Abstract
Appropriate glycosylation of recombinant therapeutic glycoproteins has been emphasized in biopharmaceutical industries because the carbohydrate component can affect safety, efficacy, and consistency of the glycoproteins. Reliable quantification methods are essential to ensure consistency of their products with respect to glycosylation, particularly sialylation. Mass spectrometry (MS) has become a popular tool to analyze glycan profiles and structures, showing high resolution and sensitivity with structure identification ability. However, quantification of sialylated glycans using MS is not as reliable because of the different ionization efficiency between neutral and acidic glycans. We report here that amidation in mild acidic conditions can be used to neutralize acidic N-glycans still attached to the protein. The resulting amidated N-glycans can then released from the protein using PNGase F, and labeled with permanent charges on the reducing end to avoid any modification and the formation of metal adducts during MS analysis. The N-glycan modification, digestion, and desalting steps were performed using a single-pot method that can be done in microcentrifuge tubes or 96-well microfilter plates, enabling high throughput glycan analysis. Using this method we were able to perform quantitative MALDI-TOF MS of a recombinant human glycoprotein to determine changes in fucosylation and changes in sialylation that were in very good agreement with a normal phase HPLC oligosaccharide mapping method.
Keywords: quantitative, sialylation, amidation, glycomics, MALDI-TOF MS
Introduction
More than two-thirds of commercialized therapeutic proteins are glycoproteins. Appropriate glycosylation of recombinant therapeutic glycoproteins has been emphasized in biopharmaceutical industries because the carbohydrate component can affect safety, efficacy, and consistency of the product.1-2 Particularly, the degree of sialylation is quite important because glycoproteins with desialylated glycans can be cleared rapidly from blood circulation.3-5 Glycosylation is an important enough issue that determining glycan structure at the clone screening stage is becoming a common practice.6-9 Also, during process development and optimization, changes in bioreactor operating parameters and downstream harvesting and purification methods can have a dramatic effect on the N-glycan structures in the final product.10-14 Storage conditions of process intermediates and even final vialed product can also affect the glycan profile.1,15-17 Therefore, it is essential to ensure consistency of glycosylation, particularly sialylated glycans. Ideally, a method for sialylated N-glycan analysis should be fast, robust and reliable, amenable to high throughput methods, provide structural information about the glycans, and yield quantitative information about what glycan structures are present in the product.
High Performance Liquid Chromatography (HPLC) analysis of fluorescently labeled glycans is commonly used for oligosaccharide mapping and quantification of glycans.18-22 Glycans can be resolved based on their size, charge, linkages, and overall structures. However, the heterogeneous and isomeric structures of N-glycans can result in chromatograms that do not have highly resolved baseline separation, and structures can not be readily determined without other orthogonal methods.
Mass spectrometry (MS) has become a popular tool to analyze glycan profiles and structures, showing high resolution and sensitivity with structure identification ability.23-25 Eukaryotic cells glycosylate with high mannose and complex structures, but for many recombinant proteins, sialylated complex glycans are critical to the target product profile. Neutral glycans such as high mannose and unsialylated complex glycans can be quantified with MS methods because the neutral charge state of the molecule does not affect ionization efficiency.23,26 However, quantification of sialylated glycans using MS is not as reliable because of the different ionization efficiency between neutral and acidic glycans.26-28 In addition, the glycosidic bonds of sialic acid easily decompose during ionization process under MALDI conditions, unless the carboxylic acid group is modified.29 Therefore, several groups have attempted neutralization of acidic glycans prior to MS quantification methods. Permethylation is a commonly used modification to neutralize acidic glycans, but degradation or loss of sialic acid residues under the harsh reaction conditions limits the reliability for quantification.30-31 Methyl esterification and amidation of the carboxyl group of sialic acid were also used to neutralize acidic glycans.28-29,32 However, neutralization of glycans is still not enough for reliable quantitative MS, particularly MALDI-TOF MS, due to the complicated mix of metal adduct such as [M+K]+ and [M+Na]+ that originate from the salts in the sample matrix.
The complicated mix of adduct ions can be simplified by introducing a permanent charge on the reducing end of neutralized glycans,33-34 showing better sensitivity and improved glycan quantification reliability in MALDI-TOF MS.26,35 Jang et al. performed the neutralization of sialylated glycan of recombinant monoclonal antibody using methyl-esterification and the resulted glycan was labeled with permanent positive charge.35 However, incomplete neutralization and degradation of sialic acid were observed on glycans containing α2-3 linked sialic acid.28 Toyota et al. reported that amidation of glycan using acetohydrazide (Ah) under mild acidic condition was free of flaws caused by the incomplete neutralization and the degradation of sialic acid.28 However, the reducing end of glycan released from protein is also subject to modification with the amidation reagent. Therefore introduction of permanent charge on the reducing end cannot be performed after the amidation.
We report here that amidation in mild acidic conditions can be used to neutralize acidic N-glycans still attached to the protein. The resulting amidated N-glycans can then be released from the protein using PNGase F, and labeled with permanent charges on the reducing end to avoid any modification and the formation of metal adducts during MS analysis. The amidated and then derivatized N-glycans were analyzed on MALDI-TOF MS and the resulting relative peak area percentages were compared with those obtained using a normal phase HPLC method (NP-HPLC). The N-glycan modification, digestion, and desalting steps were performed using a single-pot method that can be done in microcentrifuge tubes or 96-well microfilter plates, enabling high throughput glycan analysis. Using this method we were able to perform quantitative MALDI-TOF MS to determine changes in fucosylation and changes in sialylation that were in very good agreement with a normal phase HPLC oligosaccharide mapping method.
Materials and Methods
Materials
Human IgG, 2-aminobenzoic acid, iodoacetamide, and Girard’s T reagent were purchased from Sigma (St. Louis, MO). Acetohydrazide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) were obtained from TCI America (Portland, OR) and Thermo Co (Waltham, MA), respectively. N-glycosidase F (PNGase F) was obtained from New England Biolabs (Ipswich, MA). Transgenic pig derived human Factor IX was purified from pig milk as described in Gil et al 2008.36
Amidation of sialylated glycan
10-100 μg of glycoprotein was added on 96-Well Membrane-Bottom Filter Plate, 10K molecular weight cutoff (MWCO) (Harvard Apparatus, Holliston, MA) and spun down to dryness, and then reduced by adding 100 μL of 5 mM DTT and incubating at room temperature (RT) for 1 hr. Then 5 μL of 250 mM iodoacetamide and 20 μL of water were added to make 10 mM concentration and then incubated at RT in the dark for 30 min. The glycoprotein was desalted by washing with deionized water (5 × 200 μL with centrifugation at 5000 rpm). The desalted glycoprotein was reconstituted with 100 μl of 1M acetohydrazide, 20 μL of 1N HCl, and 20 μL of 2M EDC. The mixture was incubated at RT for 4 hr. The amidated glycoprotein was desalted by washing with deionized water (5 × 200 μL with centrifugation at 5000 rpm). PNGase F (substrate: enzyme = 250:1 mass ratio) in deionized water was added and incubated at 37 °C overnight. The released N-glycans were collected by centrifugation and then dried under speed vac. The dried N-glycan was reconstituted in 50/50 (v/v) Methanol/water for further modification.
Derivatization with 2-aminobenzoic acid
10 μl of amidated glycans in 50/50 (v/v) Methanol/water were mixed with 100 μL of 2-aminobenzoic acid solution (2-AA) consisting of 30 mg of 2-AA and 20 mg of sodium cyanoborohydride in 1 mL of 4% sodium acetate trihydrate (w/v) and 2% boric acid (w/v) in methanol. The mixture was heated at 80 °C for 40 min. After cooling, the mixture was diluted with 1 mL of 80 % acetonitrile in water and excess reagent was removed using C18 cartridge (Alltech, Milford, MA). The cartridge was conditioned with 2×1 mL of 95/5 (v/v) acetonitrile/water. After loading the glycan, excess reagent was removed with 2×1 mL of 95/5 (v/v) acetonitrile/water, and then the derivatized glycan was eluted with 2×0.5 mL of 20/80 (v/v) acetonitrile/water. The eluted glycan was analyzed using MALDI-TOF MS or NP-HPLC.
Derivatization with Girard’s T reagent
10 μL of N-glycan in 50/50 (v/v) Methanol/water was mixed with 100 μL of Girard’s T reagent (GT) solution (5 nmol/μL in 1/99 (v/v) acetic acid/methanol). The solution was incubated at RT for 4 hr. After reaction, the sample was dried under speed vac and then dissolved with 50/50 (v/v) methanol/water, and analyzed on MALDI-TOF MS without further purification steps.
Normal Phase-HPLC analysis
Normal phase HPLC profiling of 2-AA derivatized glycans was performed with an amine bonded polymeric column (Polymer-NH2, 5 μm, 4.6 mm ID × 250 mm L, Astec, Whippany, NJ) with a CPF10 prefilter (Vydac, Hesperia, CA) according to the method of Anumula.19 Solvent A was 2% acetic acid and 1% tetrahydrofuran (inhibited) in acetonitrile, and Solvent B 5% acetic acid, 3% triethylamine and 1% tetrahydrofuran (inhibited) in water. The gradient program started at 30% B for 2 min, followed by an increase to 50% B over 60 min and then to 95% B over 55 min. The column was isocratic with 95% B for 25 min and equilibrated with initial conditions for 15 min prior to next injection. Column temperature was 50 °C and flow rate constant at 1 mL/min. The HPLC system was a Waters 2695 Separations module and 2475 fluorescent detector (Milford, MA). The detector settings were 360 nm excitation, 425 nm emission, and 20 nm bandwidth. The peak area from a NP-HPLC profile was obtained using Empower software (Waters, Milford, MA).
MALDI-TOF MS analysis
1 μL (10-50 pmol) of a glycan sample was mixed with 1 μL of 2,5-dihydroxybenzoic acid (DHB) solution (30 mg/mL in 70/30 % (v/v) acetonitrile/water). Then 0.5 μL of the mixture was spotted on stainless steel MALDI plate (triplicate) and dried at room temperature. A Voyager DE Pro (Applied Biosystems, Foster City, CA) MALDI-TOF mass spectrometer was used to obtain MS spectra. The analysis parameters were as follows: reflectron mode, detector gain = 3.9, and laser power = 70%. 1000 shots from 20 different spots were scanned to acquire a MS spectrum. The intensity of each ion was obtained by integrating from 1st to 3rd isotopic peak of the ion. Data Explorer software (Applied Biosystems, Foster City, CA) was used to calculate the peak area. The relative percentage was the ratio of the intensity of each ion to the summed intensity of the total detected ions.
Results and Discussion
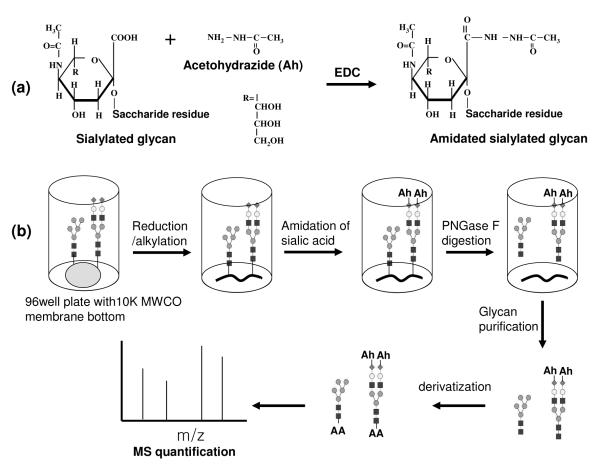
Conventional amidation and methylesterification were tried to neutralize sialylated N-glycan,37-38 but these methods were problematic with incomplete modification for α(2-3) linked sialic acid and their by products.28 A modified amidation method was reported with complete amidation of both α(2-3) and α(2-6) linked sialic acid and no by-products28 and so we adopted this method. As described in Fig 1(a), amidation was performed on the carboxyl group of sialylated N-glycans using acetohydrazide and EDC as a coupling reagent in mild acidic conditions. Acetohydrazide was selected because it can be easily coupled with carboxylic acid and there is little steric hindrance for the amidation reaction. However, the aldehyde of the reducing end of a free glycan is also likely reactive to hydrazide, so a permanent charge cannot be incorporated after the amidation reaction. To avoid this problem, we performed the amidation reaction on the sialylated N-glycan while it was still attached to the protein as shown in Fig 1(b). We note that the mildly acidic conditions may cause problems with the analysis of glycoproteins having a low pI, as proteins tend to aggregate and precipitate near their pI. The pI of the tg-FIX that we analyzed is in the range of 4-6 39, and we did not observe precipitation of our protein samples. The pI range of IgG from natural sources (such as what we used in this study) can vary widely from 5 to 940-41, whereas most recombinant monoclonal antibodies have a basic pI.42-43 If aggregation or precipitation becomes a problem, addition of denaturants or chaotropes may be necessary and should not interfere, but we did not explore if the reaction scheme we present here works with precipitating proteins. The resulting neutralized glycan can be labeled with a permanent charge on the reducing end after releasing it from protein by PNGase F, which promotes homogeneous ionization efficiency on MS, particularly in MALDI-TOF MS. In addition, the amidation reaction and the following desalting steps can be performed in a 96-well microfilter plate, so that fast and high-throughput glycan sample preparation is possible and the glycan sample loss can be minimized.
Fig 1.
(a) amidation of sialylated glycan, and (b) scheme of glycan analysis from glycoprotein using amidation and derivatization. Ah, Acetohydrazide
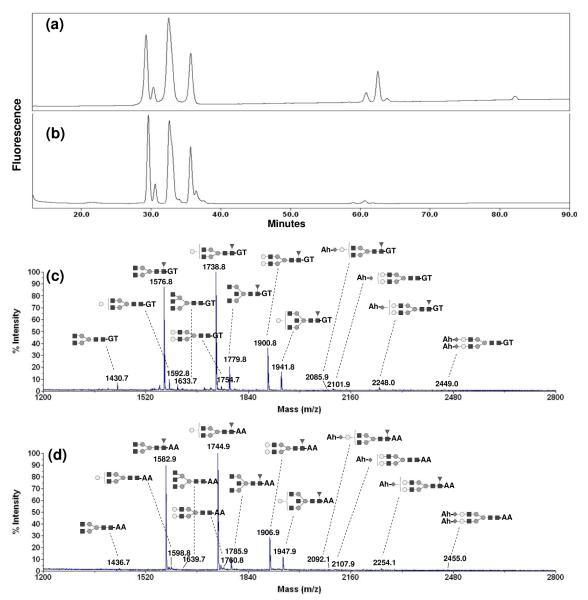
The first step of the process is neutralization of the sialic acids, while still preserving their presence and the structure of the N-glycan. We used human IgG as a model protein to demonstrate this. We compared the NP-HPLC profiles of human IgG N-glycan derivatized with 2-AA before (Fig 2(a)) and after (Fig 2(b)) amidation. After amidation, acidic glycans were clearly shifted to the neutral glycan region as shown in Fig 2(b), with less than 0.5% of the total peak area remaining for the sialylated glycan region (50-90 min in Fig 2(b)). These data showed that neutralization of acidic glycans by amidation was highly efficient. Fig 2(c) and (d) show the MALDI-TOF MS profiles of amidated human IgG N-glycan derivatized with two different reducing-end labels that impart a permanent charge on the released glycan: GT (positive) and 2-AA (negative). The GT-derivatized glycans were analyzed in positive mode and the 2-AA derivatized glycans in negative mode MALDI-TOF MS. All glycans observed on MALDI-TOF MS were the [M]+ ions for GT-derivatized glycans and [M-H]− adducts for 2-AA derivatized glycans. We did not observe any adduct associated with metal ion, for example [M+Na]+ or [M+Na-2H]− for GT or 2-AA derivatized glycans, and thus the spectra were simpler and easier to interpret. The relative peak area percentage of each structure was similar and the relative percentage difference was less than 5% between GT and 2-AA derivatized glycan from triplicate experiments. However, the MS spectrum of 2-AA derivatized glycans showed ~1.7 fold higher sensitivity than that of GT-derivatized glycans (data not shown). Therefore, we used 2-AA labeling to investigate the N-glycan structural variations of a model of a recombinant glycoprotein to show how changes in fucosylation and sialylation can be quantitatively monitored by MALDI-TOF MS. It was noted that the peak heights of large molecules were lower than those of small molecules, when the same amount of glycan were analyzed on MALDI-TOF MS.26 This is due to the lower probability for the first isotopic peak among the isotopic distributions for larger molecules. In order to minimize any bias for quantification due to those different isotopic distributions, we used the peak area calculated by integrating from the first to the third isotopic peaks of each ion rather than the first peak height. The relative percentage of sialoglycans calculated from HPLC profile (15%) was similar with that calculated from MALDI-TOF MS profile (14%).
Fig 2.
HPLC profiles of N-glycan from human IgG (a) unamidated and derivatized with 2-aminobenzoic acid (2-AA), and (b) amidated with acetohydrazide (Ah) and derivatized with 2-AA. MALDI-TOF MS spectra of N-glycan from human IgG (c) amidated with Ah and derivatized,with Girard’s T reagent (GT) and (d) amidated with Ah and derivatized with 2-AA.
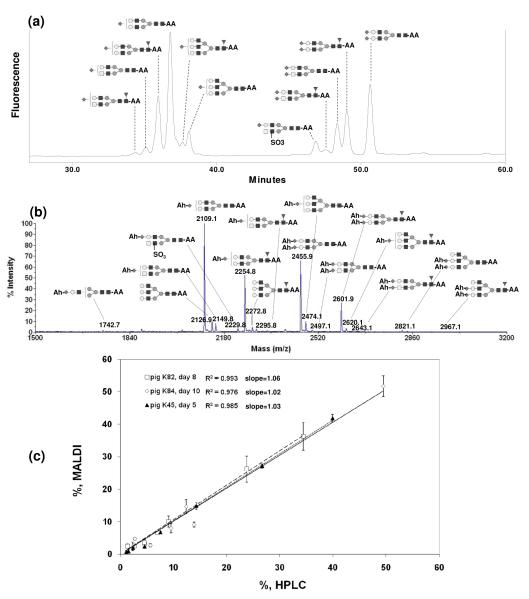
We used the MALDI-TOF MS and NP-HPLC quantification methods to analyze the N-glycans of recombinant human blood coagulation Factor IX produced in the milk of transgenic animal bioreactors (tg-FIX).36,39 Tg-FIX purified from a particular daily milk sample (day of lactation) can be viewed as a model for a different harvest timepoint from a conventional fed-batch or perfusion bioreactor. Fig 3 (a) shows the NP-HPLC profile of unamidated and 2-AA derivatized N-glycans of tg-FIX produced at lactation day 5 of animal K45. Each HPLC peak was collected and its structure was identified using MS/MS fragmentation by ESI-IonTrap MS as described in Gil et al 2008.36 The MALDI-TOF MS profile of amidated and 2-AA derivatized N-glycans of the same sample is given in Fig 3 (b). Eleven structures were identified using peaks collected from NP-HPLC, but sixteen structures including three additional low abundant species (m/z 1742, m/z 2821, and m/z 2967) were identified using MALDI-TOF MS. Fig 3(c) shows the comparison of the relative peak area percentages of the major glycans (eight most abundant structures) obtained from NP-HPLC and MALDI-TOF MS from three different tg-FIX samples (lactation day 8 of animal K82, day 10 of animal K84, and day 5 of animal K45). The R2 values from a linear fit of the data through the origin were 0.993 for day 8 of pig K82, 0.976 for day 10 of pig K84, and 0.985 for day 5 of pig K45; the averaged slope of the linear regression for these tg-FIX samples was 1.04 ± 0.02, with individual slopes ranging from 1.02 to 1.06. The relative percentage difference between MALDI-TOF and NP-HPLC peak area percentages was less than 5%, indicating that quantification using NP-HPLC corresponds to the quantification using one-pot glycan modification and MALDI-TOF MS.
Fig 3.
(a) HPLC profile of unamidated and 2-AA derivatized N-glycan, and (b) MALDI-TOF MS spectrum of amidated with Ah and 2-AA-derivatized N-glycan of tg-FIX K45 d5 (4th lactation stage). (c) Quantification comparison of major N-glycan of tg-FIX using between HPLC and MALDI-TOF MS.
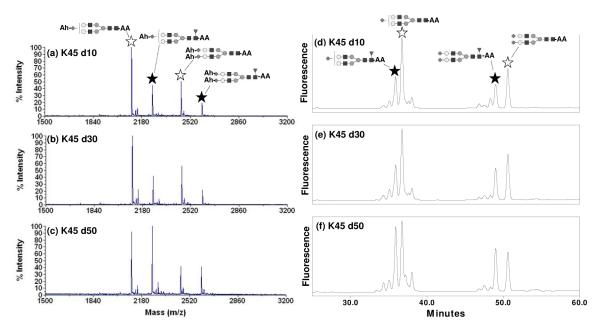
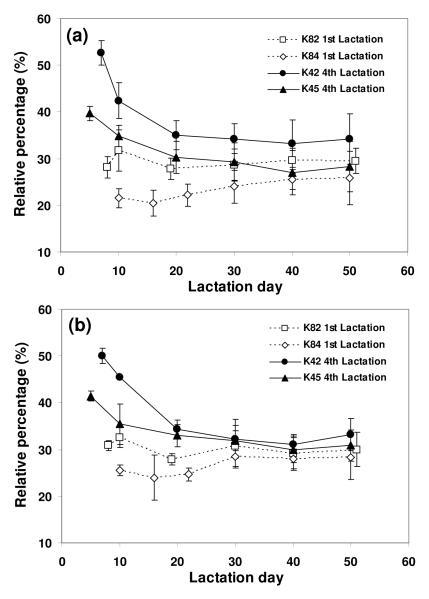
Core fucosylation is common and its effects on biological function of the glycoproteins can be significant. Previous reports showed that fucosylation on the Fc region of monoclonal antibodies strongly affected the Fc binding to its receptor.44-46 Monitoring glycan structure is therefore an activity that starts as early as clone selection with monoclonal antibodies.47-48 Therefore reliable and high throughput quantification methods would be useful to monitor fucosylation changes early on in the developmental process, and also during production. Fig 4(a), (b), and (c) show MALDI-TOF MS N-glycan profiles of tg-FIX where core fucosylation is changing (lactation day 10, 30, and 50 of animal K45, respectively). The fucosylated monosialylated and disialylated biantennary structures (indicated by a solid star) increased, while the corresponding unfucosylated structures (indicated by an open star) decreased from early to late lactation. This change in fucosylation seen in the MALDI-TOF MS spectra is mirrored in the NP-HPLC N-glycan profiles seen in Fig 4 (d), (e), and (f). Fig 5 summarizes the relative percentage change of the summed fucosylated N-glycan structures from four different transgenic animals during lactation, where fucosylated structures increased as lactation progressed. These profile changes observed by MALDI-TOF MS (Fig 5 (a)) are mirrored by changes observed by NP-HPLC (Fig 5 (b)). The MALDI-TOF MS method has the ability to discern changes in fucosylation to the same degree as the NP-HPLC method, while also providing structural information.
Fig 4.
MALDI-TOF MS spectra of amidated with Ah and 2-AA derivatized N-glycans from tg-FIX K45 (a) day10, (b) day 30, and (c) day 50. HPLC profiles of unamidated and 2-AA derivatized N-glycan from tg-FIX K45 (d) day10, (e) day 30, and (f) day 50. A solid star indicates fucosylated monosialylated and disialylated biantennary structures and an open star indicates the corresponding unfucosylated structures.
Fig 5.
Relative percentage change of fucosylated N-glycan of tg-FIX during lactation. The summed percentages of fucosylated N-glycans of tg-FIX were obtained from (a) MALDI-TOF MS and (b) NP-HPLC profiles. The data were triplicate for each sample.
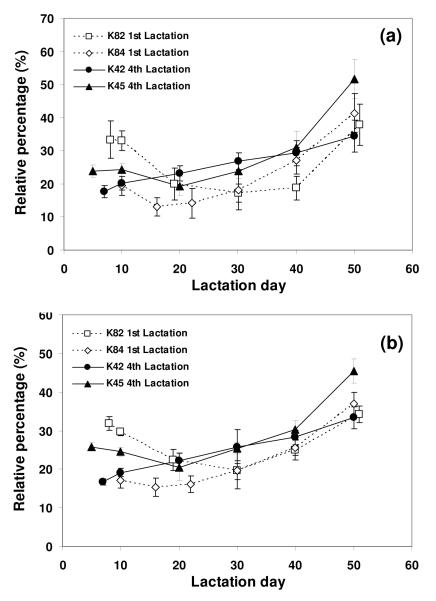
Sialylation plays an important role in glycoprotein structure and function, and can affect protein-protein interaction, pharmacokinetic properties, as well as the physical, chemical and immunogenic properties. 1,3-5,44 Sialylation of recombinant glycoproteins can also be affected by expression of sialidases which commonly exist in host cells and can remove sialic acid from glycans during bioprocessing.49-51 Bioreactor operating conditions such as dissolved oxygen, CO2, and nutrient levels also impact the degree of sialylation by altering the synthesis of CMP-sialic acids and the expression of sialyltransferases.10,12-13,51-53 Therefore, in order to ensure appropriate sialylation throughout the production of recombinant therapeutic glycoprotiens, good quantification methods for sialylated glycans are essential. Fig 6 (a) and (b) show the relative percentage of the fully sialylated glycans obtained from four transgenic animals throughout lactation using MALDI-TOF MS and NP-HPLC, respectively. Quantitation of full vs. partial sialylated glycans by MALDI-TOF MS mirrors what is observed with the NP-HPLC method. The maximum relative difference between the two methods for these samples was 5%. Thus quantitative structural information is obtained from a one-pot method that is amenable to high throughput analysis. This method could be useful for monitoring glycoprotein sialylation throughout process development and manufacturing: the clone selection stage, determination of optimal bioreactor harvest times, analysis of the effect of hold-steps in purification processing, analysis of column fractions where sialylation levels are important to fraction pooling criteria, and analysis of the stability of bulk and formulated product.
Fig 6.
Relative percentage change of fully sialylated N-glycan of tg-FIX during lactation. The summed percentages of fully sialylated N-glycan of tg-FIX were obtained from (a) MALDI-TOF MS and (b) NP-HPLC profiles. The data were triplicate for each sample.
Conclusions
Neutralization of sialylated glycans was performed in one-pot mode so that fast and high throughput glycan quantification was possible. Because the neutralization was performed while the glycan was attached to the protein, the reducing end of the glycans could be labeled with a permanent charge after releasing the glycan from protein, thus improving the sensitivity and reducing the complexity of the mass spectra. Quantification using MALDI-TOF MS method was compared with NP-HPLC oligosaccharide mapping and other orthogonal approaches including ESI-IonTrap MS/MS, and serial glycosidase digestion.26,36 The relative percentages of N-glycans from Human IgG and tg-FIX were similar between MALDI-TOF MS and NP-HPLC. We applied this MALDI-TOF MS quantification method to confirm changes in glycan fucosylation and sialylation that were observed by oligosaccharide mapping. Those data showed that the quantitative MALDI-TOF MS method can be used for monitoring and ensuring consistency of glycosylation of recombinant therapeutic glycoproteins during production. In this work, we performed quantification of the already identified glycan structure using a simple MALDI-TOF MS instrument. Our method for the neutralization of sialic acid and the purification of the resulted glycan could also be incorporated with MALDI-TOF instruments with MSn capabilities, which can provide more structural information for unknown glycans present in glycoproteins.
Acknowledgments
This work was supported by a grant from the National Heart, Lung, and Blood Institute (R01 HL078944-01) and by the University of Nebraska.
Abbreviations
- tg-FIX
transgenic pig derived Factor IX
- ESI-Ion Trap MS
electrospray ionization ion trap mass spectrometry
- NP-HPLC
normal phase high performance liquid chromatography
- 2-AA
2-aminobenzoic acid
- GT
Girard’s T reagent
- MALDI-TOF MS
matrix assisted laser desorption ionization time of flight mass spectrometry
- Ah
acetohydrazide
References
- (1).Li H, d’Anjou M. Curr. Opin. Biotechnol. 2009;20:678. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- (2).Walsh G, Jefferis R. Nat. Biotechnol. 2006;10:1241. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- (3).Jones AJS, Papac DI, Chin EH, Keck R, Baughman SA, Lin YS, Kneer J, Battersby JE. Glycobiology. 2007;17:529. doi: 10.1093/glycob/cwm017. [DOI] [PubMed] [Google Scholar]
- (4).Sinclair AM, Elliott S. J. Pharm. Sci. 2005;94:1626. doi: 10.1002/jps.20319. [DOI] [PubMed] [Google Scholar]
- (5).Hoermann R, Kubota K, Amir SM. Thyroid. 1993;3:41. doi: 10.1089/thy.1993.3.41. [DOI] [PubMed] [Google Scholar]
- (6).Higgins E. Glycoconj. J. 2010;27:211. doi: 10.1007/s10719-009-9261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Grabenhorst E, Schlenke P, Pohl S, Nimtz M, Conradt HS. Glycoconj. J. 1999;16:81. doi: 10.1023/a:1026466408042. [DOI] [PubMed] [Google Scholar]
- (8).Schroder S, Matthes F, Hyden P, Andersson C, Fogh J, Muller-Loennies S, Braulke T, Gieselmann V, Matzner U. Glycobiology. 2010;20:248. doi: 10.1093/glycob/cwp171. [DOI] [PubMed] [Google Scholar]
- (9).Burger C, Carrondo MJ, Cruz H, Cuffe M, Dias E, Griffiths JB, Hayes K, Hauser H, Looby D, Mielke C, Moreira JL, Rieke E, Savage AV, Stacey GN, Welge T. Appl. Microbiol. Biotechnol. 1999;52:345. doi: 10.1007/s002530051530. [DOI] [PubMed] [Google Scholar]
- (10).Restelli V, Wang M-D, Huzel N, Ethier M, Perreault H, Butler M. Biotechnol. Bioeng. 2006;94:481. doi: 10.1002/bit.20875. [DOI] [PubMed] [Google Scholar]
- (11).Hossler P, Khattak SF, Li ZJ. Glycobiology. 2009;19:936. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- (12).Meuwly F, Weber U, Ziegler T, Gervais A, Mastrangeli R, Crisci C, Rossi M, Bernard A, von Stockar U, Kadouri A. J. Biotechnol. 2006;123:106. doi: 10.1016/j.jbiotec.2005.10.013. [DOI] [PubMed] [Google Scholar]
- (13).Yang M, Butler M. Biotechnol. Prog. 2002;18:129. doi: 10.1021/bp0101334. [DOI] [PubMed] [Google Scholar]
- (14).Chill L, Trinh L, Azadi P, Ishihara M, Sonon R, Karnaukhova E, Ophir Y, Golding B, Shiloach J. Biotechnol. Bioeng. 2009;102:828. doi: 10.1002/bit.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Senger RS, Karim MN. Biotechnol. Prog. 2003;19:1199. doi: 10.1021/bp025715f. [DOI] [PubMed] [Google Scholar]
- (16).Butler M. Appl. Microbiol. Biotechnol. 2005;68:283. doi: 10.1007/s00253-005-1980-8. [DOI] [PubMed] [Google Scholar]
- (17).Senderoff RI, Kontor KM, Heffernan JK, Clarke HJ, Garrison LK, Kreilgaard L, Lasser GW, Rosenberg GB. J. Pharm. Sci. 1996;85:749. doi: 10.1021/js950377g. [DOI] [PubMed] [Google Scholar]
- (18).Chen X, Flynn GC. Anal. Biochem. 2007;370:147. doi: 10.1016/j.ab.2007.08.012. [DOI] [PubMed] [Google Scholar]
- (19).Anumula KR. Anal. Biochem. 2006;350:1. doi: 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- (20).Wang WT, Erlansson K, Lindh F, Lundgren T, Zopf D. Anal. Biochem. 1990;190:182. doi: 10.1016/0003-2697(90)90178-c. [DOI] [PubMed] [Google Scholar]
- (21).Hermentin P, Witzel R, Schwick-Wagner P, Blumrich M. Dev. Biol. (Basel) 2002;111:89. [PubMed] [Google Scholar]
- (22).Thim L, Bjoern S, Christensen M, Nicolaisen EM, Lund-Hansen T, Pedersen AH, Hedner U. Biochemistry. 1988;27:7785. doi: 10.1021/bi00420a030. [DOI] [PubMed] [Google Scholar]
- (23).Harvey DJ. Rapid Commun. Mass Spectrom. 1993;7:614. doi: 10.1002/rcm.1290070712. [DOI] [PubMed] [Google Scholar]
- (24).Morelle W, Canis K, Chirat F, Faid V, Michalski J-C. Proteomics. 2006;6:3993. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]
- (25).Zaia J. Chem. Biol. 2008;15:881. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gil G-C, Kim Y-G, Kim B-G. Anal. Biochem. 2008;379:45. doi: 10.1016/j.ab.2008.04.039. [DOI] [PubMed] [Google Scholar]
- (27).Naven TJP, Harvey DJ. Rapid Commun. Mass Spectrom. 1996;10:1361. doi: 10.1002/(SICI)1097-0231(199608)10:11<1361::AID-RCM642>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- (28).Toyoda M, Ito H, Matsuno Y-K, Narimatsu H, Kameyama A. Anal. Chem. 2008;80:5211. doi: 10.1021/ac800457a. [DOI] [PubMed] [Google Scholar]
- (29).Wheeler SF, Domann P, Harvey DJ. Rapid Commun. Mass Spectrom. 2009;23:303. doi: 10.1002/rcm.3867. [DOI] [PubMed] [Google Scholar]
- (30).Kang P, Mechref Y, Novotny MV. Rapid Commun. Mass Spectrom. 2008;22:721. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- (31).Kang P, Mechref Y, Kyselova Z, Goetz JA, Novotny MV. Anal. Chem. 2007;79:6064. doi: 10.1021/ac062098r. [DOI] [PubMed] [Google Scholar]
- (32).Miura Y, Shinohara Y, Furukawa J, Nagahori N, Nishimura S. Chemistry. 2007;13:4797. doi: 10.1002/chem.200601872. [DOI] [PubMed] [Google Scholar]
- (33).Naven TJP, Harvey DJ. Rapid Commun. Mass Spectrom. 1996;10:829. doi: 10.1002/(SICI)1097-0231(199608)10:11<1361::AID-RCM642>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- (34).Liu X, Li X, Chan K, Zou W, Pribil P, Li XF, Sawyer MB, Li J. Anal. Chem. 2007;79:3894. doi: 10.1021/ac070091j. [DOI] [PubMed] [Google Scholar]
- (35).Jang K-S, Kim Y-G, Gil G-C, Park S-H, Kim B-G. Anal. Biochem. 2009;386:228. doi: 10.1016/j.ab.2008.12.015. [DOI] [PubMed] [Google Scholar]
- (36).Gil G-C, Velander WH, Van Cott KE. Glycobiology. 2008;18:526. doi: 10.1093/glycob/cwn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sekiya S, Wada Y, Tanaka K. Anal. Chem. 2005;77:4962. doi: 10.1021/ac050287o. [DOI] [PubMed] [Google Scholar]
- (38).Powell AK, Harvey DJ. Rapid Commun. Mass Spectrom. 1996;10:1027. doi: 10.1002/(SICI)1097-0231(19960715)10:9<1027::AID-RCM634>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- (39).Lindsay M, Gil G-C, Cadiz A, Velander WH, Zhang C, Van Cott KE. J. Chromatogr. A. 2004;1026:149. doi: 10.1016/j.chroma.2003.11.006. [DOI] [PubMed] [Google Scholar]
- (40).Waldrep JC, Noe RL, Stulting RD. Invest. Ophthalmol. Vis. Sci. 1988;29:1538. [PubMed] [Google Scholar]
- (41).Jin Y, Luo G, Oka T, Manabe T. Electrophoresis. 2002;23:3385. doi: 10.1002/1522-2683(200210)23:19<3385::AID-ELPS3385>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- (42).Mack S, Cruzado-Park I, Chapman J, Ratnayake C, Vigh G. Electrophoresis. 2009;30:4049. doi: 10.1002/elps.200800690. [DOI] [PubMed] [Google Scholar]
- (43).He XZ, Que AH, Mo JJ. Electrophoresis. 2009;30:714. doi: 10.1002/elps.200800636. [DOI] [PubMed] [Google Scholar]
- (44).Jefferis R. Nat. Rev. Drug Discov. 2009;8:226. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- (45).Houde D, Peng Y, Berkowitz SA, Engen JR. Mol. Cell. Proteomics. 2010 doi: 10.1074/mcp.M900540-MCP200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. Mol. Immunol. 2000;37:697. doi: 10.1016/s0161-5890(00)00105-x. [DOI] [PubMed] [Google Scholar]
- (47).Lim A, Reed-Bogan A, Harmon BJ. Anal. Biochem. 2008;375:163. doi: 10.1016/j.ab.2008.01.003. [DOI] [PubMed] [Google Scholar]
- (48).Barnes C. A. Srebalus, Lim A. Mass Spectrom. Rev. 2007;26:370. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- (49).Miyagi T, Yamaguchi K. Biochemistry of glycans: sialic acid in Comrehensive Glycoscience. Elsevier BV; Amsterdam: 2007. [Google Scholar]
- (50).Miyagi T, Wada T, Yamaguchi K, Hata K. Glycoconj. J. 2004;20:189. doi: 10.1023/B:GLYC.0000024250.48506.bf. [DOI] [PubMed] [Google Scholar]
- (51).Helenius A, Aebi M. Science. 2001;291:2364. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- (52).Elhalabi JM, Rice KG. Curr. Med. Chem. 1999;6:93. [PubMed] [Google Scholar]
- (53).Tsuji S. J. Biochem. 1996;120:1. doi: 10.1093/oxfordjournals.jbchem.a021369. [DOI] [PubMed] [Google Scholar]