Abstract
Background
Cell penetrating peptides (CPPs) have been used to deliver nucleotide-based therapeutics to cells, but this approach has produced mixed results. Ionic interactions and covalent bonds between the CPPs and the cargos may inhibit the effectiveness of the CPPs or interfere with the bioactivity of the cargos.
Methods
We have created a bifunctional chimeric protein that binds DNA using the p50 domain of the NF-κB transcription factor and is functionalized for delivery with the TAT CPP. The green fluorescent protein (GFP) has been incorporated for tracking delivery. The new chimeric protein, p50-GFP-TAT, was compared to p50-GFP, GFP-TAT and GFP as controls for the ability to transduce PC12 cells with and without oligonucleotide cargos.
Results
The p50-GFP-TAT construct can deliver 30bp and 293bp oligonucleotides to PC12 cells with an optimal ratio of 1.89 protein molecules per base pair of DNA length. This correlation was validated through the delivery of a fluorescent protein transgene encoded in a plasmid to PC12 cells.
Conclusion
Self-assembling CPP-based bifunctional fusion proteins can be engineered for the non-viral delivery of nucleotide-based cargos to mammalian cells.
General significance
This work represents an important step forward in the rational design of protein-based systems for the delivery of macromolecular cargos.
Keywords: Cell penetrating peptides, TAT, p50 DNA-binding domain, oligonucleotide delivery, protein/DNA ratio
1. Introduction
The cellular plasma membrane is a formidable protective barrier, and considerable effort has been aimed at the development of delivery strategies for the introduction of exogenous biomacromolecules into mammalian cells both in vitro and in vivo [1, 2]. The search for efficient delivery vehicles has been partially motivated by the development of many large and potentially valuable mechanism-based therapeutic molecules including peptides, proteins, natural products, carbohydrates and nucleotide-based molecules. Improved targeting and delivery strategies will increase the utility of available therapeutics and help realize the development of new treatments for a variety of devastating diseases and disorders.
Cell penetrating peptides (CPPs), (also known as Trojan horse peptides, protein transduction domains, and membrane translocating sequences) are short, mostly basic peptides that have received a good deal of attention as potential delivery vehicles due to their ability to deliver various cargos to a wide variety of cell and tissue types [3-7]. One of the most well characterized CPPs is the TAT peptide which is derived from an HIV trans-activation protein [8, 9]. Despite a number of publications demonstrating the successful use of TAT for the delivery of proteins, nanoparticles, liposomes and phage particles into mammalian cells in vitro or in vivo [10–13], mixed results have been reported on the use of TAT for the delivery of nucleotide-based cargos. Several studies have reported TAT-mediated delivery of DNA oligonucleotides, antisense DNA, or plasmids, and the results varied from no obvious delivery [14–19], to no desired biological activity [20], to the observation of a fair percentage of transfected cells [21]. Modified synthetic peptides based on CPP sequences have been developed in an attempt to improve these results[14, 17, 18, 22, 23], but further improvements will be required before TAT-inspired methods can be used therapeutically.
Two major linkage strategies have been used in CPP-mediated delivery of nucleotide cargos: CPPs and nucleotides are mixed together to create non-covalent electrostatic complexes, or covalent linkages are introduced between CPPs and the nucleotide-based molecules [24–27]. In general, the non-covalently bound constructs have outperformed the covalently linked CPP-nucleotides [28]. The overall poor results observed with CPP-mediated nucleotide-based cargo delivery, however, may be caused by either charge neutralization between the CPP and the nucleotide cargo, which inhibits the transduction ability of the CPP, or interference of the CPP with the biological function of the nucleotide cargos.
When the two conjugation approaches are combined, it has been shown that the addition of non-covalently bound CPPs improves the delivery of covalently conjugated CPP-cargos [24, 25]. In addition, the introduction of other peptides, polymeric domains, or nuclear transport signals [29] with a stronger affinity for DNA may facilitate the DNA delivery. For example, when the TAT peptide has been conjugated to poly-lysine [14], poly-TAT [19] or the Mu peptide [18], the TAT fusions exhibit significantly improved DNA delivery abilities compared to the TAT peptide alone. Taken together, these results suggest that an optimal strategy may involve the creation of non-covalent protein/DNA complexes such that the DNA binding is mediated by a structured domain that is spatially separated from the cationic cell penetrating domain.
In this study, a bifunctional recombinant fusion protein was designed to test this approach. This new chimeric protein, p50-GFP-TAT (PGT) consists of a specific DNA binding domain (p50), a CPP domain (TAT), and a fluorescent labeling domain (GFP). The homodimeric p50 domain, obtained from the NF-κB transcription factor [30], exhibits a high affinity (Kd < 10pM) [31] for its target DNA-binding sequence (GGGAATTCCC), and was introduced to the construct to prevent the charges on TAT peptide from being neutralized by anionic DNA molecules. Two other recombinant fusion proteins, p50-GFP (PG) and GFP-TAT (GT), were also prepared as controls. The ability of these protein constructs to transduce cultured PC12 cells was quantitatively evaluated using flow cytometry. The efficiency of oligonucleotide delivery using these protein vehicles was also evaluated using fluorescently-labeled double stranded DNA molecules of 30bp and 293bp lengths at varying molar ratios of protein to DNA. These results were used to predict an optimal molar ratio for general DNA delivery, and this prediction was applied to the functional delivery of plasmid DNA containing the DsRed fluorescent protein transgene.
2. Materials and Methods
2.1. Materials
Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Enzymes for DNA cloning and manipulation were from New England Biolabs (Ipswich, MA). The QuickChange Site-directed mutagenesis kit was from Stratagene (La Jolla, CA). The Gel extraction kit, Miniprep kit and Midiprep kit were purchased from Qiagen (Valencia, CA). Complete protease inhibitor cocktail tablets were obtained from Roche Applied Science (Mannheim, Germany). SDS-PAGE gels and trypsin/EDTA were from Invitrogen (Carlsbad, CA). Centricon or Amicon Centrifugal Filter Units were from Millipore (Billerica, MA). The Bradford protein kit was from Pierce (Rockford, IL). Chromatography columns were obtained from GE Healthcare (Uppsala, Sweden). PC12 cells and F12K medium were obtained from American Type Culture Collection (Manassas, VA). GC5 competent E. coli cells, BL21 competent E. coli cells and all other chemicals used in the study were from Sigma-Aldrich (St. Louis, MO).
2.2. Vector preparation
The GFP gene with an N-terminus hexahistadine tag was encoded in the pRSET-S65T vector [32]. The vector for expression of GFP-TAT, named pGT, was created by adding the 14 amino acid TAT CPP sequence (NGYGRKKRRQRRRG) onto the C-terminus of the GFP gene. The DNA sequence of TAT peptide was generated by annealing two sense oligonucleotides: a-5′GAT GAA CTA TAC AAA TTT-3′ and b-5′AA CAA CGG TTA CGG TCG TAA GAA GCG TCG TCA GCG TCG TCG TGG CTA AGG TAC3′ and one antisense oligonucleotide: c-5′CTT AGC CAC GAC GAC GCT GAC GAC GCT TCT TAC GAC CGT AAC CGT TGT TAA ATT TGT ATA GTT CAT CCA TG-3′. The insert was ligated into the vector using a unique downstream KpnI site and a unique upstream SphI site which was introduced into the vector using the Quickchange kit with the following primers: d-5′GGG ATT ACA CAT GGC ATG CGA TGA ACT ATA CAA ATA-3′ and e-5′-TAT TTG TAT AGT TCA TCG CAT GCC ATG TGT AAT CCC-3′, where the insertion is shown in italics. The resultant vector, pGT was transformed into GC5 E. coli cells. Colonies were picked and used to inoculate 5ml cultures (LB broth containing 100μg/ml of ampicillin). Plasmid DNA was extracted from the cultures using Miniprep kit and each culture was screened by restriction digestion with SphI in order to verify the insertion of the TAT sequence, as the insertion destroyed the SphI site. Mutants with the correct insert were verified by DNA sequencing.
The pRES115 vector containing the p50 gene DNA-binding domain was kindly provided by Dr. Jonathan Blackburn [31]. A hexahistidine and FLAG tag sequence was generated by annealing a pair of oligonucleotides: f-5′-TAT GCA CCA TCA CCA TCA CCA TGA CTA CAA AGA TTC GGG CC-3′ and g-5′-GCT TAG AAA C AT CAG TAC CAC TAC CAC TAC CAC GT-3′. The annealed double stranded DNA contained ApaI and NdeI restriction sites. A unique ApaI site was introduced into the pRES115 vector through site-directed mutagenesis with a pair of primers: h-5′-CAT ATG GCA GAG GGC CCA TAC C -3′ and i-5′-GGT ATG GGC CCT CTG CCA TAT G-3′. The mutant pRES115 vector was sequentially digested by ApaI and NdeI and the hexahistidine and FLAG tag were ligated into the construct to create the pRES115His vector. The p50 gene with the hexahistidine and FLAG tag was amplified from the pRES115His vector using PCR with the forward primer, j-5′-CCT GTC TAG ATG CAC CAT CAC CAT CAC-3′, and the reverse primer k-5′-TCC GCC ACC TCC CGA GCT CCC C-3′. The forward primer generated an XbaI site and the reverse primer generated a tetra-glycine linker downstream of the p50 sequence. The GFP and GFP-TAT genes were amplified from pRSET-S65T or pGT vectors using PCR with the forward primer, l-5′-TCG GGA GGT GGC GGA ATG GCT AGC ATG ACT G-3′, and the reverse primer m-5′-CAG CAA AAA ACC CCT CAA GAC C-3′. The forward primer included the tetra-glycine linker used to fuse GFP or GFP-TAT to the p50 gene and the reverse primer generated a KpnI restriction site. The amplified p50 gene with the hexahistidine tag was linked to the GFP or GFP-TAT gene through the overlap extension PCR reaction. The extended p50-GFP or p50-GFP-TAT fragments were purified and amplified by PCR with primer j and primer m described above. The amplified p50-GFP or p50-GFP-TAT fragments were doubly digested with XbaI and KpnI and purified. The pRSET-S65T vector was doubly digested with XbaI and KpnI and the doubly digested p50-GFP or p50-GFP-TAT genes were ligated to the linearized pRSET-S65T vector as described above for the pGT vector. Plasmids of the correct size containing the unique ApaI site were then subjected to DNA sequencing to verify the inserted p50-GFP or p50-GFP-TAT sequences. The final plasmid constructs were named pPG and pGPT respectively.
The plasmid containing the DsRed protein (pCMV-DsRed-Express) was obtained from Clontech (Mountain View, CA). pDsRed-BD containing the target-kB binding site (5′-GGGAATTCCC-3′) was generated through two rounds of site-directed mutagenesis of the pCMV-DsRed-Express plasmid using primers, n-5′-GCTTGACGGGGAATTCCGGCGAACGTG-3′(first round forward), o-5′-CACGTTCGCCGGAATTCCCCGTCAAGC-3′ (first round reverse), p-5′-GACGGGGAATTCCCGCGAACGTGGCG-3′ (second round forward), and q-5′-CGCCACGTTC GCGGGAATTC CCCGTC-3′ (second round reverse).
2.3. Expression and purification of recombinant proteins
The G, GT, PG and PGT proteins were expressed in BL21 competent E. coli using the pRSET-S65T, pGT, pPG and pPGT vectors, respectively. Briefly, 1 L cultures were inoculated with 10mL saturated overnight cultures. The 1L cultures were grown at 37°C until the A600 of the culture reached 1.0, upon which they were induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and grown at 20°C for 20 hours. The cultures were then divided in half and harvested by centrifugation at 7,000g for 15min.
For G and GT, the bacterial pellets were re-suspended into 30 mL of resuspension buffer (50 mM sodium phosphate, 300 mM sodium chloride, 25 mM imidazole, pH=7.4) before being frozen at −80°C for later use. For the PG and PGT constructs, the re-suspension buffer (pH=7.0) was supplemented with Complete protease inhibitor cocktail tablets. Thawed bacterial suspensions were lysed by sonication for 10 min with an interval of 2 sec between sonication pulses of 5 sec. The sonicated lysate was clarified by centrifugation at 13,000g for 30 min. The supernatant was collected and loaded onto a HisTrap crude FF 5 ml column using an FPLC apparatus equilibrated with binding buffer (50 mM sodium phosphate, 300 mM sodium chloride, 25 mM imidazole, 1 mM PMSF, pH=7.4 for G and GT, pH=7.0 for PG and PGT). The column was washed with the same binding buffer until a stable A280 was observed. Protein was eluted using elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 250 mM imidazole, 1 mM PMSF, pH=7.4 for G and GT, pH=7.0 for PG and PGT).
For the G and GT proteins, the fractions exhibiting green fluorescence were assayed by SDS-PAGE gels and pooled before concentration using ultrafiltration with a YM-10 Centricon membrane. The elution buffer was exchanged to phosphate buffered saline (PBS) with 10% glycerol during the concentration process, and the protein samples were aliquoted and stored at −20°C for future use.
For the recombinant protein PG, the fractions with green fluorescence were pooled before ultrafiltration concentration using a YM-50 Centricon membrane. The concentrated samples were further purified using size-exclusion chromatography in 20 mM HEPES buffer (PH=7.0) supplemented with 2 mM EDTA, 1 mM DTT and 1 mM PMSF. The fractions with green fluorescence were assayed using SDS-PAGE and desired fractions were pooled before ultrafiltration concentration. Glycerol was added to create a final concentration of 10% (v/v), and aliquoted samples were stored at −20 °C.
For the PGT construct, the fractions with green fluorescence from the HisTrap column were pooled and loaded onto a Q-Sepharose FF ion exchange column equilibrated with PBS (pH=7.0) supplemented with 500 mM NaCl, 2 mM EDTA and 1 mM PMSF. The flow-through (non-bound) fractions with green fluorescence were pooled before ultrafiltation using a YM-50 Centricon membrane. The concentrated samples were further purified using size-exclusion chromatography and concentrated as the procedure described above for PG.
The concentrations of all of the purified proteins were determined using the Bradford method following the vendor protocol. Construct molecular weights were verified by MALDI-TOF mass spectrometry.
2.4. Dynamic light scattering
Dynamic light scattering (DLS) measurements performed using a Zetasizer ZS (Malvern, Worcestershire, United Kingdom) were used to measure the size of recombinant proteins. The sizes of proteins were determined using the particle sizing software package supplied by the manufacturer assuming the refractive index and viscosity of PBS buffer at room temperature. Particle sizes, obtained in triplicate, are reported as effective diameters.
2.5. Agarose gel electrophoresis retardation assays
Protein-DNA complexes were formed by combining 0.1nmol of an unlabeled 30bp DNA oligonucleotide with or without the target-kB binding site (0.1μmol/mL in 10mM Tris, 1mM EDTA buffer, pH=7.4) or with 0.2ug of plasmid DNA with or without the target-kB binding site (0.35 mg/mL in 20mM HEPES buffer, pH=7.0) to 0.28nmol of protein solution (5mg/ml in 20mM HEPES buffer, 10% glycerol, pH=7.0) in a microcentrifuge tube. 20mM HEPES buffer (pH=7.0) was added to the DNA solutions to obtain a final volume of 15 μL for the mixed solutions. The mixed solutions were incubated at room temperature for 20 min to ensure complexation. The resultant solutions supplemented with 16% of a 2M sucrose solution were loaded into agarose gels (1% or 1.5%) and were run at 120 V for 35min. Gels were stained with ethidium bromide, and both the DNA and fluorescent protein bands were visualized with a UV trans-illuminator.
2.6. Fluorescent-labeled DNA oligonucleotides
30bp double stranded DNA was generated by annealing a pair of complementary primers with a forward strand labeled with Alexa-647, n*-5′-CTC GTA TGT TGT GGG GAA TTC CCA GCG GAT-3′ and a reverse strand o-5′-ATC CGC TGG GAA TTC CCC ACA ACA TAC GAG-3′. Double stranded DNA of 293bp was generated by PCR using pRES115 as a template and Alexa-647 labeled primers, p*-5′-GAC CGA GCG CAG C -3′ and q*-5′-GCC CGA ATC TTT GTA GTC ATG -3′.
2.7. Cell culture
PC12 cells were cultured in F12K medium supplemented with 15% horse serum and 2.5% newborn calf serum (culture medium). The cells were harvested from tissue culture flasks using trypsin/EDTA, pelleted by centrifugation, and resuspended to 1×105 cells/ml in culture medium. One ml of the cell resuspension was added to each well of a poly-L-lysine coated 24-welled plate, and cultured for approximately five to seven days until confluency before transduction.
2.8. Samples for transduction
For each well of PC12 cells in a 24-well culture plate, a 2.8nmol protein sample was used for transduction, which was based on the dose dependent transduction efficiency of GT. To prepare the mixtures of protein and DNA, 2.8nmol protein samples were mixed with 0.005nmol, 0.01nmol, 0.025nmol, 0.05nmol, 0.1nmol, 0.25nmol and 1.4nmol of fluorescently labeled DNA in a total volume of 50ul of 20mM HEPES buffer (pH=7.0). To prepare the mixtures of protein and plasmid (pDsRed-BD), a 1ug plasmid sample was mixed with 0.7nmol, 1.4nmol or 2.8nmol of PGT or 1.4nmol of PG in a total volume of 50ul of 20mM HEPES buffer (pH=7.0). The mixtures were incubated at room temperature for 20min before dilution into 950ul of PC12 culture medium for transduction.
2.9. Transduction of PC12 cells and flow cytometry analysis
For the transduction experiments, proteins, fluorescently labeled DNA, or mixtures of protein and fluorescent labeled DNA were diluted with PC12 culture medium to a final volume of 1 ml. For the protein only transduction experiments, cells were incubated with 0.32, 1.60, 2.41, 3.21, 4.82, 6.42, and 9.64 nmol/ml of G, 0.30, 1.50, 2.26, 3.01, 4.53, 6.03 and 9.05 nmol/ml of GT, 0.30, 1.40, 2.80, 4.20, 5.60, or 8.40 nmol/ml of PG or 0.30, 1.40, 2.80, 3.20, 4.20, 5.60, or 8.40 nmol/ml of PGT. PC12 cells were incubated in the medium containing the desired protein sample for 4 hours at 37°C. After incubation, cells were washed once with PBS buffer warmed to 37°C. The cells were trypsinized and re-suspended into 200ul PBS buffer for analysis using a LSRII flow cytometer (Becton Dickinson, San Jose, CA). For cells incubated with the samples containing fluorescently labeled DNA, the cells were incubated in 200ul DNAse I digestion solution (5U DNAse I diluted into 180ul of 200mM sucrose solution supplemented with 20ul DNAse I buffer) at 37° C for 8min in order to remove the DNA extracellularly associated with the cell membranes prior to trypsinization. A total of 30,000 events per sample were counted. Viable cells were gated in forward and side scatter plots. For cells transduced with protein, the geometric mean fluorescence of the viable cells detected through the FITC channel (for GFP fluorescence, ex 488, em 515–545) was recorded. For cells transduced using protein and fluorescently labeled DNA, the geometric mean fluorescence of the viable cells detected through the Cy5 channel (for Alexa-647 fluorescence of DNA) was also recorded. The protein transduction efficiency was evaluated by relative fluorescence of FITC based on control cells. The DNA delivery efficiency was evaluated by relative fluorescence of Cy5 based on cells incubated with the same amount of fluorescently labeled DNA.
For the transfection with plasmid DNA or the mixture of plasmid and protein, the samples were diluted with culture medium to the final volume of 1ml. PC12 cells were incubated in the medium containing the desired protein/plasmid sample for 4 hours at 37°C. Then the medium was replaced with 1ml fresh culture medium. After a further 20–24 h of culture, the cells were washed once with PBS buffer warmed to 37°C, trypsinized, and resuspended into 200ul PBS buffer for flow cytometry analysis. All cells from the wells were counted. Viable cells were gated in forward and side scatter plots. The cells exhibiting DsRed fluorescence were detected through the PE-TR channel (ex 488, em 600–620). The number of red cells was recorded through the gates set based on measurements made with untreated cells. The plasmid delivery efficiency was evaluated by the percentage of red cells from the viable cells. A control experiment with cells exposed to PGT and no plasmid was also performed to confirm that the fluorescence from the GFP was not detected in the PE-TR channel.
2.10. Statistical analysis
Protein intensity results were expressed as mean relative fluorescent units (RFU) ± standard error. The differences between different protein constructs were analyzed using one-way ANOVA with a significance level of p< 0.05. Flow cytometry results were expressed as mean relative fluorescence (RF) ± standard error. Protein dose dependent transduction data were analyzed by one-way ANOVA. For the protein-mediated DNA delivery experiments, the effect of the addition of DNA on protein transduction efficiency and the effect of the addition of DNA on DNA delivery efficiency were analyzed by one-way ANOVA followed by post-hoc analysis with Tukey’s test at a significance level of p<0.05.
3. Results
3.1. Protein construction, purification and characterization
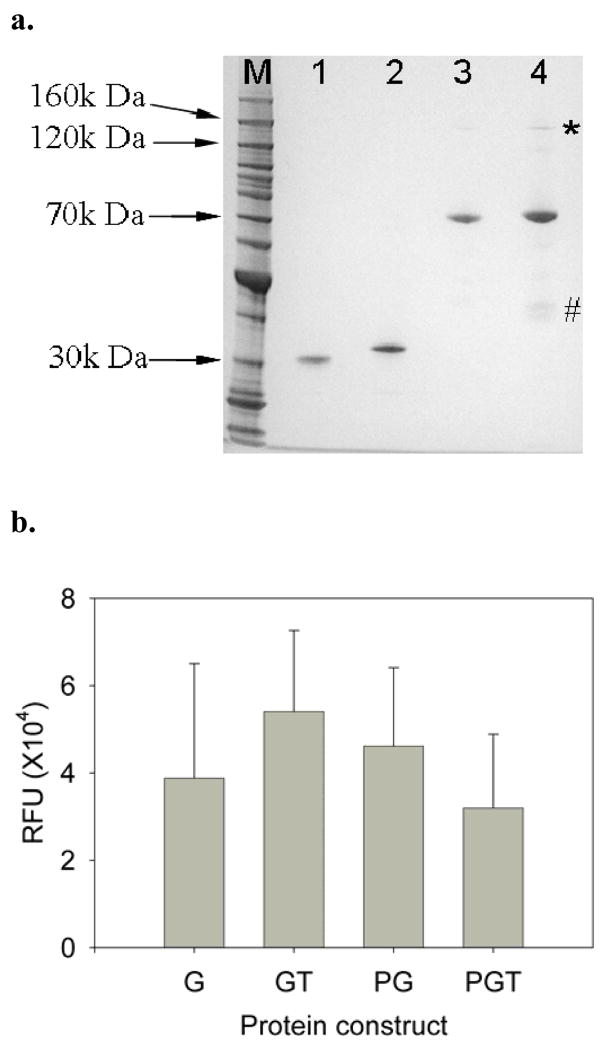
Three new chimeric fusion protein constructs (GT, PG and PGT) were designed for this work (Fig. 1). In all constructs, a hexahistadine tag was included on the N-terminus of the protein. In the constructs of GT and PGT, TAT peptide was appended to the C-terminus of the protein. In the constructs containing the p50 DNA binding protein, a linker of 4 glycine amino acids was inserted between the p50 and GFP domains. All proteins were expressed in E. coli, and their final purity was assessed by SDS-PAGE (Fig. 2a), and by Western blots using a monoclonal anti-FLAG antibody (data not shown). Molecular weights obtained by MALDI-TOF mass spectrometry, 31,313 Da for G, 33,194 Da for GT, 68,410 Da for PG and 70,407 Da for PGT, compared well to the calculated theoretical molecular weights of 31,113 Da, 33,144 Da, 68,356 Da and 70,388 Da. The hydrodynamic diameters of the recombinant proteins in PBS buffer (pH 7.0) obtained by dynamic light scattering (DLS) were 6.4±1.3 nm for G, 7.9±2.1 nm for GT, 10.8±4.1 nm for PG, and 10.5±2.9 nm for PGT. These values also indicate the correct relative sizes of the purified proteins. Since all of the fusion proteins contained the GFP fluorophore, the green fluorescence intensity per mole was compared (Fig. 2b) and the lack of a statistical difference between the samples indicates similar purities of the samples and also demonstrates that the fusion constructs do not significantly impact the bioactivity of the green fluorophores.
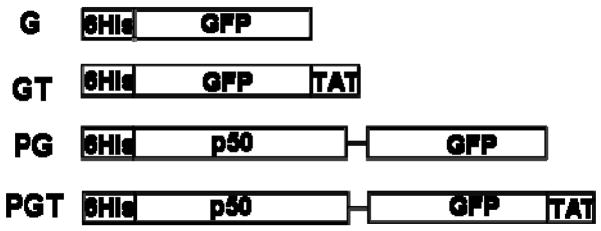
Figure 1.
Schematic of the four recombinant fusion proteins used in this study. All constructs have an N-terminal hexahistidine tag (Histag). The green fluorescent protein (GFP) contains the S65T mutation for increased fluorescence. The TAT amino acid sequence is NGYGRKKRRQRRRG. The p50 DNA binding domain is derived from the NF-κB p50 transcription factor. A 4 glycine linker was inserted between the p50 and GFP domains.
Figure 2.
(a) SDS-PAGE of the recombinant protein constructs. Molecular weight markers are in lane 1, G is in lane 2, GT is lane 3, PG is in lane 4, and PGT is in lane 5. All proteins migrated at their expected molecular weights. The * indicates possible dimer formation by PGT, and PG to a lesser extent. The # indicates possible degradation products of the PGT construct. (b) Molar fluorescent intensities of purified recombinant proteins. Error bars represent standard errors. Each experiment was performed in at least triplicate (n ≥3).
3.2. Dose response of Protein Delivery
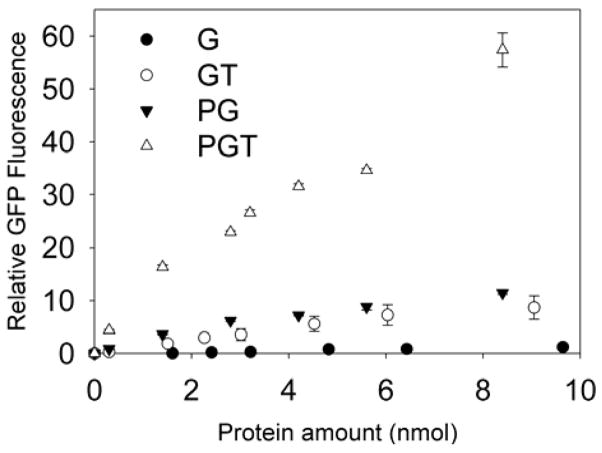
The transduction abilities of the new chimeric fusion proteins were tested using undifferentiated PC12 cells, which are a neuronal-like cell line. All proteins were dissolved in 1ml of PC12 culture medium, and cells were incubated for 4 hours at 37°C. Fig. 3 shows the relative fluorescence of the cells as compared to untreated, control cells. The control protein G was not taken up by the cells at any dose, while the uptake of GT, PG and PGT by PC12 cells displayed statistically significant dose dependencies. The fusion proteins penetrated the PC12 cells with the following efficiency order: PGT ≫ PG > GT.
Figure 3.
Dose dependent uptake of proteins by PC12 cells, G (●), GT (○), PG (▼) and PGT (△). Error bars represent standard errors. Each experiment was performed in triplicate (n = 3).
3.3. Interaction of Proteins and DNA
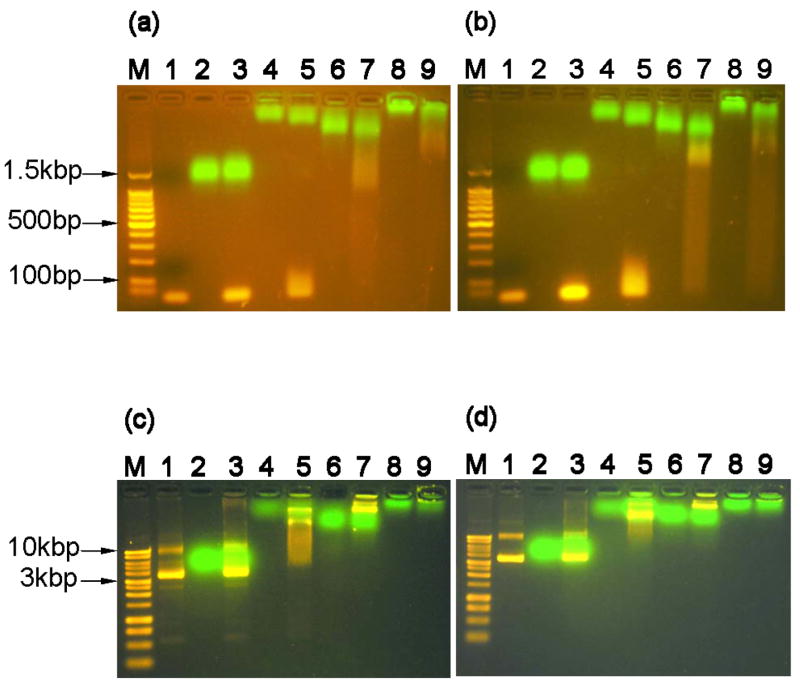
Agarose gel electrophoresis (Fig. 4) was used to assess the binding capabilities of the protein constructs to small linear oligonucleotides (Figs. 4a & 4b) and plasmid DNA molecules (Figs. 4c & 4d). Since the p50 DNA binding domain is reported to be specific for the target κB sequence, DNA molecules with the binding sequence (Figs. 4a & 4c) were compared to similar constructs without the target κB binding sequence (Figs. 4b & 4d). No obvious interaction between DNA of either size or sequence was observed with G alone (lane 3 in Figs. 4a, 4b, 4c & 4d). The GT fusion demonstrated some interaction with the small DNA samples as evidenced by a slight retardation of the oligonucleotides combined with an increased migration of the GT protein (lane 5 in Figs. 4a & 4b). This effect was independent of the presence of the target κB sequence. The interaction of the GT protein with the plasmid DNA samples was much more pronounced. The presence of the GT protein clearly retarded the migration of the plasmid DNA independently of the presence of the target κB sequence (lane 5 in Figs. 4c & 4d).
Figure 4.
Protein and DNA interactions monitored by agarose gel electrophoresis retardation. (a) mixtures of proteins and linear DNA containing the target-κB binding site (30bp in length, 0.2nmol protein to 0.01nmol DNA, 1.5% agarose gel), lane M: 100bp DNA molecular weight marker, lane 1: neat dsDNA oligonucleotide, lane 2: G, lane 3: G+DNA, lane 4: GT, lane 5: GT+DNA, lane 6: PG, lane 7: PG+DNA, lane 8: PGT, and lane 9: PGT+DNA. (b) mixtures of proteins and linear DNA without the target-κB binding site (33bp, same molar ratio and agarose gel concentration) with same lane assignments as panel A. (c) mixtures of proteins and plasmid DNA containing the target-κB binding site (~4.6kbp DNA, 0.2nmol protein : 200ng plasmid DNA, 1.0% agarose gel). Lane M: 1kb DNA molecular weight marker, all other lane assignments the same as panel A. (d) mixtures of proteins and plasmid DNA without the target-κB binding site (same plasmid size, molar ratio, and agarose gel concentration) with same lane assignments as panel C.
The addition of the p50 domain increased the interaction of the fusion proteins with the DNA for both the short oligonucleotides and the full plasmids. For the short DNA oligonucleotides, it appeared that there was a stronger interaction between PGT and the DNA as compared to PG, which was evidenced by an increased retardation of DNA in the PGT samples (lane 7 & 9 in Figs. 4a & 4b). And, in both cases, the presence of the DNA also caused the proteins to migrate farther into the gel. In the plasmid DNA samples (lane 7 in Figs. 4c & 4d), the PG protein significantly retarded the migration of the plasmid into the gel, and some protein and DNA were observed to be trapped in the loading well of the 1% gel. The interaction was even more pronounced for the PGT samples (lane 9 in Figs. 4c & 4d), as the presence of the PGT construct appeared to prevent the migration of the DNA into the gel altogether. Interestingly, there was only a minor observation of ethidium bromide-stained DNA trapped in the loading wells. It is possible that the strong interaction between PGT and the plasmid prevented the ethidium bromide from intercalating within the plasmid DNA. Similar results have also been observed in other electrophoresis retardation assays of DNA and cationic peptide or polymer complexes [14, 18, 19, 33–35].
Unexpectedly, the interaction of the protein constructs with DNA did not appear to be significantly dependent on the presence of the target κB sequence. For the small DNA oligonucleotides a small difference is possible, as there may be more pronounced smearing of DNA in the absence of the target κB sequence, but the difference, if at all, is minor (lane 7 & 9 in Figs. 4a & 4b). For the plasmid samples, the target κB sequence does not appear to have any effect on the interaction of the protein with the plasmid (lane 7 & 9 in Figs. 4c & 4d).
3.4. Protein mediated linear DNA delivery
The transduction efficiencies of the chimeric fusion proteins were evaluated using Alexa 647 labeled oligonucleotides of different lengths (30bp and 293bp) that contained the target κB sequence. In order to ensure the GFP signal was in a suitable range for detection, experiments were performed with a constant amount of protein (2.8 nmol) and varying amounts of DNA to create different DNA to protein ratios (Fig. 5). The resulting charge ratios of the protein-DNA complexes can be found in Table 1. In all cases, the GFP signal was background corrected using untreated cells, and the Alexa647 signal was background corrected against cells treated with Alexa-labeled DNA alone. These background corrections give the Relative Fluorescence (RF) values reported in Fig. 5.
Figure 5.
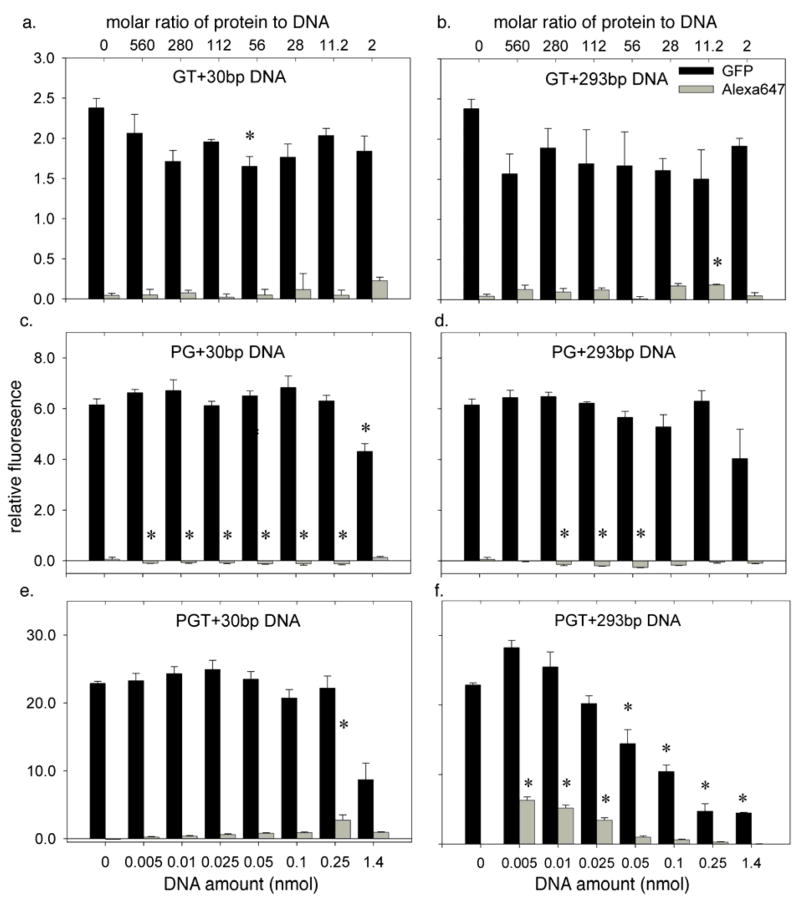
Delivery of DNA (labeled with Alexa647) and chimeric fusion proteins (containing the GFP fluorophore) to PC12 cells as monitored by flow cytometry. The protein amount was fixed in all experiments at 2.8 nmol (a) Uptake with a mixture of GT and 30bp DNA. (b) Uptake with a mixture of GT and 293bp DNA. (c) Uptake with a mixture of PG and 30bp DNA. (d) Uptake with a mixture of PG and 293bp DNA. (e) Uptake with a mixture of PGT and 30bp DNA. (f) Uptake with a mixture of PGT and 293bp DNA. In all experiments, the 30bp DNA fragment contained a single Alexa647 fluorophore, while the 293bp fragments were doubly labeled. Error bars represent standard errors. Each experiment was performed in at least triplicate (n ≥ 3). The * indicates the value is significantly different from the protein-only control (DNA amount = 0) using one-way ANOVA and post-hoc Tukey’s test (p < 0.05).
Table 1.
Charge ratios of protein-DNA complexes at corresponding molar ratios of protein to DNA (The charge ratio was calculated based on the total number of histidine, lysine, arginine, aspartic acid and glutamic acid of the fusion protein and the total number of phosphate groups of the DNA molecule.)
| Molar Ratio | 2 | 11.2 | 28 | 56 | 112 | 280 | 560 | 8600 |
|---|---|---|---|---|---|---|---|---|
| GT-30bp DNA | 0.433 | 2.43 | 6.07 | 12.1 | 24.3 | 60.7 | 121 | - |
| PG-30bp DNA | 0.533 | 2.99 | 7.47 | 14.9 | 29.9 | 74.7 | 149 | - |
| PGT-30bp DNA | 0.800 | 4.48 | 11.2 | 22.4 | 44.8 | 112 | 224 | - |
| GT-293bp DNA | 0.0444 | 0.248 | 0.621 | 1.24 | 2.48 | 6.21 | 12.4 | - |
| PG-293bp DNA | 0.0546 | 0.306 | 0.765 | 1.53 | 3.06 | 7.65 | 15.3 | - |
| PGT-293bp DNA | 0.0819 | 0.459 | 1.15 | 2.29 | 4.59 | 11.5 | 22.9 | - |
| PGT-DsRed plasmid | - | - | - | - | - | - | - | 22.4 |
The results with the GT construct suggest limited interactions between the TAT peptide and the DNA fragments. For the 30bp fragment (Fig. 5a), the transduction of the GT protein was inhibited by the presence of the oligonucleotide at a molar protein to oligonucleotide ratio of 56:1, but none of the other interactions were significant. For the 293bp oligonucleotide (Fig. 5b), inhibition of the GT transfection did not reach statistical significance. The GT construct was able to deliver the oligonucleotide to the PC12 cells at a protein to DNA ratio of 11.2:1 but none of the other ratios reached statistical significance.
A much different effect was seen with the PG construct. As was observed with the GT fragment, the DNA had little effect on the transduction of the protein. The 30bp DNA fragment (Fig. 5c) inhibited the transduction of the PG construct only at the highest DNA concentration, at which condition the molar ratio of protein to DNA is 2:1, and there was no significant inhibition effect by 293 bp fragment at any ratio (Fig. 5d). The DNA delivery results showed a much different trend. The addition of the PG protein significantly inhibited the natural uptake of the DNA oligonucleotides as compared to the protein free control experiments at almost every ratio. This effect reached statistical significance for the 30bp DNA fragment at all molar ratios except 2:1 (Fig. 5c), and for the 293 bp fragment it reached statistical significance at molar ratios of 56:1, 112:1, and 280:1 (Fig. 5d). These results suggest that the PG complex forms tight associations with the DNA molecules, and this significantly decreases their availability for natural uptake by the PC12 cells.
When PC12 cells were incubated with the mixture of PGT and DNA, significant delivery of DNA of both sizes was observed, but the presence of the DNA also inhibited the delivery of the PGT protein in both cases at certain molar ratios of protein to DNA (Figs. 5e & 5f). For the 30bp fragment (Fig. 5e), the DNA inhibited the transduction of PGT at the highest DNA concentration, a molar ratio of 2:1. The delivery of DNA increased as the molar ratio decreased, and reached a statistically significant maximum at a molar ratio of 11.2:1. The results with the 293bp fragment were more dramatic (Fig. 5f). The presence of the DNA significantly inhibited the delivery of the PGT construct at the four highest DNA concentrations (molar ratios of 2:1. 11.2:1, 28:1, and 56:1). The larger oligonucleotide, however, required a higher protein to DNA ratio for delivery than the 30bp oligonucleotide. Three ratios reached statistical significance, 112:1, 280:1, and 560:1, with the latter ratio exhibiting the highest delivery efficiency.
3.5. Protein mediated plasmid DNA delivery
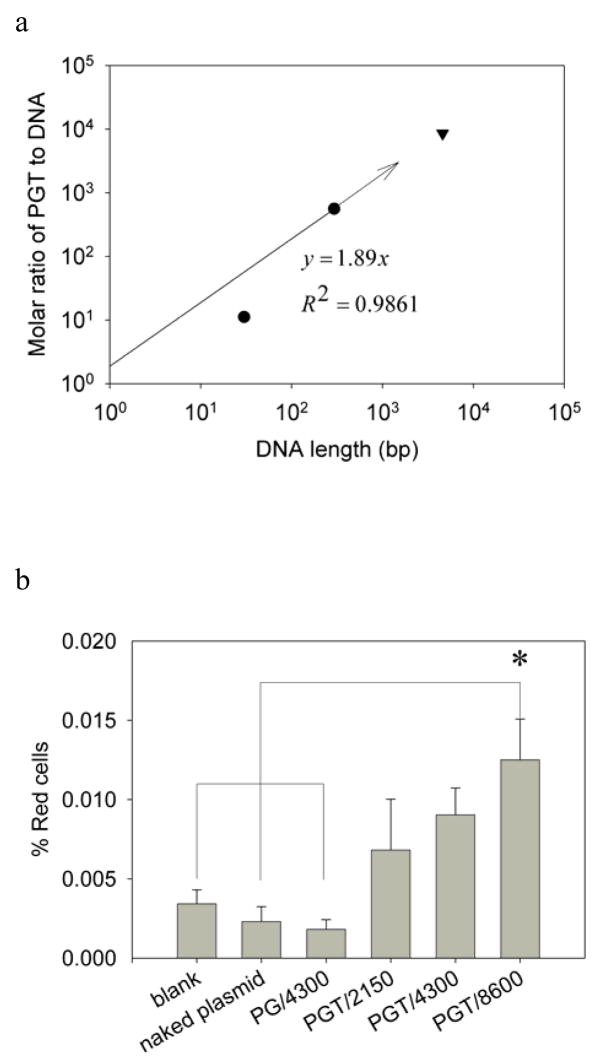
The ability of the PGT construct to deliver large plasmid DNA was also investigated. A 4.6kbp plasmid expressing the DsRed fluorescent protein was created that included the target κB sequence. A linear correlation was hypothesized to exist between the ratio of protein to DNA and the length of the DNA molecule to be delivered. Assuming an intercept of zero, this results in line with a slope of 1.89 (Fig. 6A). Therefore, it was predicted that the delivery of a 4.6kb plasmid would require a PGT to plasmid ratio of 8600:1.
Figure 6.
(a) Log-log plot of the correlation of the molar ratio needed for optimal transfection efficiency as a function of the length of the delivered DNA cargo into PC12 cells. The optimal molar ratios, 28:1 for 30bp DNA cargo (●) and 560:1 for 293bp DNA cargo (●) were obtained from PGT mediated linear DNA delivery experiments while the optimal molar ratio needed for delivering 4.6kbp plasmid (8600:1) was predicted from the correlation (—) based on 30bp and 293bp DNA cargos as the intercept of the linear fitting equation was set at the origin. (b) Delivery of the pDsRed-BD plasmid to PC12 cells using the chimeric protein constructs. Error bars represent standard errors. Each experiment was performed five times (n=5). The * indicates the value is significantly different from the untreated cells, neat plasmid transfected cells and PG plasmid complex at molar ratio of 4300:1 transfected cells using one way ANOVA and post-hoc Tukey’s test (p<0.05).
To test this prediction, the delivery of the pDsRed-BD plasmid to PC12 cells was tested with three different molar ratios of PGT to pDsRed-BD: 2150:1, 4300:1 and 8600:1. Untreated cells and cells transfected with neat pDsRed-BD were used as negative controls. Since PG was unable to deliver oligonucleotide DNA, the complex of PG and pDsRed-BD prepared at a molar ratio of 4300:1 was also used as a negative control. The pDsRed-BD plasmid was also delivered to PC12 cells using Lipofectamine 2000 as a positive control, and this resulted in a transfection efficiency of 3.97±0.41% (data not shown). The protein-mediated transfection results based on the detection of the expressed transgene using flow cytometry are shown in Fig. 6B. There was no significant difference among untreated cells, cells treated with naked plasmid, or cells treated with the PG/4300 complex. When PGT was used, all three ratios resulted in increased delivery of the transgene as compared to the negative controls, but only the PGT/8600 complex mixture reached statistical significance.
4. Discussion
In this study a chimeric fusion protein containing a cell penetrating peptide, TAT, and a DNA binding domain, p50, was evaluated for its ability to deliver oligonucleotides and plasmid DNA to cultured PC12 cells. By introducing a fluorescent protein domain to the fusion proteins and fluorescent labels or transgenes to the DNA molecules, the translocation of both the protein vehicles and nucleotide cargos to the cells were studied simultaneously. Successful delivery of nucleotide cargos to PC12 cells required the presence of both the cell penetrating domain and DNA binding domain, even though fusion proteins containing either domain alone exhibited some transduction efficiency by themselves. These results are consistent with our design goal to show that the p50 domain performs a DNA binding function and better enables the TAT domain to exhibit CPP activity. A linear correlation for the molar ratio of protein to DNA length required for the delivery of nucleotide-based cargos was developed, and this will be valuable as future protein-based delivery vehicles are created.
All of the fusion proteins created in this study were expressed in E. coli and were purified in their native states. The G and GT proteins were purified using Ni2+ affinity chromatography, and no additional purification was required. However, for the PG and PGT constructs, the addition of a protease inhibitor cocktail was necessary to reduce the degradation of fusion proteins. A size exclusion chromatography step was added for the purification of PG and PGT in order to remove impurities or hydrolyzed products at smaller molecular weights. An ion exchange chromatography step was also used to further purify PGT in order to remove genomic DNA contamination due to the strong interaction between the PGT protein and DNA. It has been reported that non-specific DNA binding of p50 cannot be detected at ionic strengths higher than 150mM. This appeared to be true for the PG fusion, since non-specific binding of DNA was removed through size exclusion chromatography with the running buffer at the ionic strength of 150mM. However, this was not the case for the purification of PGT as DNA was co-purified with the PGT protein during size exclusion chromatography, and therefore an ion-exchange column was used under higher ion strength conditions (400mM) before size exclusion chromatography in order to obtain PGT with the desired purity. From the SDS-PAGE analysis of PG and PGT (Fig. 2a, lane 3 and 4) there appears to be a heavier band at around 140 kDa and some lighter bands around 40 kDa. The heavier bands are likely dimers induced by p50 domain, and the lighter bands could be degradation products despite the presence of protease inhibitors and the use of multiple orthogonal purifications steps.
The final protein products G, GT, PG, and PGT all exhibited similar fluorescent intensities (excitation wavelength: 489nm, emission wavelength: 509nm) normalized by the molar amount of protein. The absorbance spectra show that the three fusion proteins have absorbance maxima at 489nm, which is identical to GFP (data not shown). The emission spectra at an excitation wavelength of 489nm have peaks at 508nm for G, 519nm for GT, and 503nm for both PG and PGT. The small shifts in the emission spectra upon the addition of the TAT and p50 fusions may explain the variations in the measured molar intensities of the purified proteins (Fig. 2b), as they were all measured at an emission wavelength of 509nm. These differences, however, did not reach statistical significance.
The transduction abilities of the fusion proteins were measured using PC12 cells. The cells were trypsinized before the flow cytometry assays in order to remove any fusion proteins that were associated with the outer cellular membranes. According to the dose dependent uptake behaviors shown in Fig. 3, the PG control construct was able to transfect cells with higher efficiency than the GT construct, despite the lack of an attached CPP sequence. This is not unprecedented, as it has been reported that some anti-dsDNA antibodies are capable of binding to the membranes of mammalian cells and exhibiting cellular penetrating abilities and that the residues required for binding DNA were necessary for the antibody penetration [36, 37]. Since the p50 domain used in our study is a DNA binding domain from the NF-κB transcription factor, it is possible that the p50 could also provide a similar cell binding and penetrating activity. Therefore, the p50 domain combined with the TAT domain could produce additive effects, and this was confirmed with the PGT construct exhibiting the highest cell penetrating behavior in this study.
The DNA binding behaviors of the constructs were investigated using electrophoresis retardation assays, and the interactions observed between the proteins and DNA during the purification procedures were largely seen again in the electrophoresis experiments. The GT construct did not appear to interact with any of the DNA molecules, which was expected since the dissociation constant between unmodified TAT peptides has been estimated to be 4 orders of magnitude higher than the dissociation constant between the p50 domain and its target DNA-binding sequence [31, 38]. And, surprisingly, both the PG and PGT constructs showed similar strong associations to DNA with or without the target-κB binding site.
To test the ability of the constructs to deliver DNA to cells, two labeled oligonucleotides were created. One strand of the 30bp oligonucleotide with an Alexa-647 fluorophore on the 5′ was directly hybridized to its complementary non-labeled strand in order to increase the hybridization efficiency, since the presence of a fluorophore on both strands might disrupt base-pairing and reduce the stability of the molecule. The 293bp oligonucleotide was prepared using PCR with a pair of fluorescently-labeled primers resulting in both ends of each duplex molecule being labeled with the fluorophore. Considering the fact that longer oligonucleotides are likely to exhibit decreased transduction rates as compared to smaller molecules, we anticipated that double-end labeling would facilitate fluorescence detection.
Delivery experiments with the DNA oligonucleotides were performed similarly to the protein only experiments (Fig. 5), but a DNAse I digestion was also included before the flow cytometry experiments to remove any extracellular DNA that may have been associated with the cellular membrane. As expected, the GT protein was unable to efficiently deliver DNA to the cells consistent with the results of others showing that the TAT peptide by itself exhibited only a slight ability to deliver a 21 bp oligonucleotide [15]. And the PG protein was also incapable of delivering DNA to cells even though in the absence of DNA it exhibited higher transduction efficiency than the GT protein.
The PGT fusion was the most efficient DNA delivery agent examined in this work. Because of the strong interaction between PGT and DNA, the addition of DNA inhibited the intracellular delivery of PGT, and this effect was more pronounced with the longer 293bp oligonucleotide. The optimal molar ratio for intracellular delivery was found to be 11.2:1 for the 30bp oligonucleotide and 560:1 for the 293bp oligonucleotide. Assuming a linear correlation through the origin between the optimal molar ratio for delivery and the length of the oligonucleotide, a slope of 1.89 is obtained (Fig. 6A). Although this value is close to 2, and the PGT protein is likely homodimeric, the crystal structure of the p50 domain demonstrates that the p50 domain occupies more than one DNA base pair when bound [30], and therefore this correlation indicates that an excess of PGT is required for optimal delivery. This correlation results in a charge ratio (+/−) of approximately 22.7:1 (Table 1), which is of the same order of magnitude as the charge ratios or N/P ratios reported by other researchers when the free TAT peptide or polyethyleneimine (PEI) have been used for DNA delivery [18, 21, 23, 34, 39, 40].
The correlation was used to predict that the molar ratio needed for delivery of 4.6k pDsRed-BD plasmid would be around 8600:1. The results show that this was indeed the required ratio for delivery of the functional transgene, as the lower ratios of 2150:1 and 4300:1 were not statistically different from the controls. However, the overall transfection efficiency was very low (less than 0.05%) compared to Lipofectamine 2000 mediated transfection (3.97±0.40%, data not shown).
The low efficiency may be the result of poor intracellular trafficking, as the PGT/plasmid complex could be trapped in endosomes, or the strong interaction between PGT and plasmid could inhibit the release of plasmid for transcription of the DsRed transgene. The effects of endosomal trapping may not be easily detected in linear DNA delivery experiments due to the direct fluorescent labeling of the DNA. The fusion protein construct may be improved by incorporating a domain with endosomolytic activity such as polyhistidine [41]. Polyhistidine is not able to interact with DNA at physiological pH of 7.4 but will be protonated under a slightly acidic milieu at endosomal pH levels, which results in the ‘proton sponge’ effect to facilitate emdosome escape. The transfection efficiency may also be improved by using lysosomotropic agents such as chloroquine during the transfection, which has recently been shown by Xavier et al [23]. In addition, the inclusion of the large GFP domain may also negatively impact the performance of the fusion protein constructs for DNA delivery. It is also notable that DsRed is one of the dimmest red fluorescent proteins [42] and a brighter flourophore that is better matched to the excitation wavelength the flow cytometer would likely result in larger relative estimates of the transfection efficiencies.
In previous studies using TP10 [24], TAT-Mu [18], TAT-NLS-Mu[23] or TAT-poly lysine [14] for the delivery of plasmid DNA containing a transgenic marker, PEI or lipid based agents were needed to achieve significant transfection efficiencies. Therefore, although the transfection efficiency was low, it was notable that this was achieved without the addition of cationic polymers or lipids.
5. Conclusions
We have designed and created a new bifunctional recombinant protein construct that can be used to deliver nucleotide-base cargos to PC12 cells in culture. Both the DNA binding domain (p50) and cell penetrating domain (TAT) were required for the highest observed intracellular delivery of DNA. The apparently optimal molar ratio of protein to bp of DNA length was found to be 1.89, and this ratio was used to deliver a functional transgene encoded in plasmid DNA. Several research groups have previously reported fusion proteins that incorporate the TAT peptide to enable cellular delivery. We have incorporated a specific DNA-binding domain in order to create a self-assembling system that will link the TAT peptide to DNA cargos non-covalently. There are many advantages to non-viral gene delivery [43–45], and the modular protein engineering approach can be extended to incorporate additional domains such as “smart” stimulus responsive domains for targeting [46, 47], membrane active domains for endosomal escape [48, 49], and nuclear targeting domains [50, 51] to improve the specificity and efficiency of these novel vehicles.
Supplementary Material
Supplementary data associated with this manuscript can be found online.
Acknowledgments
This work was supported by the Brain Trust (SB and BM), NIH R21 MH080024 (SB and BM), and an NSF Graduate Research Fellowship (MJS).
We thank Dr. Yasuhiro Itagaki for his assistance in MALDI-TOF mass spectrometry, Dr. Lance Kam for providing the pRSET-S65T plasmid, Dr. Jonathan Blackburn for providing the pRES115 plasmid and Dr. Zengmin Li for valuable discussions about fluorescently labeled DNA constructs. We also thank Mr. Ian Wheeldon for valuable suggestions and assistance in manuscript preparation, Mr. Mark Blenner and Mr. Elliot Campbell for valuable discussions on the manuscript. We also thank Nina Xu and Alexis King Matrka for assistance in cloning and PCR experiments. This work was supported by the Brain Trust (SB and BM), NIH R21 MH080024 (SB and BM), and an NSF Graduate Research Fellowship (MJS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Torchilin VP. Recent approaches to intracellular delivery of drugs and DNA and organelle targeting. Annu Rev Biomed Eng. 2006;8:343–75. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 2.Dietz GP, Bahr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Joliot A, Prochiantz A. Transduction peptides: from technology to physiology. Nat Cell Biol. 2004;6:189–96. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 4.Gupta B, Levchenko TS, Torchilin VP. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv Drug Deliv Rev. 2005;57:637–51. doi: 10.1016/j.addr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Kabouridis PS. Biological applications of protein transduction technology. Trends Biotechnol. 2003;21:498–503. doi: 10.1016/j.tibtech.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trehin R, Merkle HP. Chances and pitfalls of cell penetrating peptides for cellular drug delivery. Eur J Pharm Biopharm. 2004;58:209–23. doi: 10.1016/j.ejpb.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–45. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 9.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–88. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 10.Stewart KM, Horton KL, Kelley SO. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org Biomol Chem. 2008;6:2242–55. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, Hayakawa T, Takeda K, Hasegawa M, Nakanishi M. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. Journal of Biological Chemistry. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan A, Tikoo A, Kapur AK, Singh M. The taming of the cell penetrating domain of the HIV Tat: Myths and realities. J Control Release. 2007;117:148–62. doi: 10.1016/j.jconrel.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torchilin VP. Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Adv Drug Deliv Rev. 2008;60:548–58. doi: 10.1016/j.addr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Hashida H, Miyamoto M, Cho Y, Hida Y, Kato K, Kurokawa T, Okushiba S, Kondo S, Dosaka-Akita H, Katohl H. Fusion of HIV-1 Tat protein transduction domain to poly-lysine as a new DNA delivery tool. British Journal of Cancer. 2004;90:1252–1258. doi: 10.1038/sj.bjc.6601680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Andaloussi S, Jarver P, Johansson HJ, Langel U. Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: a comparative study. Biochemical Journal. 2007;407:285–292. doi: 10.1042/BJ20070507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abes S, Moulton H, Turner J, Clair P, Richard JP, Iversen P, Gait MJ, Lebleu B. Peptide-based delivery of nucleic acids: design, mechanism of uptake and applications to splice-correcting oligonucleotides. Biochemical Society Transactions. 2007;35:53–55. doi: 10.1042/BST0350053. [DOI] [PubMed] [Google Scholar]
- 17.Siprashvili Z, Scholl FA, Oliver SF, Adams A, Contag CH, Wender PA, Khavari PA. Gene transfer via reversible plasmid condensation with cysteine-flanked, internally spaced arginine-rich peptides. Hum Gene Ther. 2003;14:1225–33. doi: 10.1089/104303403767740768. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan R, Xavier J, Rangaraj N, Rao NM, Gopal V. Recombinant fusion proteins TAT-Mu, Mu and Mu-Mu mediate efficient non-viral gene delivery. J Gene Med. 2007;9:275–86. doi: 10.1002/jgm.1014. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Li M, Cui D, Fei J. Macro-branched cell-penetrating peptide design for gene delivery. J Control Release. 2005;102:699–710. doi: 10.1016/j.jconrel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Turner JJ, Arzumanov AA, Gait MJ. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Research. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignatovich IA, Dizhe EB, Pavlotskaya AV, Akifiev BN, Burov SV, Orlov SV, Perevozchikov AP. Complexes of plasmid DNA with basic domain 47–57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways. J Biol Chem. 2003;278:42625–36. doi: 10.1074/jbc.M301431200. [DOI] [PubMed] [Google Scholar]
- 22.Parker AL, Eckley L, Singh S, Preece JA, Collins L, Fabre JW. (LYS)(16)-based reducible polycations provide stable polyplexes with anionic fusogenic peptides and efficient gene delivery to post mitotic cells. Biochimica Et Biophysica Acta-General Subjects. 2007;1770:1331–1337. doi: 10.1016/j.bbagen.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Xavier J, Singh S, Dean D, Rao N, Gopal V. Designed multi-domain protein as a carrier of nucleic acids into cells. Journal of Controlled Release Article. 2008 doi: 10.1016/j.jconrel.2008.09.090. in Press. [DOI] [PubMed] [Google Scholar]
- 24.Kilk K, El-Andaloussi S, Jarver P, Meikas A, Valkna A, Bartfai T, Kogerman P, Metsis M, Langel U. Evaluation of transportan 10 in PEI mediated plasmid delivery assay. J Control Release. 2005;103:511–23. doi: 10.1016/j.jconrel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Advanced Drug Delivery Reviews. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Deshayes S, Morris M, Heitz F, Divita G. Delivery of proteins and nucleic acids using a non-covalent peptide-based strategy. Advanced Drug Delivery Reviews. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Mann A, Thakur G, Shukla V, Ganguli M. Peptides in DNA delivery: current insights and future directions. Drug Discov Today. 2008;13:152–60. doi: 10.1016/j.drudis.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 28.El-Andaloussi S, Johansson H, Magnusdottir A, Jarver P, Lundberg P, Langel U. TP10, a delivery vector for decoy oligonucleotides targeting the Myc protein. Journal of Controlled Release. 2005;110:189–201. doi: 10.1016/j.jconrel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Nakanishi M, Eguchi A, Akuta T, Nagoshi E, Fujita S, Okabe J, Senda T, Hasegawa M. Basic peptides as functional components of non-viral gene transfer vehicles. Current Protein & Peptide Science. 2003;4:141–150. doi: 10.2174/1389203033487234. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–10. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 31.Speight RE, Hart DJ, Sutherland JD, Blackburn JM. A new plasmid display technology for the in vitro selection of functional phenotype-genotype linked proteins. Chem Biol. 2001;8:951–65. doi: 10.1016/s1074-5521(01)00066-7. [DOI] [PubMed] [Google Scholar]
- 32.Yang TT, Cheng LZ, Kain SR. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Research. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooya T, Choi HS, Yamashita A, Yui N, Sugaya Y, Kano A, Maruyama A, Akita H, Ito R, Kogure K, Harashima H. Biocleavable polyrotaxane - Plasmid DNA polyplex for enhanced gene delivery. Journal of the American Chemical Society. 2006;128:3852–3853. doi: 10.1021/ja055868+. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Yadava P, Hughes J. Polyethylenimine strategies for plasmid delivery to brain-derived cells. Methods. 2004;33:144–50. doi: 10.1016/j.ymeth.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Ahn CH, Chae SY, Bae YH, Kim SW. Biodegradable poly (ethylenimine) for plasmid DNA delivery. Journal of Controlled Release. 2002;80:273–282. doi: 10.1016/s0168-3659(01)00547-8. [DOI] [PubMed] [Google Scholar]
- 36.Zack DJ, Stempniak M, Wong AL, Taylor C, Weisbart RH. Mechanisms of cellular penetration and nuclear localization of an anti-double strand DNA autoantibody. J Immunol. 1996;157:2082–8. [PubMed] [Google Scholar]
- 37.Madaio MP, Yanase K. Cellular penetration and nuclear localization of anti-DNA antibodies: mechanisms, consequences, implications and applications. J Autoimmun. 1998;11:535–8. doi: 10.1006/jaut.1998.0217. [DOI] [PubMed] [Google Scholar]
- 38.Ziegler A, Seelig J. High affinity of the cell-penetrating peptide HIV-1 Tat-PTD for DNA. Biochemistry. 2007;46:8138–8145. doi: 10.1021/bi700416h. [DOI] [PubMed] [Google Scholar]
- 39.Breunig M, Lungwitz U, Liebl R, Klar J, Obermayer B, Blunk T, Goepferich A. Mechanistic insights into linear polyethylenimine-mediated gene transfer. Biochimica Et Biophysica Acta-General Subjects. 2007;1770:196–205. doi: 10.1016/j.bbagen.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Sundaram S, Lee LK, Roth CM. Interplay of polyethyleneimine molecular weight and oligonucleotide backbone chemistry in the dynamics of antisense activity. Nucleic Acids Res. 2007;35:4396–408. doi: 10.1093/nar/gkm450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J Pharm Pharmacol. 2003;55:721–34. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 42.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 43.Nakanishi M, Eguchi A, Akuta T, Nagoshi E, Fujita S, Okabe J, Senda T, Hasegawa M. Basic peptides as functional components of non-viral gene transfer vehicles. Curr Protein Pept Sci. 2003;4:141–50. doi: 10.2174/1389203033487234. [DOI] [PubMed] [Google Scholar]
- 44.Ferrer-Miralles N, Vazquez E, Villaverde A. Membrane-active peptides for non-viral gene therapy: making the safest easier. Trends Biotechnol. 2008;26:267–75. doi: 10.1016/j.tibtech.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–7. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 46.Chockalingam K, Blenner M, Banta S. Design and application of stimulus-responsive peptide systems. Protein Eng Des Sel. 2007;20:155–61. doi: 10.1093/protein/gzm008. [DOI] [PubMed] [Google Scholar]
- 47.Massodi I, Bidwell GL, 3rd, Raucher D. Evaluation of cell penetrating peptides fused to elastin-like polypeptide for drug delivery. J Control Release. 2005;108:396–408. doi: 10.1016/j.jconrel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Lochmann D, Jauk E, Zimmer A. Drug delivery of oligonucleotides by peptides. Eur J Pharm Biopharm. 2004;58:237–51. doi: 10.1016/j.ejpb.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 49.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–5. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 50.Bremner KH, Seymour LW, Logan A, Read ML. Factors influencing the ability of nuclear localization sequence peptides to enhance nonviral gene delivery. Bioconjug Chem. 2004;15:152–61. doi: 10.1021/bc034140k. [DOI] [PubMed] [Google Scholar]
- 51.Cartier R, Reszka R. Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 2002;9:157–67. doi: 10.1038/sj.gt.3301635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this manuscript can be found online.