Abstract
During the past century, treatments for the diseases of youth and middle age have helped raise life expectancy significantly. However, cognitive decline has emerged as one of the greatest health threats of old age, with nearly 50% of adults over the age of 85 afflicted with Alzheimer’s disease. Developing therapeutic interventions for such conditions demands a greater understanding of the processes underlying normal and pathological brain ageing. Recent advances in the biology of ageing in model organisms, together with molecular and systems-level studies of the brain, are beginning to shed light on these mechanisms and their potential roles in cognitive decline.
Cognitive frailty is emerging as one of the greatest health threats of the twenty-first century. As the life expectancy of the population has increased, so too has the prevalence of cognitive decline and dementia, largely in the form of Alzheimer’s disease, which now affects almost 50% of adults over the age of 85 in the United States (1). This startling figure can only grow as the average age of the population rises, so understanding the basis of cognitive decline during ageing is critical. The greatest risk factor for cognitive decline and Alzheimer’s disease in older adults is age itself. Therefore, the development of these pathologies must be understood in the context of the molecular biology of the ageing process.
Fortunately, the past 15 years have witnessed a great increase in our knowledge of the basic molecular mechanisms of ageing. Most remarkably, functional genetic analysis has identified signalling pathways that act as master regulators of ageing and lifespan and that are conserved in yeast, nematodes, flies and mammals. Analysis of these model systems suggests that the rate of ageing is not inevitably fixed but is plastic and open to modification. Similarly, cognitive decline associated with mammalian brain ageing also seems to be variable and possibly open to modification (Table 1). An important question is whether age-related cognitive changes are mediated by any of the master regulators of ageing and lifespan identified in model organisms. Moreover, recent studies have implicated these pathways in the control of age-related brain pathology, raising the possibility that altered regulation of fundamental mechanisms of ageing may contribute to the pathogenesis of neuro degenerative disorders.
Table 1.
Signalling pathways that influence ageing in model organisms and brain ageing in mammals
| Pathway | Effects on ageing of model organism | Effects on mammalian brain ageing |
|---|---|---|
| Insulin/IGF-1 signalling | Decreased signalling promotes increased stress resistance and lifespan | Decreased signalling promotes decreased Alzheimer’s disease pathology; paradoxically, increased signalling may be neuroprotective |
| TOR signalling | Decreased signalling causes increased lifespan, increased autophagy and decreased protein translation | Regulation of autophagy and protein homeostasis may modulate toxic-protein aggregation in neurodegenerative disease |
| Mitochondrial function | Severely decreased function causes decreased lifespan, but modestly decreased function can cause increased lifespan | Progressively decreasing function during ageing contributes to decline and pathology. However, preliminary evidence suggests that modestly decreased function may engage beneficial pathways |
| Sirtuins | Can increase or decrease lifespan in different contexts | Can be neuroprotective or detrimental to neurons, depending on context |
| Caloric restriction | Optimal caloric restriction causes increased lifespan | Increased preservation of cognitive function during ageing |
IGF-1, insulin-like growth factor 1; TOR, target of rapamycin.
Two important technical advances have provided new insight into the biology of brain ageing. Microarray technology has made global gene expression analysis possible in humans and model organisms, leading to the identification of evolutionarily conserved changes during ageing. Concurrently, improvements in functional brain imaging have afforded us an unprecedented view of the workings of large-scale cognitive networks in the ageing human brain. An important challenge is to unify these two levels of analysis to obtain a more global view of brain and organismal ageing in humans.
In this Review, we explore the basic molecular mechanisms of the ageing process in the brain. We begin with a brief description of large-scale functional alterations in human brain ageing. Then we turn to a discussion of conserved mechanisms of ageing that may underlie the changes observed in the ageing brain, with a focus on mitochondrial function and oxidative stress, autophagy and protein turnover, insulin/IGF signalling, target of rapamycin (TOR) signalling and sirtuin function. To begin to understand the ageing of the brain in the context of the entire organism, we discuss how the brain might coordinate the ageing process by acting as a primary sensor of physiological and environmental stressors through the action of conserved signalling pathways. There is hope that our growing understanding of the molecular basis of brain ageing and the role of the brain in the ageing body will allow us to rise to the challenge of treating and preventing cognitive decline and Alzheimer’s disease.
Trajectory of ageing in the human brain
Ageing in humans is accompanied by stereotypical structural and neuro physiological changes in the brain and variable degrees of cognitive decline. Functional imaging studies of the human brain have recently provided an unprecedented systems view of neural activity and how it changes with ageing (Fig. 1). These studies have revealed that separate brain regions that interact to subserve higher-order cognitive functions show less-coordinated activation with ageing, suggesting a global loss of integrative function (2). Importantly, this reduced coordination of brain activity is associated with poor performance in several cognitive domains (2). In addition to being less integrated, neural activity also becomes less localized in some brain regions, particularly the prefrontal cortex, in response to executive level tasks (3,4). By contrast, young adults activate more-discrete brain regions to perform the same tasks and integrate these regions more closely with other brain regions. Aged individuals who exhibit delocalized activity show better cognitive performance than aged individuals with more localized activity, consistent with the idea that delocalization may be a compensatory response (3,5). These observations suggest that the higher-order systems biology of the brain is significantly altered by normal ageing in the absence of disease.
Figure 1. Altered functional activation of brain systems during brain ageing.
Functional imaging of brain activation during task performance shows a change in activation patterns as the human brain ages. a, Top: functional magnetic resonance imaging scans show simultaneous activation of the medial prefrontal cortex (mPFC), posterior cingulate (pC) and lateral parietal cortex (LP) in young adults, but this temporal correlation of activity is considerably reduced in aged individuals. (Images reproduced, with permission, from ref. 2.) Bottom: hypothetical connections between areas of the mPFC, pC and LP may mediate the coordinated activation in young adults, whereas declining function of such connections could underlie the observed disruption in coordinated activity in ageing brains. (Images courtesy of C. Koch, California Institute of Technology, Pasadena.) b, Positron emission tomography shows that young adults performing a memory test exhibit right-lateralized brain activation. Aged adults with poor performance in this test also had right-lateralized PFC activity, but aged adults with good performance showed bilateral activation. Thus, recruitment of additional brain areas may compensate for age-dependent functional decline in the primary areas subserving cognitive abilities (3, 5). (Images reproduced, with permission, from ref. 5.)
Age-dependent breakdown in higher-order brain systems may relate, in part, to disruption of myelinated fibres that connect neurons in different cortical regions (2). Although neuronal loss is minimal in most cortical regions of the normal ageing brain (6), changes in the synaptic physiology of ageing neurons may contribute to altered connectivity and higher-order integration. Gene expression profiling studies of ageing mouse, rat, monkey and human brains have shown significant changes in the expression of synaptic genes (7–13). In the human and rhesus macaque prefrontal cortex, many genes involved in inhibitory neurotransmission mediated by GABA (γ-aminobutyric acid) are strongly downregulated with age, potentially altering the balance between inhibitory and excitatory neurotransmission (13). This may contribute to increased neural activity in the prefrontal cortex of aged individuals, a systems-level change that may initially be compensatory but could predispose the individuals to excitotoxicity and neurodegenerative pathology.
A central question is whether these functional changes, which appear in most ageing individuals, are clearly distinct from pathological processes associated with neurodegenerative disorders such as Alzheimer’s disease. Functional magnetic resonance imaging studies suggest that changes in the activity of the hippocampus and associated cortical regions can distinguish normal ageing from pathological ageing. Normal ageing is associated with reduced metabolic activity in the subiculum and the dentate gyrus, whereas reduced activity in the entorhinal cortex may be an early indicator of Alzheimer’s disease (14). At the histopathological level, neuronal loss, beginning in the entorhinal cortex and the CA1 field of the hippocampus, together with volume loss in the medial temporal lobe, distinguishes normal ageing from the cognitive decline associated with Alzheimer’s disease (15–18). However, other pathological hallmarks of Alzheimer’s disease (such as synapse loss, amyloid plaques and neurofibrillary tangles) can correlate with cognitive decline and become extensive in Alzheimer’s disease but are detected to varying degrees in many aged individuals in the absence of cognitive decline. To understand the relationship between Alzheimer’s disease and normal brain ageing, we must gain a greater understanding of the mechanistic basis of the ageing process. The identification of ageing pathways in model organisms is beginning to shed light on this fundamental question.
Conserved pathways of ageing in the brain
Genome-wide gene expression studies during ageing of the nematode Caenorhabditis elegans, the common fruitfly (Drosophila melanogaster) and the brains of mice, rats, chimpanzees and humans have revealed a few broadly conserved functional categories of genes with age-dependent expression changes (6,19) (Table 2 and Fig. 2). In particular, most of these studies provide evidence of reduced mitochondrial function during ageing. Furthermore, reduced expression of genes involved in mitochondrial energy metabolism may become more pronounced in humans with cognitive decline and Alzheimer’s disease (20–22). Another universally conserved feature of ageing is increased expression of genes involved in stress-response pathways. In a transcriptional profiling study of the ageing cortex in mice, rhesus macaques and humans, the greatest conserved change was age-dependent upregulation of the apolipoprotein D gene (13). Apo lipoprotein D expression extends lifespan in Drosophila, and the protein functions as a lipid antioxidant conferring resistance to oxidative stress (23,24). Moreover, apolipoprotein D expression is induced in the brains of individuals with Alzheimer’s disease (25). Hence, conserved mechanisms of stress resistance during ageing may also be used by the brain to protect against the pathology of neurodegenerative disorders.
Table 2.
Evolutionary conservation of gene expression changes during brain ageing
| Gene category | Human (brain) | Rhesus macaque (brain) | Rat (brain) | Mouse (brain) | Fly (organism) | Worm (organism) |
|---|---|---|---|---|---|---|
| Stress response | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| Mitochondria | ↓ | ↓ | ↓ | ↓↑ | ↓ | ↓ |
| Neural plasticity/synaptic function | ↓ | ↓ | ↓ | ↓ | – | – |
| Inhibitory interneuron function | ↓ | ↓ | – | – | – | – |
| Ubiquitin–proteasome pathway | ↓ | ↓ | – | ↓ | – | – |
| Immune/inflammatory response | ↑ | – | ↑ | ↑ | ↑ | – |
| Metal ion homeostasis | ↑ | – | ↑ | – | – | – |
| Myelin-related proteins | ↑ | – | ↑ | – | – | – |
| Glial genes | ↑ | – | ↑ | – | – | – |
Some gene categories, such as those involved in stress responses and mitochondrial function, show conserved changes during ageing, whereas others, such as inhibitory interneuron function, exhibit primate-specific changes. An upward arrow indicates that expression increases with age; a downward arrow indicates that expression decreases with age; and a dash indicates that no change in expression with age is detected. In the ageing mouse brain, different subsets of mitochondrial genes are either age-upregulated or age-downregulated (13).
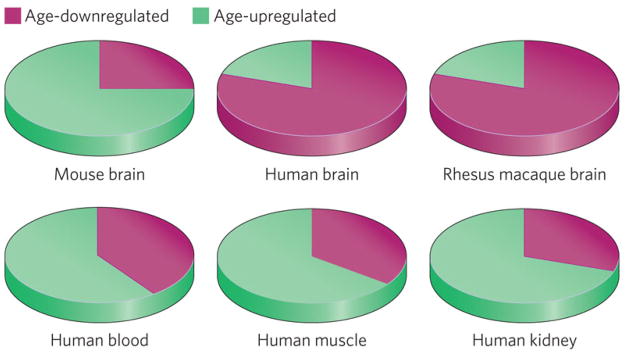
Figure 2. Evolutionary changes in gene regulation in the brain during ageing.
A broad regulatory shift in age-related gene expression appears in the primate lineage. Genes that change with ageing in the human and rhesus macaque cortex are predominantly downregulated (pink), in contrast to the mouse cortex, where most age-regulated genes are upregulated (green). This degree of gene repression is not observed in several other non-neural human tissues, including peripheral blood mononuclear cells (T.L. and B.A.Y., unpublished observations), muscle (98) and kidney (99).
Despite clear evidence of conservation of some ageing pathways and gene expression signatures, a recent study directly comparing gene expression during ageing in mouse, rhesus macaque and human brain has revealed a major evolutionary divergence (13). More than 150 genes were found to undergo age-dependent expression changes in all three organisms. Not all of these genes changed expression in the same direction in all three organisms. A significant fraction of age-regulated genes, mostly predicted to participate in neuronal functions, are upregulated with age in mice but downregulated with age in humans. This suggests an evolutionary shift since the divergence of the rodent and human lineages, resulting in coordinate repression, rather than activation, of many neuronal genes during ageing. Biochemical and stereological neuronal counting studies of the ageing human cortex suggest that this is unlikely to reflect neuronal loss (6,13). It is important to appreciate the evolution of these gene expression changes, as they may contribute to the apparent human specificity of certain neurodegenerative disorders, such as Alzheimer’s disease.
Mitochondrial dysfunction
Gene expression studies suggest that reduced expression of mitochondrial genes is a strongly conserved feature of ageing in organisms ranging from C. elegans to humans (6,19). In particular, organ-specific analysis of brain ageing has revealed a progressive decline in mitochondrial gene expression in rats, rhesus macaques and humans (7,10,13). Mitochondrial function seems to be an important modulating influence on the ageing process in all species tested, and it can have either positive or negative effects on lifespan, depending on the context (26).
Reduction of mitochondrial function would be expected to impair health and shorten lifespan. Indeed, there are examples in invertebrates and mammals where this is the case. Severe reduction of mitochondrial function in worms shortens lifespan significantly (27). Mice that have been engineered to accumulate mitochondrial DNA mutations at an elevated rate show reduced electron transport chain function, signs of accelerated ageing and shortened lifespan (28,29). Conversely, augmentation of mitochondrial function has been shown to extend lifespan. Targeted overexpression of the antioxidant enzyme catalase specifically in mitochondria is sufficient to extend mouse lifespan (30). Furthermore, artificially elevating the rate of mitochondrial respiration is sufficient to increase replicative lifespan in yeast (31). Although the actual mechanisms that underlie lifespan extension in these experimental models are not entirely clear, one hypothesis is that efficient electron transport chain function reduces the generation and release of damaging reactive oxygen species (ROS).
Brain and muscle are particularly susceptible to defective mitochondrial function. The human mitochondrial encephalomyopathies are inherited disorders caused by deletions or mutations of mitochondrial DNA. These mitochondrial defects lead to varied neurological and muscle-related impairments that depend on the number of mitochondria affected per cell (32,33). Importantly, this dependence on the number of affected mitochondria also regulates the age of onset of clinical symptoms. Hence, it has been suggested that a normal decrement in mitochondrial function may also contribute to age-dependent functional deficits in neurons and myocytes. Evidence in support of this notion comes from studies of Drosophila, in which the orthologue of the mammalian brain-specific mitochondrial uncoupling protein UCP5 functions specifically in neurons to regulate metabolism and lifespan (34). In the human brain, declining mitochondrial function may selectively affect neuronal populations with large bio energetic demands, such as the large pyramidal neurons that degenerate in Alzheimer’s disease. Thus, declining mitochondrial function may contribute to brain ageing and render neurons vulnerable to age-dependent pathology.
Unexpectedly, reduced mitochondrial function can, in some circumstances, increase lifespan. One of the earliest suggestions of this came from analysis of clk-1 mutant worms (35). CLK-1 is required for synthesis of ubiquinone, which has an important role in mitochondrial respiration, and clk-1 mutant worms have reduced respiratory rates. These worms also have long lifespans, as well as generally slow developmental and behavioural rates. Subsequent RNA interference screens found that reduction of function in many genes affecting the electron transport chain can increase lifespan (36,37). This effect seems to be crucially dose dependent, because a modest reduction in electron transport chain activity can increase lifespan, whereas a more severe reduction shortens it (27). Recent evidence suggests that this lifespan extension may be mediated by a nuclear transcriptional response to mitochondrial defects, termed the retrograde response, involving the induction of oxidative stress resistance and xenobiotic detoxification genes (38). In Drosophila, reduced expression of electron transport chain components specifically in adult neurons is sufficient to extend lifespan (39). This phenomenon may also occur in mammals, because Coq7 heterozygous mutant mice (Coq7 being the mouse orthologue of clk-1) are long lived (40). Furthermore, a mouse model with reduced activity of the cytochrome c oxidase complex, a component of the electron transport chain, shows increased lifespan (41). Intriguingly, this mouse also exhibits protection against neuronal excitotoxicity in the brain. Although the signaling mechanisms mediating increased longevity in this context are not well understood, one possibility is that ROS in a modestly increased concentration act as signalling molecules to activate survival pathways and promote longevity.
The observation that modestly reduced mitochondrial function can activate longevity pathways raises the interesting possibility that the initial decline in mitochondrial gene expression observed during brain ageing may be part of an active compensatory mechanism that increases stress resistance. Such a compensatory response might be effective in resisting transient stress. However, persistent stress associated with ageing may further reduce mitochondrial function, leading to a self-reinforcing cycle of detrimental decline (Fig. 3).
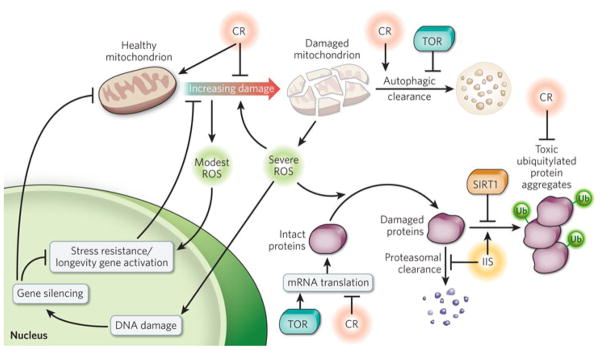
Figure 3. Conserved pathways that regulate organismal and brain ageing.
Shown are mechanisms that involve mitochondrial function, oxidative stress, autophagy, protein homeostasis, TOR signalling, insulin/IGF-1 signalling (IIS), caloric restriction (CR) and sirtuins. Modest concentrations of ROS generated by mitochondria during normal metabolism may induce stress-resistance pathways that scavenge ROS and repair damage. However, progressive mitochondrial damage may lead to pathological concentrations of ROS production, which, in turn, may contribute to further mitochondrial damage. Damaged mitochondria can be cleared by autophagy, which is promoted by CR and inhibited by TOR signalling. CR improves overall mitochondrial function, in part, by promoting mitochondrial biogenesis and reducing ROS production (100). ROS can damage other crucial macromolecules, such as DNA and proteins. Unrepaired DNA damage may give rise to epigenetic changes and gene silencing and may exacerbate mitochondrial impairment by reducing the expression of nuclear-encoded mitochondrial genes. ROS can also modify proteins, leading to protein unfolding and aggregation. Modified proteins can be removed by a number of degradative pathways, including the ubiquitin proteasome pathway. Inadequate clearance may lead to the accumulation of toxic protein aggregates. The dynamics of protein clearance and aggregate formation may be modulated by the IIS pathway and by SIRT1 and CR. The accumulation of damaged and toxic proteins may also be modified through the regulation of messenger RNA translation by TOR signalling and CR.
Oxidative stress and epigenetic changes
Multiple lines of evidence suggest that progressive oxidative damage is a conserved, central mechanism of age-related functional decline (42). Gene expression studies of whole-organism ageing in worms and flies and brain-specific ageing in mice, rats, chimpanzees and humans reveal that all six organisms show an age-dependent upregulation of oxidative stress-response genes (reviewed in ref. 6). Moreover, genes that mediate oxidative stress responses and DNA damage repair constitute the largest class of genes upregulated in the ageing human prefrontal cortex (7,11). Dietary antioxidants can suppress many age-related gene expression changes in the mouse brain (43) and can reduce cognitive decline and prevent oxidative damage to the brain in ageing rats (44).
In the ageing human brain, oxidative damage to specific gene promoters results in gene silencing (7). It may be that irreplaceable post-mitotic cells, such as neurons, respond to unrepaired DNA damage by silencing expression of the affected genomic region, rather than by undergoing apoptosis. The mechanism of silencing may be epigenetic; specifically, it may be a transition to a more repressive transcriptional state (7,13). Recent studies have shown that DNA damage can induce changes in gene expression and histone modification patterns that may be mediated, in part, by the conserved lifespan regulatory gene SIRT1 (see below), suggesting a possible mechanistic link between DNA damage, epigenomic state and ageing (45,46). Consistent with this idea, the yeast homologue of SIRT1, SIR2, is a long-established regulator of lifespan, DNA damage repair and epigenetic gene silencing (47). Furthermore, a number of chromatin-remodelling factors regulate lifespan in C. elegans (48). These considerations argue strongly for a conserved role of epigenome dynamics in the ageing process.
A recent study demonstrated the importance of altered epigenetic state in the control of brain neuronal gene expression underlying synaptic plasticity and memory. The study used a mouse model in which inducible expression of p25, an allosteric regulator of cyclin-dependent kinase 5 (CDK5), elicits neurodegeneration. A transient period of p25 expression in the adult mouse resulted in some degree of neurodegeneration and synapse loss in the hippocampus, together with memory loss (49). Environmental enrichment promoted the recovery of lost memories, which was accompanied by increased synaptic plasticity and the induction of activating histone acetylation marks. Remarkably, treatment with a pharmacological histone deacetylase inhibitor was able to mimic environmental enrichment and promote neuronal plasticity and recovery of memory function. These findings highlight the role of epigenetic changes in memory loss associated with neurodegeneration. In addition, they suggest that loss of memory storage is distinct from loss of neural pathways that access stored memory. Given that human brain ageing is accompanied by memory loss and reduced synaptic connectivity, but not significantly by neuronal loss, it is probable that loss of the ability to access stored memories underlies age-dependent memory deficits. If this is so, there is hope that pharmacological interventions affecting epigenetic state could ameliorate some of the cognitive deficits associated with ageing and neurodegenerative disorders.
Autophagy and protein turnover
Studies in model organisms have implicated autophagy as a crucial regulator of the ageing process. In worms, increased autophagy is necessary for lifespan extension by reduced insulin-like signaling (50) and dietary restriction (51). In flies, increasing autophagy in neurons alone is sufficient to extend lifespan (52), and reducing autophagy shortens lifespan and gives rise to neurodegeneration (52,53). Reduced basal autophagy in the mouse nervous system similarly leads to neurodegeneration (54,55). In autophagy-deficient flies and mice, neurodegeneration is accompanied by the accumulation of ubiquitylated protein aggregates, similar to those observed in human neurodegenerative disorders such as Huntington’s disease and Alzheimer’s disease. Consistent with this, the expression of BECN1, a key regulator of autophagy, is directly related to efficient clearance of aggregated mutant huntingtin protein in Huntington’s disease (56). Expression of BECN1, as well as a number of other key genes in the autophagic pathway, declines with ageing in the human brain (T.L. and B.A.Y., unpublished observations), potentially increasing neuronal vulnerability to the toxic effects of protein aggregates.
The signalling pathway of the kinase TOR is a central regulator of protein homeostasis that acts to inhibit autophagy and messenger RNA translation. Reduced TOR signalling has been shown to extend lifespan in yeast, worms and flies (reviewed in ref. 57). Moreover, it was recently shown that the TOR inhibitor rapamycin extends lifespan in mice even when treatment is initiated late in life (58). The extent to which TOR signalling in neurons contributes to the observed lifespan effects is unknown, but there is evidence that TOR signalling and autophagy have significant effects on pathological protein aggregates associated with age-dependent neuro degenerative diseases. For example, rapa mycin has been shown to reduce toxic-aggregate formation and disease progression by increasing autophagy in mouse and fly models of Huntington’s disease and tauopathy (59). These results suggest that the TOR pathway may modulate pathological protein aggregation associated with neurodegenerative disorders.
Accumulation of ubiquitylated protein aggregates can occur during normal human brain ageing and reaches pathological levels in neurodegenerative disorders such as Alzheimer’s disease and tauopathies. A recent study found that a mouse model of chronically reduced proteasomal activity in the brain showed elevated concentrations of several proteins, some of which had previously been found to alter expression in the brain in Alzheimer’s disease (60). Furthermore, these mice had deficits in spatial memory, consistent with a potential role for defective proteasome function in cognitive decline associated with Alzheimer’s disease. Ubiquitylated protein aggregates are not as prevalent in other ageing tissues, which may reflect the unusual longevity of post-mitotic neurons (which survive for an entire human lifetime in a metabolically active state).
Insulin/IGF signalling
Reduced signalling through the insulin/IGF-1 signalling (IIS) pathway is a strongly conserved mechanism of lifespan extension in worms, flies and mammals (reviewed in ref. 61). Polymorphisms in two IIS pathway genes, the IGF-1 receptor and the downstream FOXO3 transcription factor, have been associated with longevity and healthy ageing in humans (62–64). Importantly, reduced IIS specifically in the nervous system is sufficient to extend lifespan in several model systems. In worms, for example, reduced IIS in the nervous system accounts for a portion of the lifespan extension in IIS mutants (65,66). Furthermore, ablation in the fly brain of specific neurosecretory cells that produce insulin-like peptides is sufficient to extend lifespan (67).
In mammals, insulin and IGF-1 are neurotrophic and promote neuronal survival by inhibiting apoptosis (68). Insulin and IGF-1 can also promote learning and memory in humans and animal models (61,68). By contrast, reduction of IIS by neuron-specific knockout of the insulin receptor substrate Irs2 is sufficient to extend mouse lifespan (69). There is a dichotomy, therefore, between the neuroprotective effects of insulin and IGF-1 signalling and their apparently deleterious effects on organismal lifespan. Interestingly, the effects on lifespan parallel the effects on neurodegenerative pathology. In worms, reduced insulin signalling can ameliorate amyloid-β aggregation and cytotoxicity (70). Similarly, knockout of Irs2 or the IGF-1 receptor can reduce cognitive impairment, neurodegeneration and premature mortality in mouse models of Alzheimer’s disease (71,72). In patients with Alzheimer’s disease, there is evidence of reduced expression of components of the IGF signalling pathway (73). It is unclear, however, whether this represents an active neuroprotective response or a secondary manifestation of the neurodegenerative process.
Caloric restriction and sirtuins
Caloric restriction — that is, the reduction of calorie intake without causing malnutrition — is the only known intervention that robustly increases lifespan in many species (74). Recently, this phenomenon has been extended to primates, in a long-term experiment showing lifespan extension in calorie-restricted rhesus macaques (75). Caloric restriction has also been widely documented to have beneficial effects on the function of the brain and its vulnerability to age-dependent pathology. It prevents many age-dependent gene expression changes in the mouse brain (8,43), reduces age-related brain atrophy in rhesus macaques (75) and has significant beneficial effects on the age-dependent impairment of learning and memory in rodents (76,77) and humans (78). Unexpectedly, a three-month period of caloric restriction in healthy aged humans was sufficient to improve verbal memory by approximately 20% (ref. 78). There is also evidence that caloric restriction may confer resistance to Alzheimer’s-disease-type pathology: caloric restriction was found to reduce amyloid-β deposition and improve learning and memory in transgenic mouse models of Alzheimer’s disease (79).
The molecular basis of the beneficial effects of caloric restriction has recently begun to be elucidated (80). The NAD+-dependent deacetylase SIR2 was one of the earliest genes to be implicated in the response to caloric restriction, initially in a yeast model (81). Homologues of Sir2 (called sirtuins) have subsequently been shown to mediate some of the effects of caloric restriction in flies (82) and mammals (83,84). It is important to note, however, that the potential roles of sirtuins in caloric restriction are controversial and context dependent (see page 480), differing markedly across genetic backgrounds in yeast (85) and tissue types in mice (86). Similarly, SIRT1 can have either beneficial or detrimental effects in the brain. Evidence for the benefits of SIRT1 in brain ageing includes the finding that increased SIRT1 activity protects against axon degeneration after injury in a mouse mutant known as Wallerian degeneration slow (87). SIRT1 also protects against amyloid-β toxicity in cell culture and neurodegeneration in the p25/CDK5 mouse model, which recapitulates aspects of Alzheimer’s disease pathology and tauopathy (88). By contrast, SIRT1 may also have some detrimental effects, as Sirt1 knockout mice show reduced oxidative stress and increased neuroprotection in the ageing brain even though lifespan is shortened (89). Further studies are required to better elucidate the roles of sirtuins in brain ageing and their therapeutic potential in neurodegenerative diseases.
Potential brain regulation of organismal ageing
There is increasing evidence that the nervous system may act as a central regulator of ageing by coordinating the physiology of extraneural tissues. In worms, a number of different mutations that disrupt the function of sensory neurons extend lifespan (90). Furthermore, ablation of specific neurons can increase lifespan in worms (91) and flies (67). A notable example of the central regulatory function of the nervous system was the finding that two neurons in C. elegans, the ASI neurons, could mediate lifespan extension in response to caloric restriction (92). Lifespan extension required ASI-neuron-specific activity of the transcription factor SKN-1, a homologue of the mammalian NF-E2 related factor (NFE2L2), with known functions in activating cellular defences against oxidative and xenobiotic stress. Interestingly, the ASI neurons regulate energy metabolism in C. elegans and may thus represent a functional analogue of the mammalian hypothalamus.
The hypothalamus functions as a central regulator of metabolism and energy use, and it coordinates the physiological responses of the entire organism through hormonal signalling. In mice, reducing the growth hormone signalling from the hypothalamic–pituitary axis extends lifespan (93). Furthermore, overexpression of the mitochondrial uncoupling protein UCP2 in specific cells in the murine hypothalamus is sufficient to extend lifespan (94). These findings argue for a crucial role of hypothalamic hormonal signalling in the control of organismal ageing.
Such hypothalamic regulation of hormonal pathways might have a role in cognitive decline during brain ageing. The hypothalamus coordinates stress responses, in part through the regulation of peripheral glucocorticoid secretion (95). Glucocorticoids can be sensed directly by another part of the brain, the hippocampus, which then suppresses hypothalamic stimulation of further glucocorticoid release in a negative feedback loop (Fig. 4). However, excessive glucocorticoid production associated with chronic or severe stress may impair hippocampal neuronal function and predispose the organism to neurodegeneration (96), potentially disrupting the regulatory circuit that connects the hippocampus and the hypothalamus. Furthermore, primary hippo campal neurodegeneration in individuals with Alzheimer’s disease may also disrupt hippocampal–hypothalamic control of systemic physiological functions (97). Other brain-systemic circuits yet to be discovered may further integrate higher-order brain function with systemic physiology. Thus, primary degenerative changes in the brain could contribute to systemic breakdown in the regulation of glucocorticoids and other crucial hormones, such as IGF-1. These systemic changes may, in turn, predispose the individual to further neurodegenerative changes, setting up a progressive cycle of physiological and neurological decline (Fig. 4).
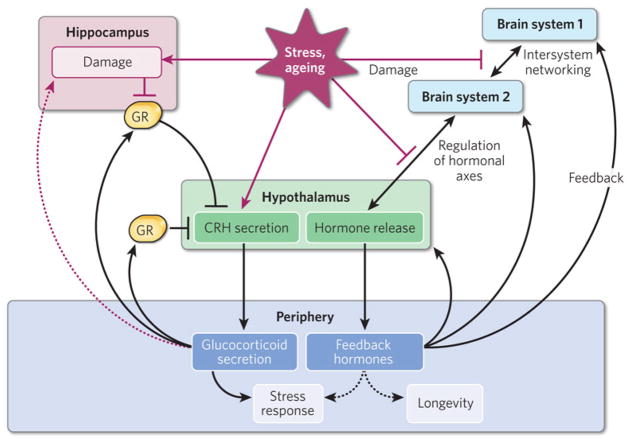
Figure 4. The brain as a potential regulator of organismal ageing.
The ageing of the brain may be coordinated with the ageing of organ systems through hormonal feedback circuits. The left-hand side of the diagram shows how glucocorticoid hormonal signalling may contribute to brain ageing. Stressors induce hypothalamic production of corticotropin-releasing hormone (CRH), leading to glucocorticoid release from the adrenal glands. The hippocampus, in turn, senses glucocorticoid concentrations through the glucocorticoid receptor (GR), resulting in feedback inhibition of glucocorticoid release through the hypothalamus. Chronically elevated glucocorticoid concentrations during ageing may be detrimental to hippocampal function, blunting the ability of the hippocampus to repress glucocorticoid release and potentially setting up a self-reinforcing process of hormonally mediated hippocampal decline. The right-hand side of the diagram shows how hormonal feedback circuits, similar to the pathways shown on the left-hand side, may be a general mechanism contributing to the decline of other functional systems in the brain. Dashed arrows indicate effects that may be indirect and/or are poorly understood mechanistically.
Future directions
The major risk factor for neurodegeneration and cognitive decline is the ageing of the brain. Conserved pathways and mechanisms that control organismal ageing, such as insulin/IGF signalling and mitochondrial function, can modulate pathology and cognitive decline in mouse, fly and worm models of Alzheimer’s disease and other neurodegenerative disorders. However, the role of these conserved pathways in the onset and progression of neurodegenerative disorders in humans is still unclear. The resolution of this basic issue will depend on future clinical interventions that target these pathways to ascertain their role in both normal age-related cognitive decline and pathological neurodegeneration.
The role of the brain as a central integrator of physiological changes during ageing is just beginning to be explored. Recent findings suggest that higher-order brain systems become less efficient with age, with some degree of disconnection between brain areas that normally function together in young adults. This may reflect, in part, gene expression changes that affect synaptic function, axonal integrity and myelination. An intriguing question is whether functional disconnection in the brain leads to disruption of brain-systemic feedback loops involving crucial hormonal and autonomic systems. Such a loss of integrated function may contribute to age-related physiological changes, such as hypertension and insulin resistance, and predispose individuals to age-related pathological changes in the brain. It will be exciting to explore the extent of these functional connections in future studies.
References
- 1.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 2.Andrews-Hanna JR, et al. Disruption of large-scale brain systems in advanced aging. Neuron. 2007;56:924–935. doi: 10.1016/j.neuron.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports that coordination of brain activity between different brain regions becomes less robust in the ageing brain, suggesting a systems-level breakdown of integrated function that correlates with poor cognitive performance
- 3.Cabeza R. Hemispheric asymmetry reduction in older adults: the HAROLD model. Psychol Aging. 2002;17:85–100. doi: 10.1037//0882-7974.17.1.85. [DOI] [PubMed] [Google Scholar]; This paper shows that spreading activation of the human prefrontal cortex from one hemisphere to both hemispheres may be a compensatory mechanism that preserves function against age-related degenerative changes
- 4.Park DC, Reuter-Lorenz P. The adaptive brain: aging and neurocognitive scaffolding. Annu Rev Psychol. 2009;60:173–196. doi: 10.1146/annurev.psych.59.103006.093656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabeza R, Anderson ND, Locantore JK, McIntosh AR. Aging gracefully: compensatory brain activity in high-performing older adults. Neuroimage. 2002;17:1394–1402. doi: 10.1006/nimg.2002.1280. [DOI] [PubMed] [Google Scholar]
- 6.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 7.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]; This paper shows that the ageing of the human cortex is characterized by a distinct transcriptional signature that includes reduced expression of genes that mediate synaptic plasticity and that correlates with age-dependent DNA damage to the promoters of these genes
- 8.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nature Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 9.Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blalock EM, et al. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser HB, Khaitovich P, Plotkin JB, Paabo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erraji-Benchekroun L, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 13.Loerch PM, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small SA, Tsai WY, DeLaPaz R, Mayeux R, Stern Y. Imaging hippocampal function across the human life span: is memory decline normal or not? Ann Neurol. 2002;51:290–295. doi: 10.1002/ana.10105. [DOI] [PubMed] [Google Scholar]
- 15.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet. 1994;344:769–772. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 16.Price JL, et al. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch Neurol. 2001;58:1395–1402. doi: 10.1001/archneur.58.9.1395. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Isla T, et al. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigue KM, Raz N. Shrinkage of the entorhinal cortex over five years predicts memory performance in healthy adults. J Neurosci. 2004;24:956–963. doi: 10.1523/JNEUROSCI.4166-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blalock EM, et al. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang WS, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–679. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez D, et al. Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Curr Biol. 2006;16:680–686. doi: 10.1016/j.cub.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Kalman J, McConathy W, Araoz C, Kasa P, Lacko AG. Apolipoprotein D in the aging brain and in Alzheimer’s dementia. Neurol Res. 2000;22:330–336. doi: 10.1080/01616412.2000.11740678. [DOI] [PubMed] [Google Scholar]
- 26.Sedensky MM, Morgan PG. Mitochondrial respiration and reactive oxygen species in mitochondrial aging mutants. Exp Gerontol. 2006;41:237–245. doi: 10.1016/j.exger.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 29.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 30.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 31.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 32.Wallace DC, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 33.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Blanco A, Fridell YW, Helfand SL. Involvement of Drosophila uncoupling protein 5 in metabolism and aging. Genetics. 2006;172:1699–1710. doi: 10.1534/genetics.105.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branicky R, Bénard C, Hekimi S. clk-1, mitochondria, and physiological rates. BioEssays. 2000;22:48–56. doi: 10.1002/(SICI)1521-1878(200001)22:1<48::AID-BIES9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 36.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 37.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]; This paper provided the first systematic demonstration of a role for the electron transport chain in metazoan lifespan control, through RNAi-mediated knockdown of several of the chain’s components
- 38.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon CA. Regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copeland JM, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dell’agnello C, et al. Increased longevity and refractoriness to Ca2+-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 42.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 43.Park SK, et al. Gene expression profiling of aging in multiple mouse strains: identification of aging biomarkers and impact of dietary antioxidants. Aging Cell. 2009;8:484–495. doi: 10.1111/j.1474-9726.2009.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, et al. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-l-carnitine and/or R-α-lipoic acid. Proc Natl Acad Sci USA. 2002;99:2356–2361. doi: 10.1073/pnas.261709299. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports that memory loss in aged rats can be reversed by restoring mitochondrial function with dietary mitochondrial substrates and antioxidants
- 45.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 48.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]; This paper shows that memory loss may be related to altered chromatin structure in a mouse model of neurodegeneration and can be reversed in part with histone deacetylase inhibitors. This raises the possibility of epigenetic approaches to the treatment of human neurodegenerative disorders
- 50.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 51.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 53.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 55.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]; References 54 and 55 describe mouse models deficient in key autophagy regulatory genes that demonstrate the essential role of autophagy in protection against age-related neurodegeneration and protein aggregation
- 56.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 57.Schieke SM, Finkel T. Mitochondrial signaling, TOR, and life span. Biol Chem. 2006;387:1357–1361. doi: 10.1515/BC.2006.170. [DOI] [PubMed] [Google Scholar]
- 58.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 60.Fischer DF, et al. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging. 2009;30:847–863. doi: 10.1016/j.neurobiolaging.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 62.Flachsbart F, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willcox BJ, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 66.Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. 2007;303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Heide LP, Ramakers GMJ, Smidt MP. Insulin signaling in the central nervous system: learning to survive. Prog Neurobiol. 2006;79:205–221. doi: 10.1016/j.pneurobio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 69.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 70.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 71.Freude S, et al. Neuronal IGF-1 resistance reduces Aβ accumulation and protects against premature death in a model of Alzheimer’s disease. FASEB J. 2009;23:3315–3324. doi: 10.1096/fj.09-132043. [DOI] [PubMed] [Google Scholar]
- 72.Cohen E, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moloney AM, et al. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol Aging. 2008;31:224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 74.Haigis MC, Guarente LP. Mammalian sirtuins — emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 75.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports that caloric restriction increases longevity in primates, delays the onset of age-related diseases and reduces age-related brain atrophy
- 76.Ingram DK, Weindruch R, Spangler EL, Freeman JR, Walford RL. Dietary restriction benefits learning and motor performance of aged mice. J Gerontol. 1987;42:78–81. doi: 10.1093/geronj/42.1.78. [DOI] [PubMed] [Google Scholar]
- 77.Stewart J, Mitchell J, Kalant N. The effects of life-long food restriction on spatial memory in young and aged Fischer 344 rats measured in the eight-arm radial and the Morris water mazes. Neurobiol Aging. 1989;10:669–675. doi: 10.1016/0197-4580(89)90003-1. [DOI] [PubMed] [Google Scholar]
- 78.Witte AV, Fobker M, Gellner R, Knecht S, Flöel A. Caloric restriction improves memory in elderly humans. Proc Natl Acad Sci USA. 2009;106:1255–1260. doi: 10.1073/pnas.0808587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halagappa VK, et al. Intermittent fasting and caloric restriction ameliorate age-related behavioral deficits in the triple-transgenic mouse model of Alzheimer’s disease. Neurobiol Dis. 2007;26:212–220. doi: 10.1016/j.nbd.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 80.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 81.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 82.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 85.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:e296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 88.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 91.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 92.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]; This paper demonstrates the essential role of two C. elegans neurons in coordinating the organismal response to caloric restriction and argues for a central role of the nervous system in this form of increased longevity
- 93.Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Conti B, et al. Transgenic mice with a reduced core body temperature have an increased life span. Science. 2006;314:825–828. doi: 10.1126/science.1132191. [DOI] [PubMed] [Google Scholar]
- 95.Jankord R, Herman JP. Limbic regulation of hypothalamo-pituitary-adrenocortical function during acute and chronic stress. Ann NY Acad Sci. 2008;1148:64–73. doi: 10.1196/annals.1410.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Conrad CD. Chronic stress-induced hippocampal vulnerability: the glucocorticoid vulnerability hypothesis. Rev Neurosci. 2008;19:395–411. doi: 10.1515/revneuro.2008.19.6.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bao AM, Meynen G, Swaab DF. The stress system in depression and neurodegeneration: focus on the human hypothalamus. Brain Res Rev. 2008;57:531–553. doi: 10.1016/j.brainresrev.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 98.Zahn JM, et al. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodwell GE, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maalouf M, Rho JM, Mattson MP. The neuroprotective properties of calorie restriction, the ketogenic diet, and ketone bodies. Brain Res Rev. 2009;59:293–315. doi: 10.1016/j.brainresrev.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]