Abstract
Here we describe the components of a histone deacetylase (HDAC) complex that we term the CoREST-HDAC complex. CoREST-HDAC is composed of polypeptides distinct from previously characterized HDAC1/2-containing complexes such as the mSin3 and nucleosome remodeling and deacetylating (NRD, also named NURD, NuRD) complex. Interestingly, we do not observe RbAp46 and RbAp48 in this complex, although these proteins have been observed in all previously identified complexes and are thought to be part of an HDAC1/2 core. We identify the transcriptional corepressor CoREST and a protein with homology to polyamine oxidases as components of CoREST-HDAC. The HDAC1/2-interacting region of CoREST is mapped to a 179-aa region containing a SANT domain, a domain found in other HDAC1/2-interacting proteins such as NCoR, MTA1, and MTA2. Furthermore, we demonstrate that the corepressor function of CoREST depends on this region. Although CoREST initially was cloned as a corepressor to REST (RE1 silencing transcription factor/neural restrictive silencing factor), we find no evidence for the existence of the eight-zinc finger REST transcription factor as an interacting partner in this complex; however, we do find evidence for association of the putative oncogene ZNF 217 that contains eight zinc fingers.
Mammalian human histone deacetylases (HDACs) have been observed to exist in large, multisubunit protein complexes (1). HDAC1 and HDAC2, in particular, have been well characterized and previously found predominantly in either an mSin3-containing complex or in a complex containing the ATP-dependent chromatin remodeling protein CHD4 (also known as Mi-2) and MTA2 (2–5). The former complex is referred to as the mSin3 complex and the latter, as the nucleosome remodeling and deacetylating (NRD) complex. The mSin3 complex is recruited by DNA-binding transcription factors such as the unliganded nuclear hormone receptors and the Mad/Max heterodimer, whereas the NRD complex has been shown to be recruited by the transcription factors Ikaros and hunchback (6, 7). The emerging model of mSin3 function is one in which the complex is recruited as a repressor under one set of conditions and released and exchanged for histone acetyltransferase coactivators under a different set of conditions. In contrast, less is currently understood about the regulation and function of the NRD complex at the transcriptional level.
In the course of purifying HDAC1/2-associated proteins, we obtained peptide sequence corresponding to a hypothetical protein of unknown function (KIAA0071). The partially translated product possesses homology to MTA1 and MTA2 in the region of the SANT domain, and we hypothesized that the full-length protein derived from KIAA0071 might be a component of an uncharacterized HDAC1/2-containing complex. Antibodies were generated against KIAA0071 and used to purify associated polypeptides. At the same time, an effort was undertaken to clone the full-length cDNA of KIAA0071. While this work was in progress, Andres et al. (8) reported the cloning of full-length KIAA0071 as CoREST, a protein found as a corepressor to the REST (RE1 silencing transcription factor/neural restrictive silencing factor) transcription factor via interaction with its carboxyl terminus. Interestingly, REST has been shown to contain two repression domains: the N-terminal domain recruits the mSin3 HDAC complex, and the C-terminal domain recruits CoREST (9–11). Here, we demonstrate that CoREST is a component of an HDAC complex distinct from the mSin3 and NRD complexes. We describe a polypeptide complex containing CoREST and HDAC1/2 and demonstrate that CoREST repressor function depends on a domain required for the interaction of CoREST with HDAC1/2.
Experimental Procedures
Antibodies.
Antibodies against RbAp48, HDAC1, and HDAC2 have been described (12, 13). Antibodies against MTA2 and CoREST were generated as follows. Synthetic peptides corresponding to an internal fragment of MTA2 and the C terminus of CoREST were conjugated covalently to keyhole limpet hemocyanin (Pierce) and used to immunize rabbits. Antibodies were purified from the antisera on peptide affinity columns, prepared by using the Sulfolink system (Pierce). For immunoprecipitation purposes, 3 mg of antibody was crosslinked to 2 ml of protein A-agarose bead slurry (GIBCO/BRL) by using dimethyl pimelimidate (Pierce). Antibodies to mSin3 and RbAp46 were purchased from Santa Cruz Biotechnology. FLAG M2 antibody was purchased from Sigma.
Purification of CoREST-Associated Polypeptides.
HeLa cell extract was prepared by lysis of 1.5 × 109 cells in 30 ml of ice-cold JLB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/10% glyercol/0.5% Triton X-100) plus a complete protease inhibitor mixture (Boehringer Mannheim). The lysate was incubated for 30 min on ice with occasional rocking and subsequently clarified by centrifugation at 20,000 × g at 4°C for 30 min. To 15 ml of the extract, 200 μl of anti-CoREST beads was added and allowed to incubate with rotation at 4°C for 3.5 h. As a negative control, a similar immunoprecipitation was performed with anti-CoREST beads that had been preincubated with 500 μg of peptide for 15 min. Beads were quickly washed twice with 15 ml of MSWB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1 mM EDTA/0.1% Nonidet P-40) followed by three 5-min washes of 15 ml of MSWB at room temperature. Beads were eluted by incubation with 50 μl of 1.2× SDS/PAGE loading buffer followed by 10 μl of double-distilled water to extract any remaining protein. The combined eluate was adjusted to 10% 2-mercapoethanol, boiled for 3 min, and separated by 10% SDS/PAGE followed by staining with colloidal Coomassie blue (NOVEX, San Diego). Bands were excised and submitted for peptide mass spectrometric sequencing (Harvard Microchemistry Facility, Cambridge, MA).
DNA Constructs.
A partial clone of CoREST (KIAA0071) was generously provided by the Kazusa DNA Research Institute (Chiba, Japan). The 5′ fragment of KIAA0071 (nucleotides 1–331) was PCR-amplified from a λ ZAP HeLa cDNA library (Stratagene) and ligated to KIAA0071 through a unique AvaII site to generate full-length CoREST, which subsequently was cloned as a HindIII–EcoRI fragment into pBluescript (SK−) (Stratagene). FLAG-epitope-tagged CoREST constructs were generated by PCR with a 3′ primer containing the FLAG-epitope coding sequence. All PCR-generated CoREST expression constructs were subcloned into pCDNA1.1amp (Invitrogen) as HindIII–EcoRI fragments.
The Gal4-VP16 expression plasmids containing CoREST and various deletions were constructed by subcloning PCR-amplified, FLAG-epitope-tagged fragments into the HindII–EcoRI sites of pSP/GAL(1–147)-VP16. The GAL4-luc reporter and GAL4-VP16 plasmids were described (20).
Cloning of p110b.
A blast search of the GenBank databases with the peptide sequence obtained from the p110b polypeptide resulted in the identification of a 2,985-bp mRNA sequence containing an ORF of 2,559 bp (GenBank accession number AB011173). This clone was generously provided by the Kazusa DNA Research Institute and used to generate a C-terminally FLAG-epitope-tagged p110 expression construct by PCR. This construct was inserted in the NotI and EcoRI sites of the pBJ5 mammalian expression vector.
Transfections, Immunoprecipitation, and Western Blotting.
FLAG-epitope-tagged CoREST and p110 constructs were transiently transfected by electroporation into simian virus 40 large T antigen (T-Ag) Jurkats. Forty-eight hours posttransfection, cells were lysed in JLB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/10% glycerol/0.5% Triton X-100) containing a complete protease inhibitor mixture (Boehringer Mannheim). Lysis proceeded for 15 min at 4°C, after which the cellular debris was pelleted by centrifugation at 20,000 × g for 2 min. Recombinant proteins were immunoprecipitated from the supernatant by incubation with α-FLAG M2 agarose affinity gel (Sigma) for 2 h at 4°C. For Western blot analysis, the beads were washed three times for 5 min at room temperature with MSWB, and the proteins were separated by 10% SDS/PAGE. For enzyme activity assays, the beads were washed three times with JLB at 4°C.
HDAC Activity Assays.
3[H]Acetate-incorporated histones were isolated from trichostatin A (TSA)-treated HeLa cells by hydroxyapatite chromatography as described (3). Immunoprecipitates were incubated with 1.3 mg (10,000 dpm) of histones for 2 h at 37°C. HDAC activity was determined by scintillation counting of the ethyl-acetate-soluble 3[H]acetic acid (12).
Luciferase Assays.
For each sample, 107 simian virus 40 T-Ag Jurkat cells were transfected in duplicate or triplicate by electroporation with 100 ng of luciferase reporter, 100 ng of cytomegalovirus—β-galactosidase to serve as a control for protein expression levels, 100 ng of expression construct, and 3.7 μg of carrier DNA. Twenty-four hours posttransfection, the samples were harvested and lysed after a 15-h treatment of 0 or 50 nM TSA. Luciferase activity was determined by using a standard luciferase assay (Promega). Luciferase values were normalized for transfection efficiency by dividing by the β-galactosidase activity. These assays were performed three times with similar results.
Results
Identification of HDAC2-Associated Proteins.
In a previous study, HDAC2-associated polypeptides were purified from HeLa cell extract by immunoaffinity chromatography (3). Specifically bound polypeptides were separated by SDS/PAGE and visualized by silver staining. Bands were specifically excised and submitted for peptide mass spectrometric sequencing. Several polypeptides identified were components of the then-described NRD complex, whereas others corresponded to proteins or expressed sequence tags (ESTs) of unknown function. In particular, peptide sequences were obtained for a polypeptide of apparent molecular mass of 60 kDa that corresponded to the hypothetical protein KIAA0071. Sequence features of KIAA0071 include two SANT domains separated by a highly charged intervening domain. KIAA0071 since has been fully cloned by Andres et al. (8) and subsequently will be referred to as CoREST.
CoREST Is a Component of an HDAC1/2 Complex.
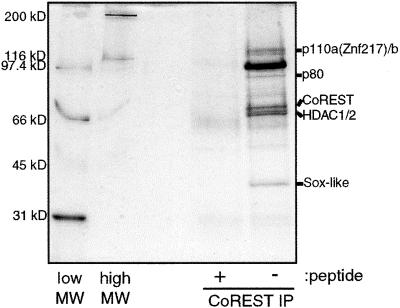
In an effort to characterize CoREST further, antibodies were raised to a C-terminal peptide epitope of CoREST and used to immunopurify CoREST-associated polypeptides from HeLa cell extract. The polypeptides specifically bound to agarose-immobilized anti-CoREST antibodies were eluted with the peptide antigen, separated by SDS/PAGE, and visualized by silver staining (Fig. 1). The pattern of associated proteins is distinct from that of the mSin3 and NRD complexes. To characterize these proteins further, bands from the gel were excised and sequenced by mass spectrometry. In total, seven bands that appeared to be stoichiometric with CoREST were excised and sequenced. As expected, sequences derived from CoREST, HDAC1, and HDAC2 were obtained. Of the remaining four polypeptides, none have been demonstrated previously to interact with histone deacetylases. p40 is a Sox-like protein, p110b contains homology to polyamine oxidases, p110a is ZNF217, an eight-zinc finger protein, and p80 is a hypothetical protein of unknown function (14). Sequences derived from RbAp46 and RbAp48 were not observed; nor were signature components such as mSin3, MTA2, or CHD4 of the mSin3 and NRD complexes.
Figure 1.
Silver stain of CoREST complex. CoREST-associated polypeptides were immunoprecipitated from HeLa extract in the presence or absence of a competitor peptide and separated by SDS/PAGE. Lanes marked “low MW” and “high MW” are protein molecular weight standards.
CoREST Interacts with HDAC1 and HDAC2 and Is a Component of a Distinct HDAC Complex.
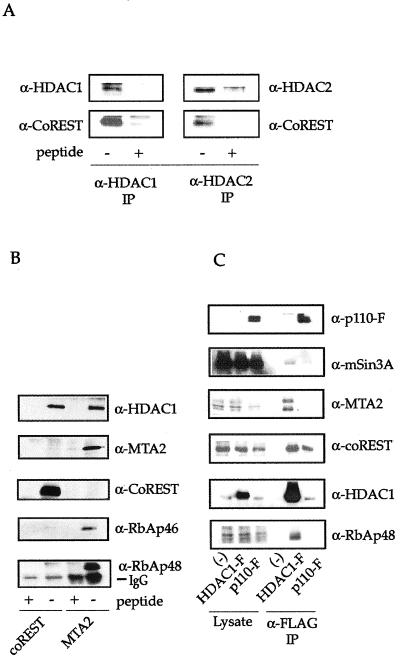
To confirm the association between CoREST and HDAC1/2, coimmunoprecipitation–Western blotting experiments were performed. HDAC1/2 were immunoprecipitated from T-Ag Jurkat extract and Western blotted with antisera raised against CoREST. As expected, a 60-kDa band was specifically recognized in both HDAC1 and HDAC2 immunoprecipitations (Fig. 2A). To establish further that CoREST-associated polypeptides are components of an HDAC complex distinct from mSin3 and NRD, CoREST immunoprecipitations were analyzed for the presence of mSin3 and MTA2, two components of the mSin3 and NRD complexes, respectively (Fig. 2B and data not shown). Neither was detected by Western blotting, suggesting that the CoREST-HDAC complex exists as a distinct entity.
Figure 2.
Coimmunoprecipitation–Western blot experiments. (A) HDAC1 and HDAC2 were immunoprecipitated from T-Ag Jurkat extracts and Western blotted for CoREST. (B) Polypeptides specific to CoREST and MTA2 were immunoprecipitated from HeLa extract and Western blotted. (C) Polypeptides associated with recombinant p110-FLAG were immunoprecipitated and Western blotted for components of various HDAC complexes.
p110b Interacts with the CoREST-HDAC Complex.
Peptide sequence analysis of the p110b polypeptide immunoprecipitated by the CoREST antibody revealed that it corresponded to the previously identified p110b polypeptide of the immunoprecipitated HDAC2 complex (3). A blast search of the GenBank databases with this peptide sequence resulted in the identification of a 2,985-bp mRNA sequence containing an ORF of 2,559 bp (GenBank accession number AB011173). This clone was provided by the Kazusa DNA Research Institute and used to generate a C-terminally FLAG-epitope-tagged p110b expression construct by PCR.
Coimmunoprecipiation and Western blot analysis were performed to determine whether p110b is a specific component of the CoREST-HDAC1 complex or if it is also present in the mSin3 and NRD complexes. FLAG-epitope-tagged p110b and HDAC1 were transiently expressed in T-Ag Jurkat cells, immunopurified by using anti-FLAG agarose, and subjected to Western blot analysis with antibodies to FLAG, mSin3A, MTA2, CoREST, HDAC1, RbAp48 (Fig. 2C), and CHD4 (data not shown). HDAC1 specifically coimmunoprecipitates with all of the proteins of the mSin3, NRD, and coREST-HDAC1 complexes, as expected. p110b coimmunoprecipitates strongly with CoREST and HDAC1 and only very weakly with mSin3A. This suggests that although it is a component of the CoREST/HDAC complex, p110b may also associate with mSin3A-containing complexes. However, that p110b also does not coimmunoprecipitate RbAp48 further demonstrates that it is not a component of previously identified histone deacetylase complexes. Interestingly, p110b appears as a doublet, suggesting that it contains posttranslational modifications. This electromobility shift does not appear to be due to phosphorylation, and both forms of p110b associate with HDAC1 (data not shown).
A Region Containing the N-Terminal SANT Domain of CoREST Is Required for HDAC1/2 Association.
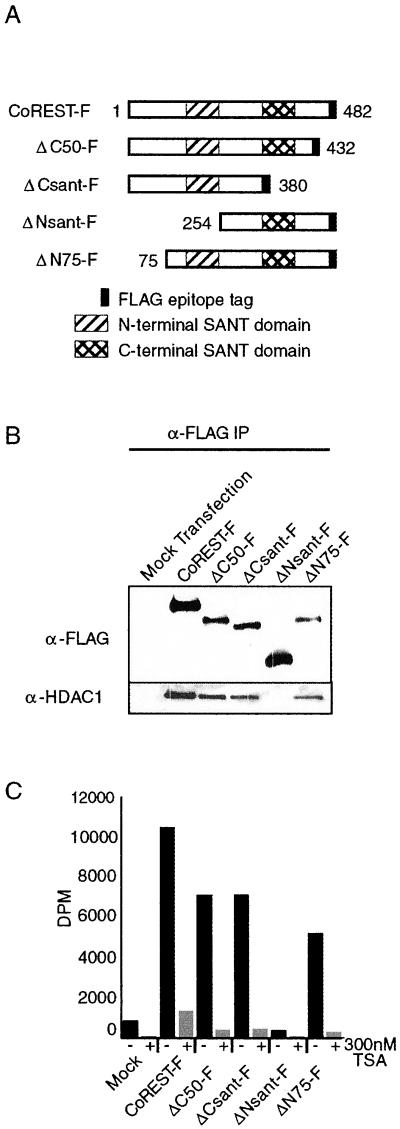
To identify the HDAC1/2-interacting region of CoREST, a series of FLAG-epitope-tagged deletions of CoREST was constructed (Fig. 3A) and transiently expressed in T-Ag Jurkat cells. Anti-FLAG immunoprecipitates were separated by SDS/PAGE and analyzed by Western blotting for the presence of HDAC1. FLAG-tagged full-length CoREST coimmunoprecipitated endogenous HDAC1 specifically. The immunoprecipitation of the deletion mutants of CoREST revealed that a 179-aa region containing the N-terminal SANT domain is critical for interaction, because the deletion of this region resulted in loss of both HDAC1 association (Fig. 3B) and HDAC activity (Fig. 3C), as measured by a tritiated histone deacetylase assay.
Figure 3.
A 179-aa region containing the N-terminal SANT domain of CoREST is required for interaction with HDAC1. (A) Four different deletion constructs were generated to map potential HDAC1-interacting regions. (B) Coimmunoprecipitation–Western blot experiments demonstrate that a 179-aa region containing the N-terminal SANT domain of CoREST is required for association with HDAC1. (C) ΔNsant-F lacks associated histone deacetylase activity.
Transcriptional Repression by CoREST Requires the 179-aa Region Containing the N-Terminal SANT Domain.
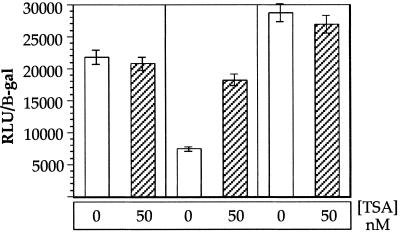
It has been shown previously that CoREST can repress transcription from a reporter gene (9). To investigate whether the CoREST-specific repression is mediated by an interaction with HDAC, we deleted the 179-aa region containing the N-terminal SANT domain (ΔNsant-F) and constructed a fusion protein containing the DNA-binding domain of GAL4 and the VP16 transactivator. A similar fusion was made with the full-length CoREST. To test the ability of ΔNsant-F to repress transcription, T-Ag Jurkat cells were transfected with a GAL4-luciferase reporter and either full-length CoREST or GAL4-ΔNsantF-VP16 fusions. Twenty-four hours posttransfection, the samples were harvested and lysed after a 15-h treatment of 0 or 50 nM TSA. As expected, GAL4-VP16 alone activated transcription efficiently. Full-length GAL4-CoREST-VP16 was a potent repressor relative to GAL4-VP16. Conversely, deletion of the 179-aa region important for interaction with HDAC resulted in a loss of an ability to repress transcription relative to full-length CoREST (Fig. 4). In addition, 50 nM TSA treatment derepressed the activity of GAL4-COREST-VP16, whereas it had little effect on the activity of GAL4-ΔNsantF-VP16 or GAL4-VP16. The remaining CoREST deletions that maintained the capacity to interact with HDAC also exhibited repressor activity in the same luciferase reporter gene assay (data not shown).
Figure 4.
A 179-aa region containing the N-terminal SANT domain is required for transcriptional repression. Cells were transfected with Gal4-VP16 expression constructs fused to nothing, full-length CoREST, or ΔNsant-F. Empty Gal4-VP16 is a potent transcriptional activator, and full-length CoREST is a potent transcriptional repressor. The repression by full-length CoREST is derepressed with TSA treatment. Relative to full-length CoREST, ΔNsant-f is derepressed, demonstrating that the HDAC-interacting region of the N-terminal SANT domain is required for full transcriptional repression.
Discussion
Here, we identify a complex of proteins associated with HDAC1 and HDAC2. Interestingly, these proteins represent a novel HDAC1/2 complex distinct and separate from the previously characterized mSin3 and NRD complexes. Whereas the mSin3 and NRD complexes both contain the HDAC1/2-RbAp46/48 core complex, CoREST-HDAC is noticeably lacking RbAp46 and RbAp48 as determined by coimmunoprecipitation studies with CoREST and p110. Although it is possible that RbAp46/48 are displaced by the antibodies to CoREST and the FLAG tag of p110, we find this to be unlikely, as RbAp46 and RbAp48 are thought to interact with HDAC1/2 in a direct fashion and, hence, should be relatively unaffected by interactions with CoREST or p110 (12, 15). A component of the CoREST-HDAC complex, CoREST, was identified previously as a corepressor for the REST/NRSF transcription factor. It is known that REST is an eight-zinc finger transcription factor that contains two independent repression domains: an N-terminal domain that recruits mSin3 and a C-terminal domain that recruits CoREST. Combined with the data presented in this paper that CoREST is part of the CoREST-HDAC histone deacetylase complex, it is now apparent that REST has the potential to mediate transcriptional repression via two distinct HDAC complexes. Although CoREST first was identified as a REST-interacting protein, we believe that CoREST forms a stable histone deacetylase complex that is recruited by transcription factors, of which REST is perhaps one of many. Whereas CoREST immunoprecipitations yielded seemingly stoichiometric quantities of HDAC1/2, p110, and other associated proteins by silver staining, we failed to detect REST by either silver staining or Western blotting (data not shown). However, we detected near-stoichiometric amounts of ZNF217, another eight-zinc finger transcription factor that is a candidate oncogene.
CoREST contains two 50-aa SANT domains that resemble the DNA-binding domains of Myb-related DNA-binding proteins that are also present in the HDAC-interacting proteins MTA1 and MTA2 as well as the transcription factors SWI3, ADA3, NCoR, and TFIIIB, for which SANT is named (16). In this study, we have identified a region containing the N-terminal SANT domain of CoREST, as necessary for interaction with HDAC1. Deletion of this region results in loss of associated HDAC1 and histone deacetylase activity. Furthermore, when CoREST is fused to the GAL4 DNA-binding domain and VP16 transactivator, the N-terminal SANT domain-containing region of CoREST is necessary for efficient transcriptional repression. Removal of this region causes transcriptional derepression relative to the full-length protein, consistent with a role for histone deacetylase activity in the repressive properties of CoREST.
Sequence database searches reveal copies of the SANT domain in a number of proteins that participate in transcriptional regulation. In a blastp search, CoREST displayed significant similarity with two ESTs from the human brain (ESTs BAA91872 and BAA92581). These ESTs are identical except for their C termini, and both contain dual SANT domains that are nearly identical to the those found in CoREST. The high degree of identity in the N-terminal SANT domains between CoREST and the unknown ESTs suggests the possibility that the proteins encoded by the unknown ESTs potentially could interact with histone deacetylases in a context similar to that of the HDAC-CoREST complex.
p110b shares weak homology to several FAD-containing enzymes and exhibits the highest degree of similarity to polyamine oxidases. However, immunoprecipitated p110-F does not appear to bind to FAD-agarose, nor does it display any FAD-, NAD-, or NADH-dependent histone deacetylase activity. In a recent report of a biochemical purification of a histone deacetylase complex (cI) containing CoREST and p110b, it was demonstrated that p110b contains FAD by in vivo radiolabeling HeLa cells with [3H]riboflavin (17). The role of p110 in the coREST-HDAC1 complex is unclear, although it may contribute a novel enzymatic activity.
The acetylation status of histones plays an important role in transcriptional regulation. The identification in recent years of histone deacetylases and histone acetyltransferases and their associated protein complexes has shed light on the mechanisms of repression and activation of numerous transcription factors. Yet the identification of multiple and distinct complexes of the same histone deacetylases suggests a transcriptional regulation process more complex than the mere recruitment of histone deacetylase activity. It is still unclear which role, if any, accessory proteins in the complex may play other than serving as protein–protein contacts. In vitro, the ATP-dependent chromatin-remodeling activity of NRD facilitates histone deacetylation of chromatinized templates (3, 4). In the yeast Sir2 protein, histone deacetylase activity is stimulated by the redox cofactor NAD (18), possibly by acting as an electrophilic, oxocarbenium ion-based catalyst of N-acetyl amide hydrolysis (19). Of interest, p110 in the CoREST-HDAC complex possesses homology to enzymes involved in redox processes, although no enzymatic activity has been demonstrated yet. A speculative model for the role of different HDAC complexes is that the mSin3 complex is recruited for simple deacetylation of dynamically regulated promoters, whereas the NRD and CoREST-HDAC complexes are recruited to promoters that require stable repression (e.g., tissue-specific silencing) or that are heritable states (e.g., genomic imprinting). The associated non-HDAC enzymatic activities may play a role in determining the nature of the repression.
Acknowledgments
We thank Chris Hassig and Mary Kay Pflum for helpful discussions and Bosiljka Tasic for help in producing antibodies against CoREST and MTA2. We thank William Lane and the Harvard Microchemistry Facility for help in peptide sequencing and the National Institutes of Health Cell Culture Center and Sera/Source for technical assistance. This work was supported by the National Institute of General Medical Sciences. Predoctoral fellowships from the National Science Foundation to J.K.T, A.Y., and C.M.G. are gratefully acknowledged. S.L.S. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations
- HDAC
human histone deacetylase
- NRD
nucleosome remodeling and deacetylating
- T-Ag
large T antigen
- TSA
trichostatin A
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB011173).
References
- 1.Ng H H, Bird A. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 2.Hassig C A, Schreiber S L. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 3.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 4.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 6.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison J A, Bienz M, Muller J. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 8.Andres M E, Burger C, Peral-Rubio M J, Battaglioli E, Anderson M E, Grimes J, Dallman J, Ballas N, Mandel G. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimes J A, Nielsen S J, Battaglioli E, Miska E A, Speh J C, Berry D L, Atouf F, Holdener B C, Mandel G, Kouzarides T. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, Myers S J, Dingledine R. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 11.Roopra A, Sharling L, Wood I C, Briggs T, Bachfischer U, Paquette A J, Buckley N J. Mol Cell Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 13.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins C, Rommens J M, Kowbel D, Godfrey T, Tanner M, Hwang S I, Polikoff D, Nonet G, Cochran J, Myambo K, et al. Proc Natl Acad Sci USA. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aasland R, Stewart A F, Gibson T. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 17.Humphrey, G. W., Wang, Y., Russanova, V. R., Hirai, T., Qin, J., Nakatani, Y. & Howard, B. H. (2000) J Biol. Chem., in press. [DOI] [PubMed]
- 18.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 19.Tanner K G, Landry J, Sternglanz R, Denu J M. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. . (First Published December 5, 2000, 10.1073/pnas.250422697) [DOI] [PMC free article] [PubMed] [Google Scholar]