robust Multi-Chip Average
- ROD
Relative Optical Density
- S100B
S100 calcium binding protein
- SLC1A2
Solute carrier family 1, member 2
- SLC1A3
Solute carrier family 1, member 3
- SLC17A6
Solute carrier family 17, member 6
- TH
Tyrosine hydroxylase
- TrkB
Tyrosine kinase B receptor
- VGLUT
Vesicular glutamate transporter
References
- 1.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 2.Rivelli S, Jiang W. Depression and ischemic heart disease: what have we learned from clinical trials? Curr Opin Cardiol. 2007;22(4):286–291. doi: 10.1097/HCO.0b013e3281ead011. [DOI] [PubMed] [Google Scholar]
- 3.Stimmel GL, Dopheide JA, Stahl SM. Mirtazapine: an antidepressant with noradrenergic and specific serotonergic effects. Pharmacotherapy. 1997;17(1):10–21. [PubMed] [Google Scholar]
- 4.Wong EH, Sonders MS, Amara SG, Tinholt PM, Piercey MF, Hoffmann WP, et al. Reboxetine: a pharmacologically potent, selective, and specific norepinephrine reuptake inhibitor. Biol Psychiatry. 2000;47(9):818–829. doi: 10.1016/s0006-3223(99)00291-7. [DOI] [PubMed] [Google Scholar]
- 5.Berman RM, Narasimhan M, Miller HL, Anand A, Cappiello A, Oren DA, et al. Transient depressive relapse induced by catecholamine depletion: potential phenotypic vulnerability marker? Arch Gen Psychiatry. 1999;56(5):395–403. doi: 10.1001/archpsyc.56.5.395. [DOI] [PubMed] [Google Scholar]
- 6.Brunello N, Blier P, Judd LL, Mendlewicz J, Nelson CJ, Souery D, et al. Noradrenaline in mood and anxiety disorders: basic and clinical studies. Int Clin Psychopharmacol. 2003;18(4):191–202. doi: 10.1097/00004850-200307000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358(1):55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- 8.Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. 2006;163(11):1905–1917. doi: 10.1176/ajp.2006.163.11.1905. [DOI] [PubMed] [Google Scholar]
- 9.Arango V, Underwood MD, Mann JJ. Fewer pigmented locus coeruleus neurons in suicide victims: preliminary results. Biol Psychiatry. 1996;39(2):112–120. doi: 10.1016/0006-3223(95)00107-7. [DOI] [PubMed] [Google Scholar]
- 10.German DC, Manaye KF, White CL, 3rd, Woodward DJ, McIntire DD, Smith WK, et al. Disease-specific patterns of locus coeruleus cell loss. Ann Neurol. 1992;32(5):667–676. doi: 10.1002/ana.410320510. [DOI] [PubMed] [Google Scholar]
- 11.Loughlin SE, Foote SL, Bloom FE. Efferent projections of nucleus locus coeruleus: topographic organization of cells of origin demonstrated by three-dimensional reconstruction. Neuroscience. 1986;18(2):291–306. doi: 10.1016/0306-4522(86)90155-7. [DOI] [PubMed] [Google Scholar]
- 12.Loughlin SE, Foote SL, Fallon JH. Locus coeruleus projections to cortex: topography, morphology and collateralization. Brain Res Bull. 1982;9(1-6):287–294. doi: 10.1016/0361-9230(82)90142-3. [DOI] [PubMed] [Google Scholar]
- 13.Mason ST, Fibiger HC. Regional topography within noradrenergic locus coeruleus as revealed by retrograde transport of horseradish peroxidase. J Comp Neurol. 1979;187(4):703–724. doi: 10.1002/cne.901870405. [DOI] [PubMed] [Google Scholar]
- 14.Kim MA, Lee HS, Lee BY, Waterhouse BD. Reciprocal connections between subdivisions of the dorsal raphe and the nuclear core of the locus coeruleus in the rat. Brain Res. 2004;1026(1):56–67. doi: 10.1016/j.brainres.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 15.Singewald N, Philippu A. Release of neurotransmitters in the locus coeruleus. Prog Neurobiol. 1998;56(2):237–267. doi: 10.1016/s0301-0082(98)00039-2. [DOI] [PubMed] [Google Scholar]
- 16.Foote SL, Bloom FE, Aston-Jones G. Nucleus locus ceruleus: new evidence of anatomical and physiological specificity. Physiol Rev. 1983;63(3):844–914. doi: 10.1152/physrev.1983.63.3.844. [DOI] [PubMed] [Google Scholar]
- 17.Ordway GA, Schenk J, Stockmeier CA, May W, Klimek V. Elevated agonist binding to alpha2-adrenoceptors in the locus coeruleus in major depression. Biol Psychiatry. 2003;53(4):315–323. doi: 10.1016/s0006-3223(02)01728-6. [DOI] [PubMed] [Google Scholar]
- 18.Ordway GA, Smith KS, Haycock JW. Elevated tyrosine hydroxylase in the locus coeruleus of suicide victims. J Neurochem. 1994;62(2):680–685. doi: 10.1046/j.1471-4159.1994.62020680.x. [DOI] [PubMed] [Google Scholar]
- 19.Klimek V, Stockmeier C, Overholser J, Meltzer HY, Kalka S, Dilley G, et al. Reduced levels of norepinephrine transporters in the locus coeruleus in major depression. J Neurosci. 1997;17(21):8451–8458. doi: 10.1523/JNEUROSCI.17-21-08451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci U S A. 2005;102(43):15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci U S A. 2004;101(43):15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, et al. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods. 2007 doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, et al. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13(6):609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 24.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones EG, Hendry SH, Liu XB, Hodgins S, Potkin SG, Tourtellotte WW. A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J Neurosci Methods. 1992;44(2-3):133–144. doi: 10.1016/0165-0270(92)90006-y. [DOI] [PubMed] [Google Scholar]
- 26.Bernard R, Kerman IA, Meng F, Evans SJ, Amrein I, Jones EG, et al. Gene expression profiling of neurochemically-defined regions of the human brain by in situ hybridization-guided laser capture microdissection. J Neurosci Methods. 2009;178(1):46–54. doi: 10.1016/j.jneumeth.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eymin C, Charney Y, Greggio B, Bouras C. Localization of noradrenaline transporter mRNA expression in the human locus coeruleus. Neurosci Lett. 1995;193:41–44. doi: 10.1016/0304-3940(95)11663-h. [DOI] [PubMed] [Google Scholar]
- 28.Paxinos G, Huang X-F. Altas of the human brainstem. Academic Press; San Diego: 1995. [Google Scholar]
- 29.Kerman IA, Buck BJ, Evans SJ, Akil H, Watson SJ. Combining laser capture microdissection with quantitative real-time PCR: effects of tissue manipulation on RNA quality and gene expression. J Neurosci Methods. 2006;153(1):71–85. doi: 10.1016/j.jneumeth.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Schoor O, Weinschenk T, Hennenlotter J, Corvin S, Stenzl A, Rammensee HG, et al. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques. 2003;35(6):1192–1196. doi: 10.2144/03356rr01. 1198-1201. [DOI] [PubMed] [Google Scholar]
- 31.Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344(1):141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 33.Libus J, Storchova H. Quantification of cDNA generated by reverse transcription of total RNA provides a simple alternative tool for quantitative RT-PCR normalization. Biotechniques. 2006;41(2):156, 158, 160. doi: 10.2144/000112232. passim. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Figueroa AL, Norton CS, Lopez-Figueroa MO, Armellini-Dodel D, Burke S, Akil H, et al. Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry. 2004;55(3):225–233. doi: 10.1016/j.biopsych.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 36.Rage F, Silhol M, Biname F, Arancibia S, Tapia-Arancibia L. Effect of aging on the expression of BDNF and TrkB isoforms in rat pituitary. Neurobiol Aging. 2007;28(7):1088–1098. doi: 10.1016/j.neurobiolaging.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds J, Logan A, Berry M, Dent RG, Gonzales AM, Toescu EC. Age-dependent changes in Fibroblast growth factor 2 (FGF-2) expression in mouse cerebellar neurons. J Cell Mol Med. 2005;9(2):398–406. doi: 10.1111/j.1582-4934.2005.tb00365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster MJ, Herman MM, Kleinman JE, Shannon Weickert C. BDNF and trkB mRNA expression in the hippocampus and temporal cortex during the human lifespan. Gene Expr Patterns. 2006;6(8):941–951. doi: 10.1016/j.modgep.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY, et al. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45(9):1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 40.Valentine GW, Sanacora G. Targeting glial physiology and glutamate cycling in the treatment of depression. Biochem Pharmacol. 2009;78(5):431–439. doi: 10.1016/j.bcp.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata E, Sasaki M, Tohyama K, Otsuka K, Sakai A. Reduced signal of locus ceruleus in depression in quantitative neuromelanin magnetic resonance imaging. Neuroreport. 2007;18(5):415–418. doi: 10.1097/WNR.0b013e328058674a. [DOI] [PubMed] [Google Scholar]
- 42.Zhu MY, Klimek V, Dilley GE, Haycock JW, Stockmeier C, Overholser JC, et al. Elevated levels of tyrosine hydroxylase in the locus coeruleus in major depression. Biol Psychiatry. 1999;46(9):1275–1286. doi: 10.1016/s0006-3223(99)00135-3. [DOI] [PubMed] [Google Scholar]
- 43.Karolewicz B, Szebeni K, Stockmeier CA, Konick L, Overholser JC, Jurjus G, et al. Low nNOS protein in the locus coeruleus in major depression. J Neurochem. 2004;91(5):1057–1066. doi: 10.1111/j.1471-4159.2004.02792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bissette G, Klimek V, Pan J, Stockmeier C, Ordway G. Elevated concentrations of CRF in the locus coeruleus of depressed subjects. Neuropsychopharmacology. 2003;28(7):1328–1335. doi: 10.1038/sj.npp.1300191. [DOI] [PubMed] [Google Scholar]
- 45.Auer DP, Putz B, Kraft E, Lipinski B, Schill J, Holsboer F. Reduced glutamate in the anterior cingulate cortex in depression: an in vivo proton magnetic resonance spectroscopy study. Biol Psychiatry. 2000;47(4):305–313. doi: 10.1016/s0006-3223(99)00159-6. [DOI] [PubMed] [Google Scholar]
- 46.Michael N, Erfurth A, Ohrmann P, Arolt V, Heindel W, Pfleiderer B. Neurotrophic effects of electroconvulsive therapy: a proton magnetic resonance study of the left amygdalar region in patients with treatment-resistant depression. Neuropsychopharmacology. 2003;28(4):720–725. doi: 10.1038/sj.npp.1300085. [DOI] [PubMed] [Google Scholar]
- 47.Pfleiderer B, Michael N, Erfurth A, Ohrmann P, Hohmann U, Wolgast M, et al. Effective electroconvulsive therapy reverses glutamate/glutamine deficit in the left anterior cingulum of unipolar depressed patients. Psychiatry Res. 2003;122(3):185–192. doi: 10.1016/s0925-4927(03)00003-9. [DOI] [PubMed] [Google Scholar]
- 48.Aston-Jones G, Ennis M, Pieribone VA, Nickell WT, Shipley MT. The brain nucleus locus coeruleus: restricted afferent control of a broad efferent network. Science. 1986;234(4777):734–737. doi: 10.1126/science.3775363. [DOI] [PubMed] [Google Scholar]
- 49.Jodo E, Aston-Jones G. Activation of locus coeruleus by prefrontal cortex is mediated by excitatory amino acid inputs. Brain Res. 1997;768(1-2):327–332. doi: 10.1016/s0006-8993(97)00703-8. [DOI] [PubMed] [Google Scholar]
- 50.Noriega NC, Garyfallou VT, Kohama SG, Urbanski HF. Glutamate receptor subunit expression in the rhesus macaque locus coeruleus. Brain Res. 2007;1173:53–65. doi: 10.1016/j.brainres.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singewald N, Kaehler ST, Philippu A. Noradrenaline release in the locus coeruleus of conscious rats is triggered by drugs, stress and blood pressure changes. Neuroreport. 1999;10(7):1583–1587. doi: 10.1097/00001756-199905140-00035. [DOI] [PubMed] [Google Scholar]
- 52.Meldrum B, Garthwaite J. Excitatory amino acid neurotoxicity and neurodegenerative disease. Trends Pharmacol Sci. 1990;11(9):379–387. doi: 10.1016/0165-6147(90)90184-a. [DOI] [PubMed] [Google Scholar]
- 53.Sanacora G, Rothman DL, Mason G, Krystal JH. Clinical studies implementing glutamate neurotransmission in mood disorders. Ann N Y Acad Sci. 2003;1003:292–308. doi: 10.1196/annals.1300.018. [DOI] [PubMed] [Google Scholar]
- 54.Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16(3):675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, et al. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276(5319):1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 56.Levy LM, Warr O, Attwell D. Stoichiometry of the glial glutamate transporter GLT-1 expressed inducibly in a Chinese hamster ovary cell line selected for low endogenous Na+-dependent glutamate uptake. J Neurosci. 1998;18(23):9620–9628. doi: 10.1523/JNEUROSCI.18-23-09620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rothstein JD, Jin L, Dykes-Hoberg M, Kuncl RW. Chronic inhibition of glutamate uptake produces a model of slow neurotoxicity. Proc Natl Acad Sci U S A. 1993;90(14):6591–6595. doi: 10.1073/pnas.90.14.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanacora G, Kendell SF, Levin Y, Simen AA, Fenton LR, Coric V, et al. Preliminary evidence of riluzole efficacy in antidepressant-treated patients with residual depressive symptoms. Biol Psychiatry. 2007;61(6):822–825. doi: 10.1016/j.biopsych.2006.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mineur YS, Picciotto MR, Sanacora G. Antidepressant-like effects of ceftriaxone in male C57BL/6J mice. Biol Psychiatry. 2007;61(2):250–252. doi: 10.1016/j.biopsych.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 60.Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, ffrench-Mullen J, et al. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry. 2009;14(2):175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- 61.Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1(8):623–634. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 62.Zink M, Vollmayr B, Gebicke-Haerter PJ, Henn FA. Reduced expression of glutamate transporters vGluT1, EAAT2 and EAAT4 in learned helpless rats, an animal model of depression. Neuropharmacology. 2009 doi: 10.1016/j.neuropharm.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Miller G. Neuroscience. The dark side of glia. Science. 2005;308(5723):778–781. doi: 10.1126/science.308.5723.778. [DOI] [PubMed] [Google Scholar]
- 64.Bowley MP, Drevets WC, Ongur D, Price JL. Low glial numbers in the amygdala in major depressive disorder. Biol Psychiatry. 2002;52(5):404–412. doi: 10.1016/s0006-3223(02)01404-x. [DOI] [PubMed] [Google Scholar]
- 65.Miguel-Hidalgo JJ, Wei J, Andrew M, Overholser JC, Jurjus G, Stockmeier CA, et al. Glia pathology in the prefrontal cortex in alcohol dependence with and without depressive symptoms. Biol Psychiatry. 2002;52(12):1121–1133. doi: 10.1016/s0006-3223(02)01439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ongur D, Drevets WC, Price JL. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc Natl Acad Sci U S A. 1998;95(22):13290–13295. doi: 10.1073/pnas.95.22.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Webster MJ, Knable MB, Johnston-Wilson N, Nagata K, Inagaki M, Yolken RH. Immunohistochemical localization of phosphorylated glial fibrillary acidic protein in the prefrontal cortex and hippocampus from patients with schizophrenia, bipolar disorder, and depression. Brain Behav Immun. 2001;15(4):388–400. doi: 10.1006/brbi.2001.0646. [DOI] [PubMed] [Google Scholar]
- 68.Giaume C, McCarthy KD. Control of gap-junctional communication in astrocytic networks. Trends Neurosci. 1996;19(8):319–325. doi: 10.1016/0166-2236(96)10046-1. [DOI] [PubMed] [Google Scholar]
- 69.Nagy JI, Patel D, Ochalski PA, Stelmack GL. Connexin30 in rodent, cat and human brain: selective expression in gray matter astrocytes, co-localization with connexin43 at gap junctions and late developmental appearance. Neuroscience. 1999;88(2):447–468. doi: 10.1016/s0306-4522(98)00191-2. [DOI] [PubMed] [Google Scholar]
- 70.Sen J, Belli A. S100B in neuropathologic states: the CRP of the brain? J Neurosci Res. 2007;85(7):1373–1380. doi: 10.1002/jnr.21211. [DOI] [PubMed] [Google Scholar]
- 71.Bennett MV, Contreras JE, Bukauskas FF, Saez JC. New roles for astrocytes: gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26(11):610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arolt V, Peters M, Erfurth A, Wiesmann M, Missler U, Rudolf S, et al. S100B and response to treatment in major depression: a pilot study. Eur Neuropsychopharmacol. 2003;13(4):235–239. doi: 10.1016/s0924-977x(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 73.Rothermundt M, Arolt V, Wiesmann M, Missler U, Peters M, Rudolf S, et al. S-100B is increased in melancholic but not in non-melancholic major depression. J Affect Disord. 2001;66(1):89–93. doi: 10.1016/s0165-0327(00)00321-9. [DOI] [PubMed] [Google Scholar]
- 74.Wallraff A, Odermatt B, Willecke K, Steinhauser C. Distinct types of astroglial cells in the hippocampus differ in gap junction coupling. Glia. 2004;48(1):36–43. doi: 10.1002/glia.20040. [DOI] [PubMed] [Google Scholar]
- 75.Figiel M, Allritz C, Lehmann C, Engele J. Gap junctional control of glial glutamate transporter expression. Mol Cell Neurosci. 2007;35(1):130–137. doi: 10.1016/j.mcn.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 76.Zeng XN, Sun XL, Gao L, Fan Y, Ding JH, Hu G. Aquaporin-4 deficiency down-regulates glutamate uptake and GLT-1 expression in astrocytes. Mol Cell Neurosci. 2007;34(1):34–39. doi: 10.1016/j.mcn.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 77.Turner CA, Akil H, Watson SJ, Evans SJ. The fibroblast growth factor system and mood disorders. Biol Psychiatry. 2006;59(12):1128–1135. doi: 10.1016/j.biopsych.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 78.Siuciak JA, Lewis DR, Wiegand SJ, Lindsay RM. Antidepressant-like effect of brain-derived neurotrophic factor (BDNF) Pharmacol Biochem Behav. 1997;56(1):131–137. doi: 10.1016/S0091-3057(96)00169-4. [DOI] [PubMed] [Google Scholar]
- 79.Duman RS. Role of neurotrophic factors in the etiology and treatment of mood disorders. Neuromolecular Med. 2004;5(1):11–25. doi: 10.1385/NMM:5:1:011. [DOI] [PubMed] [Google Scholar]
- 80.Manji HK, Duman RS. Impairments of neuroplasticity and cellular resilience in severe mood disorders: implications for the development of novel therapeutics. Psychopharmacol Bull. 2001;35(2):5–49. [PubMed] [Google Scholar]
- 81.Maragnoli ME, Fumagalli F, Gennarelli M, Racagni G, Riva MA. Fluoxetine and olanzapine have synergistic effects in the modulation of fibroblast growth factor 2 expression within the rat brain. Biol Psychiatry. 2004;55(11):1095–1102. doi: 10.1016/j.biopsych.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 82.Monteggia LM, Barrot M, Powell CM, Berton O, Galanis V, Gemelli T, et al. Essential role of brain-derived neurotrophic factor in adult hippocampal function. Proc Natl Acad Sci U S A. 2004;101(29):10827–10832. doi: 10.1073/pnas.0402141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turner CA, Gula EL, Taylor LP, Watson SJ, Akil H. Antidepressant-like effects of intracerebroventricular FGF2 in rats. Brain Res. 2008;1224:63–68. doi: 10.1016/j.brainres.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez JA, Clinton SM, Turner CA, Watson SJ, Akil H. A new role for FGF2 as an endogenous inhibitor of anxiety. J Neurosci. 2009;29(19):6379–6387. doi: 10.1523/JNEUROSCI.4829-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60(8):804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- 86.Deuschle M, Blum WF, Strasburger CJ, Schweiger U, Weber B, Korner A, et al. Insulin-like growth factor-I (IGF-I) plasma concentrations are increased in depressed patients. Psychoneuroendocrinology. 1997;22(7):493–503. doi: 10.1016/s0306-4530(97)00046-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.