Abstract
Juvenile rheumatoid arthritis (JRA) is mediated by Th1-immune responses. In children with JRA, synovial T-cells express high levels of the Th1-chemokine receptor CCR5, which has been implicated in susceptibility to rheumatoid arthritis. To test the hypothesis that genetic variation in CCR5 is associated with susceptibility to JRA, we analyzed patterns of variation in the 5'cis-regulatory region of CCR5 in 124 multiplex families from a JRA affected-sibpair registry. After sequencing the upstream region of CCR5, variants were tested for association with JRA by transmission disequilibrium testing. A single nucleotide polymorphism, C-1835T, was significantly under-transmitted to children with early-onset JRA (p<0.01). C-1835T was genotyped in 424 additional simplex and multiplex families. CCR5-1835T allele was under-transmitted in the cohort of all probands with JRA (p<0.02), as well as in those with early-onset (p<0.01) or pauciarticular JRA (p<0.05). Another variant, a 32-bp deletion in the open reading frame of CCR5, (CCR5-Δ32), was also tested in ~700 simplex and multiplex families. CCR5-Δ32 was also significantly under-transmitted to probands with early-onset JRA (p<0.05). Both variants are in regions under natural selection, and result in functional consequences. Our results suggest these CCR5 variants are protective against early-onset JRA.
Keywords: Juvenile arthritis, CCR5, association, TDT, autoimmunity, chemokines
Introduction
Juvenile rheumatoid arthritis (JRA) is a heterogeneous collection of chronic arthritides in children.1 The subtypes of JRA are similar to the major subtypes of Juvenile Idiopathic Arthritis (JIA) described by the International League of Associations for Rheumatology.2 Both genetic and environmental factors are believed to play a role in the pathogenesis of JRA. Numerous associations have been reported between JRA and polymorphisms in genes that encode the human leukocyte antigens (HLA). However, HLA associations account only in part for the overall genetic susceptibility to JRA3, suggesting that other, non-HLA variants also influence susceptibility to JRA.4 The search for non-HLA variants that influence susceptibility to JRA has been challenging for several reasons, including the large number of candidate genes and access to relatively small JRA cohorts that has limited statistical power to detect associations. These challenges can be overcome, in part, by performing adequately powered studies of candidate genes that play a compelling role in the pathophysiology of JRA.
Direct and indirect evidence suggests that the pathogenesis of JRA is mediated, in part, by T-cells. The inflamed synovial tissue in JRA is characterized by a prominent infiltrate of T-cells, plasma cells, macrophages and proliferating synoviocytes.5,6 The recruitment of pro-inflammatory cells into the synovium of children with JRA is mediated by chemokines that selectively attract Th1 T-cells.7–9 These T-cells are characterized by the production of interleukin-2, interferon-γ, and tumor necrosis factor-β. Several studies have demonstrated that Th1 cytokines also predominate in the synovial fluid samples from patients with JRA.10–12
A complex network of chemokines and chemokine receptors orchestrates the homing of T-cells to sites of inflammation. CC chemokine receptor 5 (CCR5) is a receptor preferentially expressed on the surface of Th1-cells. The predominance of CCR5-positive synovial mononuclear cells among patients with different types of inflammatory arthritis suggests that CCR5 plays an important role in synovial inflammation.13 Expression of CCR5 is also increased in synovial T-cells from children with JRA.7 Furthermore, two of the ligands for CCR5, CCL3 and CCL5, have been shown to be elevated in synovial fluid samples from adults with rheumatoid arthritis (RA), as well as in children with JIA.14 CCR5 blockade in collagen induced arthritis, an animal model of RA results in the inhibition of arthritis.15, 16 Together, these data suggest that genetic variation in CCR5 could affect the phenotype of inflammatory arthritis including JRA.
A 32-bp deletion in the open reading frame (exon 3) of the gene encoding CCR5 (CCR5-Δ32), has been tested for association with RA in adults with conflicting results.17–20 Homozygosity for CCR5-Δ32 results in absent expression of CCR5 on the cell surface. This allele is associated with protection against RA in adults.21 However, a systematic investigation of this and other CCR5 polymorphisms in children with JRA has not been reported to our knowledge. The objective of the present study was to test the hypothesis that genetic variants at the CCR5 locus are associated with susceptibility to JRA or subtypes of JRA.
Results
Sequence variation at the CCR5 locus
The complete sequence of the 5’ cis-regulatory region of CCR5 was determined in 507 individuals from 124 multiplex JRA families. A total of 10 single nucleotide polymorphisms (SNPs) were identified (Table 1), most of which have been described previously.22 The frequencies of these alleles were comparable to published frequencies in individuals of European ancestry. Two SNPS (G-2248A, G-1939A) were seen in one family each, and one SNP (C-2132T) was observed in two families. All SNPs were in Hardy Weinberg Equilibrium (HWE).
Table 1.
Single nucleotide polymorphisms in the 5’cis-regulatory region of CCR5
| SNP Name1 | Variant | Minor allele frequency |
|---|---|---|
| −2852 | A / G | 0.48 |
| −2733 | A / G | 0.15 |
| −2554 | G / T | 0.32 |
| −2459 | A / G | 0.41 |
| −2248 | G / A | 0.002 |
| −2135 | C / T | 0.41 |
| −2132 | C / T | 0.005 |
| −2086 | A / G | 0.31 |
| −1939 | G / A | 0.002 |
| −1835 | C / T | 0.105 |
SNP: Single nucleotide polymorphism. The positions are relative to the starting of CCR5 transcriptional start site. Minor allele frequencies were calculated from unaffected, unrelated parents of the multiplex JRA cohort.
The allele frequency of CCR5-Δ32 among unrelated, unaffected parents was ~10 %, similar to figures published for individuals of European ancestry23. The genotype frequencies of the wild type CCR5 and CCR5-Δ32 in unaffected, unrelated individuals did not deviate from HWE. The allele and genotype frequencies among unrelated, unaffected parents, siblings and probands are shown in Table 2. None of more than 2000 individuals from ~700 families that were typed for both CCR5 −1835 and CCR5-Δ32 were homozygous for the minor alleles of both loci.
Table 2.
Distribution of CCR5 genotypes in JRA families
| Genotype / Allele | Parents | Siblings | Probands |
|---|---|---|---|
| CCR5 -1835 C/C | 715 (79.4 %) | 368 (82.9%) | 546 (81.7%) |
| CCR5-1835 C/T | 158 (17.6 %) | 58 (13.1%) | 104 (15.6%) |
| CCR5-1835 T/T | 5 (0.6 %) | 2 (0.5%) | 4 (0.6%) |
| CCR5-1835 C allele frequency | 90. 4 % | 92.8 % | 91.4 % |
| CCR5-1835 T allele frequency | 9.6 % | 7. 2% | 8.6 % |
| CCR5 WT/WT | 948 (82.0 %) | 446 (80.8 %) | 692 (83.1 %) |
| CCR5 WT/ Δ32 | 190 (16.4 %) | 98 (17.8 %) | 135 (16.2 %) |
| CCR5 Δ32 /Δ32 | 18 (1.6 %) | 8 (1.4 %) | 6 (0.7 %) |
| CCR5 WT allele frequency | 90.2 % | 89.7 % | 91.2 % |
| CCR5 Δ32 allele frequency | 9.8 % | 10.3 % | 8.8 % |
Allele and genotype frequencies were calculated from parents, unaffected siblings and cases from the simplex and multiplex JRA cohorts.
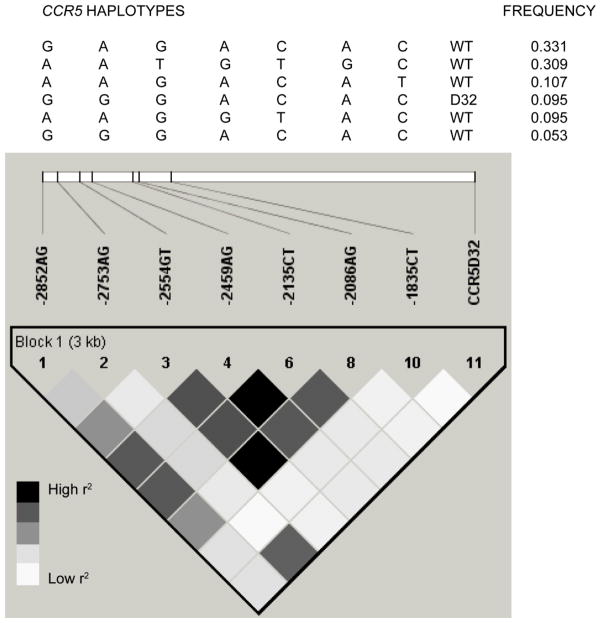
Examination of pairwise linkage disequilibrium (LD) between the CCR5 variants revealed that although this region comprised a single haplotype block using the definition of Gabriel et al,24 several pairs of polymorphisms showed low levels of LD using r2, another measure of LD (Figure 1). A total of 9 haplotypes were observed, of which four were rare, being detected in < 1% of the samples. CCR5 -1835T and CCR5-Δ32 were not in LD (r2 = 0.012), and these variants were on two different haplotypes, each with a frequency of ~10%. (Figure 1).
Figure 1. Linkage disequilibrium and haplotypes at the CCR5 locus.
An LD plot is depicted in the bottom part of the figure based on the measured r2. Each square represents the magnitude of LD for a single pair of markers, with black color indicating high r2. A key is provided below the LD plot. Analysis of this LD plot suggests the presence of a single haplotype block. Haplotypes spanning this block are shown above the LD plot, along with their frequency among unrelated, unaffected parents. Only haplotypes with a frequency >5 % are shown. The haplotypes were formed by SNPs at positions −2852, −2753, −2554, −2459, −2135, - 2086, and −1835 relative to the translational start site, and the CCR5-Δ32 polymorphism in the open reading frame on exon 3 of the CCR5 gene. Unrelated, unaffected parents of the multiplex families were used for the haplotype and LD analysis of all pairwise comparisons, with the exception of the comparison between the CCR5 C-1835T and CCR5-Δ32 polymorphisms, for which the unrelated, unaffected parents of all the simplex and multiplex families were used. CCR5 C-1835T and CCR5-Δ32 were 2.3 kb apart, and had a D′ of 1, but r2 of only 0.012. The frequencies of the CCR5-1835C-CCR5 WT, CCR5-1835T-CCR5 WT, CCR5-1835C-CCR5 Δ32, and CCR5-1835T-CCR5 Δ32, two-locus haplotypes were 9.9 %, 80.4 %, 9.7 % and 0 % respectively.
Association between JRA and CCR5 variants
Analysis of the 124 multiplex families revealed no statistically significant associations with any of the SNPs or haplotypes and the JRA cohort as a whole. However after stratification of the JRA cohort into the sub-phenotypes, one allele, CCR5 -1835T was significantly under-transmitted to probands with early-onset JRA (p < 0.01) by transmission disequilibrium test (TDT). A CCR5-haplotype that contained this SNP was also was under-transmitted to children with disease onset before 6 years of age, but this result was not statistically significant. CCR5 C-1835T was genotyped in 23 additional multiplex families and 401 simplex trios with JRA. Examination of the combined cohort confirmed that the −1835T allele was significantly under-transmitted in the cohort of all patients with JRA (p < 0.02), as well as patients with pauciarticular JRA (p < 0.05) and early-onset JRA (p < 0.01) (Table 3). The family-based estimate of the odds ratio for the level of protection conferred by the transmission of −1835T was 0.5 (95 % confidence interval 0.3–0.8, p <0.001) in 254 early-onset JRA trios where there were 37 transmissions (34 %) of −1835T, while the −1835 C allele was transmitted 52 % of the time. Because 21 of the simplex families were ascertained in Utah, whereas the other families were part of a Cincinnati Cohort, we repeated the analyses excluding the Utah cases. The result of the TDT was virtually the same and the frequency of the minor allele among unrelated, unaffected parents was not significantly different between the Cincinnati and Utah simplex cohorts.
Table 3.
Results of transmission disequilibrium testing for transmission of the −1835 T allele
| JRA phenotype | Number of families | Number of affected | −1835C | −1835T | P1 | ||
|---|---|---|---|---|---|---|---|
| Observed | Expected | Observed | Expected | ||||
| All JRA2 | 548 | 651 | 1190 | 1172 | 112 | 130 | 0.02 |
| Pauciarticular | 269 | 313 | 573 | 562 | 53 | 64 | 0.05 |
| Polyarticular | 225 | 240 | 440 | 435 | 40 | 45 | 0.25 |
| Systemic | 83 | 83 | 151 | 149 | 15 | 17 | 0.33 |
| Early onset3 | 317 | 350 | 643 | 630 | 57 | 70 | 0.01 |
| Late onset3 | 224 | 241 | 441 | 433 | 41 | 49 | 0.07 |
| Unaffected siblings4 | 269 | 4244 | 779 | 787 | 61 | 69 | 0.13 |
P values were calculated by bootstrap simulation based on 10,000 samples.
JRA: Juvenile rheumatoid arthritis. All JRA refers to the combined cohort of simplex and multiplex families.
Early onset was defined as onset before age 6, and late onset was defined as onset at 6 years or later
Children with JRA were excluded from this analysis, and only families with unaffected siblings of children with JRA were used for this analysis.
Examination of the combined simplex and multiplex cohort of ~700 families by TDT revealed no significant deviation of transmission of CCR5-Δ32 in the combined cohort of probands with JRA (Table 4). However, after stratification, CCR5-Δ32 was significantly under-transmitted to probands with early onset JRA (p< 0.05). The family-based estimate of the odds ratio for the level of protection conferred by the transmission of CCR5-Δ32 was 0.8 (95 % confidence interval 0.5–1.2, p < 0.2) in 330 early-onset JRA trios where there were 57 transmissions (44.5%) of CCR5Δ32, while the CCR5 WT was transmitted 51 % of the time. The lack of statistical significance of the odds ratio might reflect lack of power since only ~70 % of the cohort had both parents’ genotypes available.
Table 4.
Results of transmission disequilibrium testing for transmission of the CCR5-Δ32 allele
| JRA phenotype | Number of families | Number of affected | CCR5 WT | CCR5-Δ 32 | P1 | ||
|---|---|---|---|---|---|---|---|
| Observed | Expected | Observed | Expected | ||||
| All JRA | 697 | 819 | 1493 | 1486 | 145 | 152 | 0.36 |
| Pauciarticular | 346 | 409 | 746 | 738 | 72 | 80 | 0.14 |
| Polyarticular | 283 | 303 | 543 | 547 | 63 | 59 | 0.47 |
| Systemic | 104 | 104 | 198 | 194 | 10 | 14 | 0.08 |
| Early onset2 | 405 | 459 | 850 | 839 | 68 | 79 | 0.05 |
| Late onset2 | 327 | 355 | 634 | 637 | 76 | 74 | 0.66 |
| Unaffected siblings3 | 348 | 5483 | 984 | 989 | 112 | 107 | 0.46 |
P values were calculated by bootstrap simulation based on 10,000 samples.
Early onset was defined as onset before age 6, and late onset was defined as onset at 6 years or later
Children with JRA were excluded from this analysis, and only families with unaffected siblings of children with JRA were used for this analysis.
In addition, we applied TDT only to the unaffected siblings of the probands with JRA in order to detect if there was a possible transmission distortion of CCR5 −1835T and CCR5-Δ32 in these families. An allele-specific segregation bias, which is defined as an altered transmission of an allele independent of its role in disease, could falsely inflate the statistical significance. Hence, we tested if these alleles could also be preferentially transmitted to unaffected siblings of the JRA probands. There was no significant under or over-transmission of either the CCR5 -1835T or CCR5-Δ32 to the unaffected siblings in these families (Tables 3 and 4). Thus, these data provide evidence that our findings are not a result of segregation bias.
We also performed TDT analysis for the transmission of the CCR5 C-1835T and CCR5-Δ32 haplotypes (Table 5). The haplotype containing the rare alleles of both loci was not observed. However, the haplotype containing the major alleles at both loci (−1835C and CCR5 wild type in the open reading frame), was significantly over-transmitted to all probands with JRA (p < 0.03), and to those with early onset (<0.003), or pauciarticular subtypes (< 0.02). TDT analysis of the unaffected siblings showed no significant deviation of transmission of this haplotype. The results of the two other haplotypes containing one minor allele at each locus were similar to the results of the TDT analysis for the individual polymorphisms, and generally trended in the direction of under-transmission of the minor alleles to the probands with JRA, early onset or pauciarticular subtypes.
Table 5.
Results of transmission disequilibrium testing for transmission of CCR5 C-1835T and CCR5-Δ32 haplotypes
| JRA Phenotype | Number of families | Number of affected | −1835C −CCR5 WT | −1835C −CCR5 Δ32 | −1835T −CCR5 WT | −1835T-CCR5 Δ32 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obs1 | Exp1 | P2 | Obs | Exp | P2 | Obs | Exp | P2 | Obs | Exp | P2 | |||
| All JRA | 537 | 660 | 1082 | 1062 | 0.03 | 120 | 136 | 0.03 | 117 | 121 | ns | 0 | 0.6 | ns |
| Pauciarticular | 273 | 337 | 549 | 533 | 0.019 | 62 | 72 | ns | 63 | 68 | ns | 0 | 0.24 | ns |
| Polyarticular | 201 | 221 | 360 | 360 | ns | 37 | 41 | ns | 46 | 41 | ns | - | - | - |
| Systemic | 99 | 99 | 170 | 165 | ns | 19 | 20 | ns | 9 | 14 | 0.04 | - | - | - |
| Early onset 3 | 325 | 379 | 633 | 614 | 0.003 | 66 | 78 | 0.02 | 59 | 66 | ns | 0 | 0.6 | ns |
| Late onset 3 | 247 | 276 | 444 | 441 | ns | 50 | 56 | ns | 58 | 55 | ns | - | - | - |
| Unaffected Siblings4 | 272 | 4304 | 707 | 704 | ns | 64 | 71 | ns | 89 | 85 | ns | 0 | 0.25 | ns |
Obs: Observed; Exp: Expected
P values were calculated by bootstrap simulation based on 10,000 samples.
Early onset was defined as onset before age 6, and late onset was defined as onset at 6 years or later.
Children with JRA were excluded from this analysis, and only families with unaffected siblings of children with JRA were used for this analysis.
Discussion
These results demonstrate that two variants (CCR5-1835T and CCR5-Δ32) in the gene encoding CCR5 are associated with JRA, especially in children with disease onset before the age of 6 years. Both variants are believed to have functional consequences, and it should be noted that both effects on JRA were in the same direction, i.e., protective against JRA.
Case-control association studies are commonly used to investigate associations between phenotypes of interest and genetic variants. However, case-control studies can produce spurious associations due to population stratification. Several approaches have been suggested to account for population stratification.25 Many of these approaches use family-based controls to reduce the effect of population stratification. The TDT provides a joint test of linkage and association and eliminates the effects of stratification when applied to probands and parents. Although TDT based methods are more robust to population stratification, they are thought to be less powerful than conventional case-control studies at detecting associations.25 While failure to detect statistically significant associations by TDT could be due to type 2 statistical errors, positive results obtained by TDT are meaningful. We think that the demonstration of significant deviation of transmission of two CCR5 variants to children with JRA likely represents a true association. This conclusion is further supported by the observation that there is no deviation of transmission of these alleles to unaffected children in these families, although ideally a larger cohort of unaffected siblings would have likely given more robust results.
To minimize statistical artifacts, our strategy was to first test the 5’ cis-regulatory CCR5 variants in only the multiplex cohort, and then to confirm the significant results by genotyping a large cohort of simplex families. Thus, the only SNP we tested in the entire cohort was C-1835T, and we demonstrated a significant association not only with the entire JRA cohort, but also some JRA subtypes, which had been designated a priori. The other polymorphism tested, CCR5-Δ32 was chosen based on its known functional consequence and confirmed associations in adults with RA. Again we had designated the subtypes to be tested a priori. While the JRA subtypes have unique clinical and genetic features, they cannot be thought of as being entirely independent. Similarly variants in LD with one another are also not independent. For these reasons, we did not think it appropriate to apply the conventional Bonferroni correction to our results. Moreover, the Bonferroni correction tends to be very conservative, increasing the possibility of a type-2 statistical error. Our combined JRA cohort is one of the largest reported among studies looking at genetic associations in JRA, likely further minimizing type-2 statistical errors. Our study had sufficient power (> 0.8) to detect an association under most models for a marker allele frequency of 0.1, both for JRA as a whole, and for most sub-phenotypes examined.26 Ultimately, like other genetic association studies the present observations need to be validated by replication in independent cohorts.
The SNP CCR5 C-1835T, located in the 5’ cis-regulatory region of CCR5 has been shown to result in loss of novel nuclear factor binding.27 Investigations of the transcriptional activity of different haplotypes in the cis-regulatory region of CCR5 have demonstrated that CCR5-haplotypes containing -1835T (HHF) have significantly different promoter efficiency compared to other haplotypes.27 Haplotypes containing −1835T are associated with different disease modifying effects among patients with acquired immune deficiency syndrome.28 These data suggest the possibility that −1835T is protective against developing early onset JRA, conceivably by altering the expression level of CCR5. Although one would expect these haplotypes to decrease the expression of CCR5, haplotypes containing −1835T appear to increase the efficiency of the CCR5 promoter in vitro. It is possible that −1835T may have other effects in vivo. Also, this effect of −1835T is on gene expression, and the effect if any, of −1835T on cell surface expression of CCR5 is unknown. Furthermore the effects observed could be due to complex interactions with other polymorphisms on these haplotypes. Ideally, experiments addressing the genotype-phenotype correlations of CCR5 variants and CCR5 expression in subjects with JRA would better address the role played by this polymorphism and others in the development or phenotype of inflammatory arthritis. It is also possible that another variant in LD with this SNP influences the protective effect observed here. For instance −1835T has been reported to be in LD with a SNP in the coding region of another chemokine receptor gene, CCR2 (CCR2-V64I), located ~9kb 5’ of the CCR5 gene on chromosome 3p21.31.22 The CCR2-64I allele has also been shown to have protective influences against human immunodeficiency virus (HIV) disease progression.29
It has been proposed that genetic variants subjected to natural selection are likely to harbor functional polymorphisms and hence are good candidates for association studies.30 The 5’ cis-regulatory region of CCR5 has been demonstrated to be under balancing selection, where diverse haplotypes have been maintained.22 The 5’cis-regulatory region of CCR5 has several polymorphisms that regulate the expression of CCR5, and different CCR5 SNPs and haplotypes have been associated with phenotypic differences in HIV infection.28 It has also been suggested that the CCR5-Δ32 allele has been subjected to recent positive selection.31, 32 (But see 33). The fact that our associations have been discovered in regions affected by natural selection strengthens the evidence that they might have functional significance.
Studies investigating the association between CCR5-Δ32 and RA in adults have reported conflicting results.17, 20 However, a meta-analysis of the published association studies between CCR5-Δ32 and RA suggests that some of these studies were underpowered, and confirms that this polymorphism is strongly and negatively associated with RA (p < 0.0001).21 This suggests that CCR5-Δ32 is associated with protection against the development of RA. Until now, this polymorphism has not been formally investigated in association studies of individuals with JRA, although two children with JRA who were homozygous for this mutation have been described.7, 9 The demonstration of under-transmission of CCR5-Δ32 to probands with early onset JRA is consistent with these observations and suggests that JRA and RA share some genetic risk factors.
To answer the question whether these association results could be due to LD between CCR5 - 1835T and CCR5-Δ32, we examined the haplotypes and the LD pattern in CCR5. The minor alleles, −1835T and CCR5-Δ32 were not in LD, and they occur on two different haplotypes. Examination of the published data on Great Apes, and Old and New world monkeys, shows that neither allele is present in these species and that both are newly derived in humans, suggesting that both of these variants are derived from the ancestral haplotype of CCR5-1835C-CCR5 WT.27 Thus, the protective effects observed at these loci appear to be independent of each other and not due to LD. As anticipated, we did not find haplotypes that had the minor alleles at both loci. However, the haplotype containing CCR5 −1835C and CCR5 WT (i.e., lacking the protective variants) did show evidence of excessive transmission to all children with JRA and those with early onset JRA by TDT, further strengthening our results. Together these observations support the notion that CCR5 might play important roles in the pathogenesis of inflammatory arthritis, suggesting that CCR5 blockade could modify the phenotype of inflammatory arthritis.
Methods
DNA samples were obtained from children with JRA and one or both parents. DNA from unaffected siblings was also available in several families. The diagnosis of JRA was made according to the American College of Rheumatology (ACR) criteria.1 As most subjects had been enrolled before adoption of the more recently proposed criteria for the diagnosis of JIA, the ACR criteria for a diagnosis of JRA was used. Samples from 147 multiplex families with two or more affected siblings with JRA from around the USA, and 540 simplex families with one affected child with JRA from Cincinnati OH were available from a JRA Registry sponsored by the National Institutes of Arthritis and Musculoskeletal and Skin diseases, and the Cincinnati Children’s Hospital Medical Center. The multiplex families were ascertained and enrolled from around the USA through a JRA Registry as described earlier.34 Additionally, 21 simplex JRA families were ascertained from Salt Lake City UT. In all, DNA from both parents was available in 105 of the multiplex families, 370 of the simplex families from Cincinnati, and all 21 families from Utah. DNA from at least one parent was available in most of the remaining families. DNA from unaffected siblings was available in 38 multiplex and 313 Cincinnati simplex families. Institutional review board approvals were obtained from participating institutions, and parents and/or children provided informed consent. In all, there were 419 children with pauciarticular onset (< 5 joints in the first 6 months of disease), 331 with polyarticular onset (5 or more joints in the first 6 months of disease), and 107 with systemic onset JRA. The median age of onset of JRA in the simplex cohort was 5.8 years, and the median age on onset in the multiplex cohort was 4.5 years. The children with JRA were predominantly female, similar to other studies, with 72% and 73 % of the cases from simplex and multiplex cohorts, respectively, being female. Most (85% of the simplex and 88% of the multiplex) families were of European Ancestry.
CCR5 sequencing
A 1122 bp region corresponding to human CCR5 −2867 to −1745 was amplified by polymerase chain reaction (PCR) in 507 individuals from 124 multiplex families, and sequenced on both strands as previously described.22 Briefly, cycle sequencing was carried out in 5μl reaction volumes using ABI BigDye (v3.1) Terminator chemistry. The sequence ladders were precipitated with 62.5% EtOH/1M KOAc, resuspended in 15 μl of dH2O and electrophoresed on an ABI3700 DNA analyzer prepared with POP-5 capillary gel matrix. The sequenced region includes part of exon 1, intron 1, exons 2A and 2B, and the 5’ end of intron 2. All of these exons are non-coding. This region, defined in earlier publications as the 5’ cis regulatory region, contains several unique CCR5-SNPs and haplotypes that have been shown to have functional consequences.22, 27 The primers used for PCR amplification were CCR5L2 (5’ - CCA AAC TGT GAC CCT TTC C – 3’) and CCR5R3: (5’ - AGT AGC TCT CTG CTG TCT TCT CA – 3’). The programs PHRED, PHRAP and CONSED were used to evaluate sequence trace files35. Potential heterozygotes were identified by using the program POLYPHRED version 3.5. Polymorphisms were verified by examining the individual sequence trace files manually, and for most polymorphisms both the forward and reverse sequences were evaluated. CCR5 re-sequencing allowed us to successfully identify ~ 99% of the polymorphisms in this region.
Genotyping
DNA samples from 23 multiplex families and 401 simplex families were genotyped for the SNP C-1835T, using a PCR restriction fragment length polymorphism based assay using EcoRV as described previously.28 This SNP results in a C to T change at position −1835 in the 5’ cis-regulatory region of CCR5, and has been shown to result in loss of novel nuclear factor binding.27 The restriction site for EcoRV was created by changing an A to G in the antisense primer at position −1832. The enzyme digests the amplicons that contain the T allele at position − 1835. The following primers were used for PCR amplification: CCR5927_S: (5’ GTT GGT TTA AGT TGG CTT 3’) and CCR5927_AS: (5’ ATC TTA AAG ATT ATA TTT TAA GAT AAT TGT ATG AGC ACT TGG TGT TTG CCA GAT -3’). The genotyping success rate was 95%. Several representative samples that had been sequenced were also genotyped by the RFLP method for quality control, and the genotyping calls were consistent.
Genotyping for the CCR5-Δ32 polymorphism was carried out by performing a PCR reaction in 697 multiplex and simplex families with JRA. PCR was performed with the following primers spanning the region of the CCR5-Δ32 deletion: CCR5_d32_R: 5’ TTC AGG AGA AGG ACA ATG TTG TAG G 3’ and CCR5_d32_L: 5’ ATT ACA CCT GCA GCT CTC ATT TTC 3’. The observed wild type and deletion fragments were 250 bp and 218 bp respectively. The genotyping success rate was 99%.
Statistical analysis
Pairwise LD between the different SNPs in the promoter region and the CCR5-Δ32 polymorphism was assessed using the program Haploview, which also was used to infer haplotypes and haplotype blocks.36 Only variants with frequency > 5 % were used for inferring haplotypes and haplotype blocks. The haplotype block definition proposed by Gabriel et al was used, wherein pairs of SNPs in strong LD are defined as those in which the one-sided upper 95% confidence bound on D′ (a normalized measure of allelic variation) is >98% and the lower bound is >70%.24 LD was also assessed using an alternate measure, r2, which unlike D' is not strongly biased by low frequency variants or missing haplotypes.
All SNPs were tested for HWE using unrelated, unaffected individuals (i.e., parents). SNPs and haplotypes were tested for association with JRA by TDT, which tests for preferential transmission of specified alleles from heterozygous parents to the affected offspring. TDT analysis was performed using TRANSMIT 2.5.4.37 TRANSMIT infers missing parental haplotypes using HWE, thus allowing the inclusion of all families in the analysis.
The multiplex JRA cohort was analyzed for an association with the CCR5 SNPs as well as the haplotypes. The combined JRA cohort was analyzed for an association with C-1835T by TDT. Additionally, 697 simplex and multiplex families were analyzed for an association with CCR5- Δ32 polymorphism by TDT. All analyses were repeated after stratifying the JRA cohort by onset type (pauciarticular, polyarticular, or systemic), as well as by age of onset (early onset < 6 years, or late onset > 6 years). The division into early/late onset JRA included all subtypes. These JRA sub-phenotypes were designated a priori based on reports suggesting that JRA comprises a collection of clinically and genetically heterogeneous phenotypes38, 39, and by the demonstration of age-specific effects of JRA associated HLA alleles.40 Furthermore the various subtypes of juvenile arthritis differ in their outcomes.41 The approach of stratifying the JRA cohorts in the present study was similar to the stratification scheme employed in the genome-wide scan for JRA, which identified several regions of suggestive linkage on stratification of the JRA cohort into sub-phenotypes.42 These observations suggest that the various subtypes of JRA likely represent distinct phenotypes that share the feature of inflammatory arthritis in children. In addition TDT was also performed using only the unaffected siblings to determine if there was preferential transmission of CCR5 alleles or haplotypes to unaffected siblings as well in these families.
We also calculated the family-based estimate of the odds ratio for the level of protection conferred by CCR5 −1835T and by CCR5-D32 in families of probands with early onset JRA using the methods described by Ackerman et al, where transmitted versus non-transmitted alleles from the parental matings are compared.43 They proposed that in the absence of segregation distortion, and in families in HWE, the non-transmitted chromosomes can be used as population controls to estimate relative risk. Both of these variants were in HWE in our cohort, and there was no significant deviation of transmission of these variants to unaffected siblings, suggesting a lack of segregation distortion. Only trios where both parental genotypes where available were used for these calculations. Thus, 254 and 330 trios with early onset JRA were used for the analyses of C-1835T and CCR5-Δ32 respectively.
Acknowledgments
Supported in part by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (K23-AR-50177, N01-AR-42272, P01-AR-048929, P30-AR-47363), National Center for Research Resources (M01-RR-00064); The Cincinnati Pediatric Rheumatology Tissue Repository, and The Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; The Val A Browning Charitable Foundation, The Primary Children’s Medical Center Foundation, The Clinical Genetics Research Program, and The Children’s Health Research Center Salt Lake City, UT. We would also like to thank the families that participated in this project.
References
- 1.Cassidy JT, et al. A study of classification criteria for a diagnosis of juvenile rheumatoid arthritis. Arthritis Rheum. 1986;29:274–81. doi: 10.1002/art.1780290216. [DOI] [PubMed] [Google Scholar]
- 2.Petty RE, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 3.Prahalad S, et al. Juvenile rheumatoid arthritis: linkage to HLA demonstrated by allele sharing in affected sibpairs. Arthritis Rheum. 2000;43:2335–8. doi: 10.1002/1529-0131(200010)43:10<2335::AID-ANR22>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 4.Rosen P, Thompson S, Glass D. Non-HLA gene polymorphisms in juvenile rheumatoid arthritis. Clin Exp Rheumatol. 2003;21:650–6. [PubMed] [Google Scholar]
- 5.Wynne-Roberts CR, Anderson C. Light- and electron-microscopic studies of normal juvenile human synovium. Semin Arthritis Rheum. 1978;7:279–86. doi: 10.1016/0049-0172(78)90026-4. [DOI] [PubMed] [Google Scholar]
- 6.Wynne-Roberts CR, Anderson CH, Turano AM, Baron M. Light-and electronmicroscopic findings of juvenile rheumatoid arthritis synovium: comparison with normal juvenile synovium. Semin Arthritis Rheum. 1978;7:287–302. doi: 10.1016/0049-0172(78)90027-6. [DOI] [PubMed] [Google Scholar]
- 7.Wedderburn LR, Robinson N, Patel A, Varsani H, Woo P. Selective recruitment of polarized T cells expressing CCR5 and CXCR3 to the inflamed joints of children with juvenile idiopathic arthritis. Arthritis Rheum. 2000;43:765–74. doi: 10.1002/1529-0131(200004)43:4<765::AID-ANR7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Wedderburn LR, Woo P. Type 1 and type 2 immune responses in children: their relevance in juvenile arthritis. Springer Semin Immunopathol. 1999;21:361–74. doi: 10.1007/BF00812262. [DOI] [PubMed] [Google Scholar]
- 9.Thompson SD, et al. Chemokine receptor CCR4 on CD4+ T cells in juvenile rheumatoid arthritis synovial fluid defines a subset of cells with increased IL-4:IFN-gamma mRNA ratios. J Immunol. 2001;166:6899–906. doi: 10.4049/jimmunol.166.11.6899. [DOI] [PubMed] [Google Scholar]
- 10.Eberhard BA, Laxer RM, Andersson U, Silverman ED. Local synthesis of both macrophage and T cell cytokines by synovial fluid cells from children with juvenile rheumatoid arthritis. Clin Exp Immunol. 1994;96:260–6. doi: 10.1111/j.1365-2249.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray KJ, et al. Contrasting cytokine profiles in the synovium of different forms of juvenile rheumatoid arthritis and juvenile spondyloarthropathy: prominence of interleukin 4 in restricted disease. J Rheumatol. 1998;25:1388–98. [PubMed] [Google Scholar]
- 12.Ozen S, Tucker LB, Miller LC. Identification of Th subsets in juvenile rheumatoid arthritis confirmed by intracellular cytokine staining. J Rheumatol. 1998;25:1651–3. [PubMed] [Google Scholar]
- 13.Mack M, et al. Predominance of mononuclear cells expressing the chemokine receptor CCR5 in synovial effusions of patients with different forms of arthritis. Arthritis Rheum. 1999;42:981–8. doi: 10.1002/1529-0131(199905)42:5<981::AID-ANR17>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Pharoah DS, et al. Expression of the inflammatory chemokines CCL5, CCL3 and CXCL10 in juvenile idiopathic arthritis, and demonstration of CCL5 production by an atypical subset of CD8+ T cells. Arthritis Res Ther. 2006;8:R50. doi: 10.1186/ar1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plater-Zyberk C, Hoogewerf AJ, Proudfoot AE, Power CA, Wells TN. Effect of a CC chemokine receptor antagonist on collagen induced arthritis in DBA/1 mice. Immunol Lett. 1997;57:117–20. doi: 10.1016/s0165-2478(97)00075-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang YF, et al. A non-peptide CCR5 antagonist inhibits collagen-induced arthritis by modulating T cell migration without affecting anti-collagen T cell responses. Eur J Immunol. 2002;32:2124–32. doi: 10.1002/1521-4141(200208)32:8<2124::AID-IMMU2124>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.Cooke SP, Forrest G, Venables PJ, Hajeer A. The delta32 deletion of CCR5 receptor in rheumatoid arthritis. Arthritis Rheum. 1998;41:1135–6. doi: 10.1002/1529-0131(199806)41:6<1135::AID-ART24>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Garred P, et al. CC chemokine receptor 5 polymorphism in rheumatoid arthritis. J Rheumatol. 1998;25:1462–5. [PubMed] [Google Scholar]
- 19.Gomez-Reino JJ, et al. Association of rheumatoid arthritis with a functional chemokine receptor, CCR5. Arthritis Rheum. 1999;42:989–92. doi: 10.1002/1529-0131(199905)42:5<989::AID-ANR18>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Pokorny V, et al. Evidence for negative association of the chemokine receptor CCR5 d32 polymorphism with rheumatoid arthritis. Ann Rheum Dis. 2005;64:487–90. doi: 10.1136/ard.2004.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prahalad S. Negative association between the chemokine receptor CCR5-Delta32 polymorphism and rheumatoid arthritis: a meta-analysis. Genes Immun. 2006;7:264–8. doi: 10.1038/sj.gene.6364298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamshad MJ, et al. A strong signature of balancing selection in the 5' cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens JC, et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507–15. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 26.Chen WM, Deng HW. A general and accurate approach for computing the statistical power of the transmission disequilibrium test for complex disease genes. Genet Epidemiol. 2001;21:53–67. doi: 10.1002/gepi.1018. [DOI] [PubMed] [Google Scholar]
- 27.Mummidi S, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–61. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez E, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96:12004–9. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostrikis LG, et al. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–3. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 30.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 31.Galvani AP, Novembre J. The evolutionary history of the CCR5-Delta32 HIV-resistance mutation. Microbes Infect. 2005;7:302–9. doi: 10.1016/j.micinf.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biol. 2005;3:e339. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabeti PC, et al. The case for selection at CCR5-Delta32. PLoS Biol. 2005;3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroldo MB, Tague BL, Shear ES, Glass DN, Giannini EH. Juvenile rheumatoid arthritis in affected sibpairs. Arthritis Rheum. 1997;40:1962–6. doi: 10.1002/art.1780401107. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–51. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2004 doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65:1170–7. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fink CW, Fernandez-Vina M, Stastny P. Clinical and genetic evidence that juvenile arthritis is not a single disease. Pediatr Clin North Am. 1995;42:1155–69. doi: 10.1016/s0031-3955(16)40057-x. [DOI] [PubMed] [Google Scholar]
- 39.Prahalad S, Glass DN. Is juvenile rheumatoid arthritis/juvenile idiopathic arthritis different from rheumatoid arthritis? Arthritis Res. 2002;4:303–310. [Google Scholar]
- 40.Murray KJ, et al. Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum. 1999;42:1843–53. doi: 10.1002/1529-0131(199909)42:9<1843::AID-ANR8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 41.Prahalad S. Subtype-specific outcomes in juvenile idiopathic arthritis: a systematic review. Current Medical Literature: Rheumatology. 2006;25:1–9. [Google Scholar]
- 42.Thompson SD, et al. A genome-wide scan for juvenile rheumatoid arthritis in affected sibpair families provides evidence of linkage. Arthritis Rheum. 2004;50:2920–30. doi: 10.1002/art.20425. [DOI] [PubMed] [Google Scholar]
- 43.Ackerman H, et al. A comparison of case-control and family-based association methods: the example of sickle-cell and malaria. Ann Hum Genet. 2005;69:559–65. doi: 10.1111/j.1529-8817.2005.00180.x. [DOI] [PubMed] [Google Scholar]