Abstract
Present investigations suggest that approximately 30% of colorectal cancer (CRC) cases arise on the basis of inherited factors. We hypothesize that the majority of inherited factors are moderately penetrant genes, common in the population. We use an affected sibling pair approach to identify genetic regions that are coinherited by siblings with CRC. Individuals from families with at least two siblings diagnosed with colorectal adenocarcinoma or high grade dysplasia were enrolled. Known familial CRC syndromes were excluded. A genome-wide scan on 151 DNA samples from 70 kindreds was completed using deCODE's 1100 short tandem repeat marker set at an average 4 cM density. Fine mapping on a total of 184 DNAs from 83 kindreds was done in regions suggesting linkage. Linkage analysis was accomplished with MERLIN analysis package. Linkage analysis revealed three genetic regions with NPL LOD scores ≥ 2.0: Ch. 3q29, LOD 2.61 (p=0.0003); Ch. 4q31.3, LOD 2.13 (p=0.0009); and Ch. 7q31.31, LOD 3.08 (p=0.00008). Affected siblings with increased sharing at the 7q31 locus have an 3.8 year (±3.5) earlier age of CRC onset although this is not statistically significant (p=0.11). No significant linkage was found near genes causing known syndromes or, regions previously reported (8q24, 9q22, and 11q23). The chromosome 3q21-q24 region reported to be linked in CRC relative pairs, is supported by our study, albeit a minor peak (LOD 0.9, p=0.02). No known familial cancer genes reside in the 7q31 locus, thus the identified region may contain a novel susceptibility gene responsible for common familial CRC.
Keywords: sibpair, genetic linkage, colorectal cancer, 7q31
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer death and is one of the most familial malignancies 1. Approximately 10% of the U.S. population has at least one first-degree relative with colorectal cancer 2. Kindred and twin studies estimate that as much as 35% of the risk for colorectal adenocarcinoma is attributable to heredity 3,4. The known inherited colon cancer syndromes, however, account for less than 5% of CRC, leaving the genetic etiology unknown for the majority of familial CRC cases.
Association studies have reported a few low-penetrance genetic variants associated with colon cancer risk that could account for a yet to be defined proportion of the familial CRC cases 5,6. Affected-relative-pair studies have also reported genetic regions that are coinherited more often in first-degree relatives with CRC than those without. These include 9q22.33, 3q21-24, and 11q23 7-10. A continued combination of association and linkage approaches will be important to defining new genes that predispose to common familial CRC and will in turn allow the synthesis of more risk directed CRC prevention and mortality reduction efforts.
Here we report the results of a genome-wide linkage analysis using 83 kindreds where at least two siblings were diagnosed with colorectal cancer (invasive or high grade dysplasia). A set of 1100 highly-polymorphic short tandem repeat markers were used for genotyping (deCODE 4cM set) of the affected sibships. The results provide strong evidence for linkage of CRC to a locus at 7q31 and support linkage to the previously reported 3q21-q24 locus 9.
Materials and Methods
Ascertainment and collection of colon cancer sibling pairs
Subjects were enrolled in the Inheritance of Colon Cancer: A Sibling Pair Study following review and approval by the Institutional Review Boards of the eleven participating centers. Informed consent was obtained from all research participants. Kindreds were ascertained from Cancer Genetics Network members, population-based county-wide or state-wide cancer registries, clinical referral and direct-to-patient marketing. Each family had at least two individuals diagnosed with colorectal adenocarcinoma, or a polyp with high-grade dysplasia (also reported as carcinoma in situ, which is synonymous with the previous term) over the age of 20. Diagnosis was confirmed by pathology reports in most cases or cancer registry data. A total of 274 individuals were enrolled in the study, however 88 individuals were excluded from linkage analysis because a sibling was not enrolled (deceased or not interested) or they a known colon cancer syndrome. In the end, 186 DNAs constituted a complete sibship and were used for linkage analysis.
Ruling out of known syndromes
Families with known colon cancer syndromes including familial adenomatous polyposis, Juvenile Polyposis, Puetz-Jeghers, and Cowdens syndrome were excluded by medical record review. A small possibility exists that families with attenuated or MYH polyposis could enter the study, but given the rarity of these conditions and the chance that at least one sibling would have a clinically recognizable condition (≥ 10 adenomas) resulting in study exclusion, their inclusion would be very minor effect. When tumor blocks were available on one or more colon cancer cases, microsatellite instability (MSI) analysis was performed as previously described 11,12 to rule out Hereditary nonpolyposis colorectal cancer (HNPCC of Lynch Syndrome). Genomic DNA from cases that demonstrated MSI-high was sequenced for mutations in MLH1 and MSH2. Five mutation positive families were excluded from the analysis. Individuals who were confirmed to have a known syndrome were referred to a genetic counselor and clinical genetic testing was offered.
Genotyping and linkage analysis
A genome scan was performed on 138 affected and 13 unaffected siblings composing 70 kindreds (64 complete affected sibships of pairs (60) or trios (4)) using deCODE's 1100 short tandem repeat (STR) marker set at an average 4 cM density 13. Fine mapping included 40 STR markers run on 184 individuals from 83 kindreds (163 affected and 21 unaffected). This contained 75 complete affected sibships of pairs (71) or trios (4). Unaffected siblings were available on 16 kindreds, however there was insufficient data for discordant analysis (32 discordant relationships; 5 concordant unaffected relationships).
Genome-wide nonparametric linkage analysis was performed with Merlin 14 using the Kong and Cox linear model 15, and Whittemore and Halpern's Sall sharing statistic 16. Parametric dominant model was run with a disease allele frequency of 0.001 and an absolute disease risk of 0.025 for non-carriers and 0.5 for carriers. Parametric recessive model was run with an allele frequency of 0.05 and disease risk of 0.025 for noncarriers and heterozygotes, and 0.05 for homozygous carriers. Individuals were coded as affected if they had a confirmed diagnosis of invasive CRC or a colonic adenoma with high grade dysplasia or carcinoma in situ. Linkage analysis was run using two sources of allele frequencies for each marker; both are represented in Figure 1. The first was 157,810 genotypes generated from the complete data set (sibpair allele frequencies). The second was a set of 8000 genotypes from 13 CEPH parents who were part of HapMap. The CEPH DNAs were collected in 1980 from U.S. residents with northern and western European ancestry by the Centre d'Etude du Polymorphisme Humain (CEPH) and have been used as a reference panel for establishing genetic maps of the human genome including the HapMap, a catalog of common genetic variants that occur in human beings.
Figure 1. Linkage analysis of genome scan.
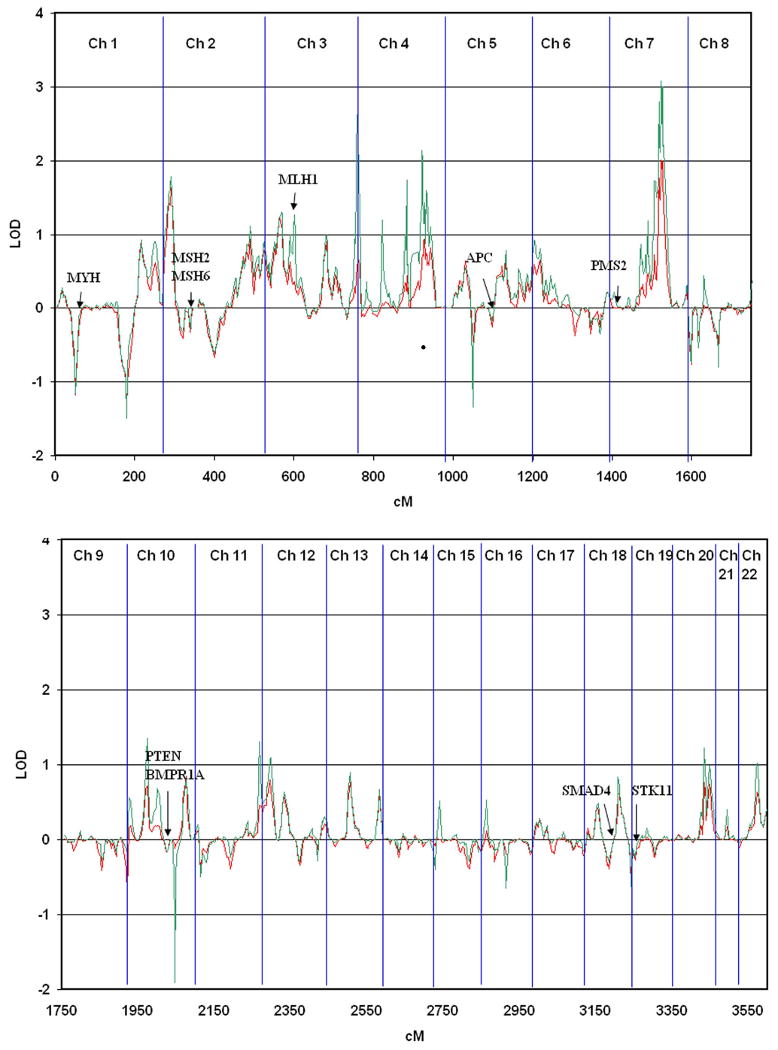
Multipoint nonparametric linkage analyses using sibpair study allele frequencies (red line) or CEPH Caucasian control allele frequencies (green line) are shown. The cumulative chromosome length is based on the DeCode genetic map. Each chromosome boundary is indicated by a blue vertical line. Genes known to cause colon cancer syndromes are noted on the map.
To assess the association of allele sharing with variation in quantitative pathology traits we used the Variance Components module of Merlin which regresses the pairwise covariation of trait values against the identical by descent (IBD) sharing for each marker. Age at diagnosis, tumor grade (grade 1 is well differentiated, grade 2 is moderately differentiated, grade 3 is poorly differentiated), tumor location (right-sided cancers are defined as proximal to the splenic flexure), tumor size at largest dimension, lymph node involvement, and cancer stage (0-4) were modeled as continuous traits. MSI status was available on too few cases to use this as a covariate.
Results
Characteristics of affected individuals
The mean age of colorectal cancer diagnosis was 58.3 (std. dev. ± 12.5) with a range of 28 to 91 years and a median of 60 years. Carcinoma in situ or high-grade dysplasia was the diagnosis in 14% of the cases, lymph node involvement was found in 30% of the cancers, and 12% had metastasized to other tissues. Cancer staging was available on 76% of the cases. Of these, 33% were Stage 1, 30% were Stage 2, 27% were Stage 3 and 10% were Stage 4. The racial make-up of the participants was 87% white, 4% black, 1% Native American, and 8% other. The ratio of affected males to affected females was nearly equal (82:81). MSI analysis was performed on at least one colorectal tumor from 46 of the 83 families. Of the 46 families, 39 were microsatellite stable or low, four had microsatellite unstable cancers, and three had both stable and unstable cancers. Germline mutations in MLH1 and MSH2 were not identified in five of the seven MSI unstable cases. Two did not undergo sequencing.
Genotyping and linkage analysis
A genome-wide scan on 151 samples from 70 kindreds was completed using deCODE's 1100 STR marker set at an average 4 cM density. All DNA samples successfully amplified, however two samples had low yield. Excluding these two samples, 91.9% of the genotypes were successfully obtained. Fine mapping on a total of 184 DNAs (163 affected and 21 unaffected) from 83 kindreds was done using 40 additional deCODE markers at peaks identified in the genome scan on chromosomal regions 4q24-4q26, 4q31-4q32, 7q11-7q22, and 7q31-7q33. In this 40 marker fine mapping run, 96.5% of the genotypes were successful.
Multipoint nonparametric linkage analysis identified three regions with NPL LOD scores greater than 2.0 on chromosomes 3q29 (tel), 4q31.3, and 7q31 (Figure 1 and Table 1). Of these, the 7q31.31 region spanned 13.6 cM (22.63 Mbp) with a maximum NPL LOD of 3.08 and a p=0.00008 (CEPH allele frequencies) or 2.01 and p=0.001 (study allele frequencies) at D7S643. The peak contains 96 RefSeq genes. No known familial cancer genes reside within this region, thus the identified region may contain a novel susceptibility gene responsible for common familial colorectal cancer. Two genes in this region are involved in cancer development 17: MET, a proto-oncogene and hepatocyte growth factor receptor involved in hepatocellular carcinoma and SMO, a G protein-coupled receptor involved in basal cell carcinoma. Candidate cancer genes in this region were also identified from a list of genes hypothetically involved in cancer based on similarity in Gene Ontology (GO term) and sequence properties to a set of 291 genes known to be involved cancer development 17,18. Seven of these genes were predicted to have greater than 80% probability of being involved in cancer development: PAX4, DOCK5, TFEC, FOXP2, NRF1, and IRF5. Other interesting candidate genes in this region are GPR37, G protein-coupled receptor 37 which is an endothelin receptor type B-like, and two WNT ligands involved in the APC/beta-catenin signaling pathway, WNT2 and WNT16.
Table 1. Genetic regions showing multipoint NPL LOD ≥ 2.0.
| Location | Study allele frequency | CEPH allele frequency | |||||
|---|---|---|---|---|---|---|---|
| Marker | cM | cyto | Mbp | LOD | p-value | LOD | p-value |
| D3S240 | 219.97 | 3q29 | 195.6 | 0.58 | 0.05 | 2.61 | 0.0003 |
| D4S2999* | 152.70 | 4q31.3 | 155.0 | 0.69 | 0.04 | 2.13 | 0.0009 |
| D7S2459 | 120.05 | 7q22.3 | 107.1 | 1.44 | 0.005 | 2.02 | 0.0012 |
| D7S525* | 121.35 | 7q31.1 | 109.4 | 1.76 | 0.002 | 2.78 | 0.0002 |
| D7S2418 | 122.15 | 7q31.1 | 109.8 | 1.63 | 0.003 | 2.49 | 0.0004 |
| D7S471* | 122.82 | 7q31.1 | 111.8 | 1.51 | 0.004 | 2.37 | 0.0005 |
| D7S486 | 124.82 | 7q31.2 | 115.7 | 1.46 | 0.005 | 2.14 | 0.0009 |
| D7S633* | 125.99 | 7q31.2 | 116.8 | 1.82 | 0.002 | 2.97 | 0.00011 |
| D7S643* | 126.81 | 7q31.31 | 120.5 | 2.01 | 0.0012 | 3.08 | 0.00008 |
| D7S685* | 127.87 | 7q31.32 | 121.1 | 2.00 | 0.0012 | 2.95 | 0.00011 |
| D7S2529* | 128.64 | 7q31.32 | 122.2 | 1.89 | 0.002 | 2.75 | 0.0002 |
| D7S2520* | 129.44 | 7q31.32 | 123.4 | 1.90 | 0.002 | 2.51 | 0.0003 |
| D7S648 | 129.45 | 7q31.33 | 123.6 | 1.90 | 0.002 | 2.50 | 0.0003 |
| D7S2203* | 130.56 | 7q31.33 | 125.1 | 1.99 | 0.0012 | 2.99 | 0.0001 |
| D7S1873* | 131.07 | 7q31.33 | 125.9 | 1.99 | 0.0012 | 2.77 | 0.0002 |
| D7S680* | 131.23 | 7q31.33 | 126.5 | 1.91 | 0.002 | 2.57 | 0.0003 |
| D7S514 | 131.55 | 7q31.33 | 126.8 | 1.73 | 0.002 | 2.36 | 0.0005 |
| D7S530 | 133.20 | 7q32.1 | 129.0 | 1.44 | 0.005 | 2.12 | 0.0009 |
| D7S2519* | 133.67 | 7q32.2 | 129.8 | 1.42 | 0.005 | 2.20 | 0.0007 |
indicates markers used for fine mapping. The location (cM) is derived from published deCODE data and used for linkage analysis in this study 26. The cytogenetic and physical location is from UCSC Genome Browser March 2006 assembly. Bonferroni-corrected p-value for multiple comparisons for the full genome scan at D7S643 is 0.086 (0.00008*1081 markers).
The results of parametric analysis under dominant and recessive models using study allele frequencies are presented in the supplementary data (Figure S1). The parametric models do not fit the data very well, suggesting that the parameter values supplied are not very realistic. The recessive model identifies a novel peak with a LOD of 2.7 (p=0.0004) at D2S2358. The gene for epidermal growth factor receptor 4 (ERBB4) resides close to this peak and has been associated with a small percentage of colorectal cancers 19-21.
There were no major peaks near genes know to cause colon cancer syndromes (MLH1, MSH2, MSH6, PMS2, APC, MYH, SMAD4, BMPR1A, PTEN, STK11). However a minor peak is observed near MLH1 (62.7cM) at D3S1612 (60.05 cM) with a LOD of 1.25 (unadjusted p=0.008). This is only present when CEPH allele frequency is used, which may be an artifact or may represent a D3S1612 allele segregating with a common, mildly penetrant MLH1 variant predisposing siblings to developing colorectal cancer. Linkage analysis was also run excluding the 7 families with at least one microsatellite unstable cancer (supplementary data, Figures S2 and S3). The results of this analysis appear very similar to the full dataset and the peak near MLH1 is not eliminated (supplementary data, Figure S4).
The 8q24 region recently associated with colorectal cancer risk was not supported by this study 6,22,23. The chromosome 9q22.33 region previously reported to be linked in affected relative pairs with advanced adenomas and colon cancers, was also not confirmed by this study 7,9. Each of the markers in the 9q22.33 region gave negative LOD scores. However, the chromosome 3q21-q24 region reported to be linked in colorectal cancer relative pairs is supported by our study, albeit a minor peak at D3S1292 (LOD 0.9 p=0.02) 8.
Characteristics of siblings that share the 7q31.31 locus
Pathology features of the colorectal cancers from the affected sibling pairs were analyzed to determine if coinheritance at the 7q31 locus was associated with any particular trait. Pairwise covariation for age of CRC diagnosis, histologic grade (1-3), location of cancer (right vs. left sidedness), size of tumor, and lymph node involvement, and stage of CRC was regressed against the IBD sharing at this locus. Only age of diagnosis and grade of cancer contributed to the heritability, however, neither was significant (Table 2). Poorly differentiated tumors were more likely to be seen with increased sharing at 7q31. Of note is the trend toward younger age of onset of CRC with increased sharing at 7q31. Linear regression of age of diagnosis versus identity by descent (IBD) showed that the mean age of diagnosis decreases by 3.8 years as IBD goes from 0 to 1 alleles (std. err. ±3.5).
Table 2. Variance of cancer traits versus IBD value at 7q31 (D7S643).
| TRAIT | # individuals | # sibships | Heritability | LOD | p-value |
|---|---|---|---|---|---|
| Age of CRC diagnosis | 155 | 85 | 29% | 0.33 | 0.11 |
| Histology grade | 68 | 34 | 12% | 0.07 | 0.30 |
| Right-sided cancers | 137 | 73 | 0% | 0 | 0.5 |
| Size of tumor | 114 | 60 | 0% | 0 | 0.5 |
| Lymph node involvement | 102 | 55 | 0% | 0 | 0.5 |
| Stage | 84 | 46 | 0% | 0 | 0.5 |
Number of individuals and number of sibships available for each trait are listed. The estimated locus specific heritability, LOD score and corresponding p-value are also reported.
Discussion
Using the affected sibling-pair approach to identify genetic regions involved in common familial colorectal, a major locus has been identified on chromosome 7q31. This region clearly stands out from the rest of the genome demonstrating linkage at multiple genetic markers over a 13.6 cM region when either study allele frequencies or CEPH allele frequencies are used in the analysis. A region on chromosome 7 with linkage (NPL LOD 1.85, p=0.032) is also reported by Kemp et al. when analysis was restricted to 38 families with colorectal cancer as the diagnosis 8. The reported peak spans approximately 80 to 110 Mbp, whereas our peak spans 107 to 129 Mbp on chromosome 7.
Interestingly, LOH of one or more markers in the 7q31.1-31.2 around marker D7S522 region has been reported in a series of 11 of 18 colon carcinoma cases, with the maximum LOH at D7S522 (115.9 Mbp), within the region identified in this study. The authors suggest that a tumor suppressor gene relevant to the development of epithelial cancers is present in this region 24. Certainly, the MET gene is in this region of LOH at 116.1 Mbp.
Additionally, the ERBB4 gene is near a peak identified through the recessive parametric model on 2q33.3. A functional polymorphism at -782G>T in this gene was recently reported as a risk factor in colorectal cancer (OR 2.21, 95% CI 1,22-3.99) and will be important to examine in this sibling pair population 19.
The study was originally designed to be much larger based on modeling and power estimates to detect linkage as described by Kerber et al., 2008 25(manuscript in press). Considering the minimum observed p-value of 0.0001 as a critical threshold, and taking the relatively conservative value of 0.2 as an estimated attributable risk for inherited susceptibility to colon cancer, we see reasonable (≥ 10%) power only if this locus accounts for the majority of the heritable effect, either because of high penetrance or high prevalence. It is not clear that the majority of heritable CRCs can be explained by variation at this locus, so we might wish to consider alternative explanations. Two details of the modeling may have resulted in over-conservative power estimates when compared to the study as we have actually carried it out: 1) the markers employed are far more heterogeneous than the 4-allele markers simulated; and 2) the multipoint methods employed in the present study using intervals of 1 cM may have considerably more power than the simulated tests. These two factors probably interact to increase power, so estimates may be quite conservative. Nevertheless, the p-values we have observed are surprisingly small considering the study design and sample size.
Although this is one of the largest colorectal cancer sibling-pair studies reported thus far, a major limitation is nonetheless its size, 75 families with DNA from two or more affected individuals. A strength as well as a challenge of our study was to restrict enrollment to individuals with invasive colorectal cancer or an adenoma with high grade dysplasia or carcinoma in-situ. This was a challenge due to the high mortality of colorectal cancer; it was difficult to enroll two siblings surviving colorectal cancer and willing to participate in research. Other groups have done similar studies, but included fewer families and less penetrant phenotypes, such as the presence of advanced adenomas (≥1cm) 7 or any colonic polyp including benign hyperplastic polyps 8. As Kemp et al. report, distinct genetic loci are identified when the study set is restricted to colorectal cancer.
Use of the CEPH allele frequencies for linkage analysis consistently results in higher LOD scores (vs. the study allele frequencies). This is most likely due to minor alleles being over represented in the CEPH data set. Since the CEPH allele set is limited to 13 individuals and 5% the size of the study allele set, this is not surprising. The one peak on chromosome 3, using only CEPH allele frequencies, is interesting in that it is close to MLH1 and might represent a marker commonly found in the study population and linked to a pathogenic MLH1 variant.
Further confirmatory studies as well as expansion of the study population size are needed to confirm this 7q31 locus and identify other genetic loci contributing to hereditary colorectal cancer risk in the population. Investigation of genes within this region will be equally important, especially regulatory or coding SNPs that may be enriched in the colorectal cancer population. Finally, evaluation of LOH in tumor blocks in regions showing linkage, especially the 7q31 region which has demonstrated LOH in sporadic colorectal cancers, would provide further evidence for region containing a tumor suppressor gene responsible for familial colorectal cancer.
Supplementary Material
Acknowledgments
Additional support was provided by a University of Utah General Clinical Research Center Grant M01-RR00064 and N01-PC-67000, Utah Cancer Registry grant N01-PC-35141 from the National Cancer Institute's SEER program with additional support from the Utah State Department of Health and the University of Utah, the Huntsman Cancer Foundation. We also acknowledge University of Colorado Comprehensive Cancer Center for the infrastructure and the information technology group for data collection, and Alicia Salkowski at the Karmanos Cancer Institute for protocol development, piloting, and study coordination.
Financial Support: Project is funded by NIH grant numbers U24-CA78174 (G.P.M.), PO1-CA073992 (R.W.B.), U24-CA78134 (H.A.), U01-CA78285 (H.A.), N01-PC35145 (A.G.S.), U24-CA78148(C.A.G.), CA74799 (D.JA.), U24-CA78142 (L.C.S), CA78157-05S1 (J.E.S.), and U01-CA078284 (D.M.F.).
References
- 1.Burt RW. Colon cancer screening. Gastroenterology. 2000;119:837–53. doi: 10.1053/gast.2000.16508. [DOI] [PubMed] [Google Scholar]
- 2.Ivanovich JL, Read TE, Ciske DJ, et al. A practical approach to familial and hereditary colorectal cancer. Am J Med. 1999;107:68–77. doi: 10.1016/s0002-9343(99)00168-0. [DOI] [PubMed] [Google Scholar]
- 3.Cannon-Albright LA, Skolnick MH, Bishop DT, et al. Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med. 1988;319:533–7. doi: 10.1056/NEJM198809013190902. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.de Jong MM, Nolte IM, te Meerman GJ, et al. Low-penetrance genes and their involvement in colorectal cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2002;11:1332–52. [PubMed] [Google Scholar]
- 6.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24. 21 Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 7.Wiesner GL, Daley D, Lewis S, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci U S A. 2003;100:12961–5. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemp Z, Carvajal-Carmona L, Spain S, et al. Evidence for a colorectal cancer susceptibility locus on chromosome 3q21-q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet. 2006;15:2903–10. doi: 10.1093/hmg/ddl231. [DOI] [PubMed] [Google Scholar]
- 9.Kemp ZE, Carvajal-Carmona LG, Barclay E, et al. Evidence of linkage to chromosome 9q22.33 in colorectal cancer kindreds from the United Kingdom. Cancer Res. 2006;66:5003–6. doi: 10.1158/0008-5472.CAN-05-4074. [DOI] [PubMed] [Google Scholar]
- 10.Djureinovic T, Skoglund J, Vandrovcova J, et al. A genome wide linkage analysis in Swedish families with hereditary non-familial adenomatous polyposis/non-hereditary non-polyposis colorectal cancer. Gut. 2006;55:362–6. doi: 10.1136/gut.2005.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Research. 1998;58:5248–57. [PubMed] [Google Scholar]
- 12.Samowitz WS, Slattery ML. Microsatellite instability in colorectal adenomas. Gastroenterology. 1997;112:1515–9. doi: 10.1016/s0016-5085(97)70032-5. [DOI] [PubMed] [Google Scholar]
- 13.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 14.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 15.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–88. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994;50:118–27. [PubMed] [Google Scholar]
- 17.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furney SJ, Higgins DG, Ouzounis CA, et al. Structural and functional properties of genes involved in human cancer. BMC Genomics. 2006;7:3. doi: 10.1186/1471-2164-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rokavec M, Justenhoven C, Schroth W, et al. A novel polymorphism in the promoter region of ERBB4 is associated with breast and colorectal cancer risk. Clin Cancer Res. 2007;13:7506–14. doi: 10.1158/1078-0432.CCR-07-0457. [DOI] [PubMed] [Google Scholar]
- 20.Kountourakis P, Pavlakis K, Psyrri A, et al. Prognostic significance of HER3 and HER4 protein expression in colorectal adenocarcinomas. BMC Cancer. 2006;6:46. doi: 10.1186/1471-2407-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soung YH, Lee JW, Kim SY, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer. 2006;118:1426–9. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- 22.Gruber SB, Moreno V, Rozek LS, et al. Genetic Variation in 8q24 Associated with Risk of Colorectal Cancer. Cancer Biol Ther. 2007;6:1143–7. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 23.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 24.Zenklusen JC, Thompson JC, Klein-Szanto AJ, et al. Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res. 1995;55:1347–50. [PubMed] [Google Scholar]
- 25.Kerber R, Amos C, Yeap B, et al. Design Considerations in a Sib-pair Study of Linkage for Susceptibility Loci in Cancer. BMC Medical Genetics. 2008 doi: 10.1186/1471-2350-9-64. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–8. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


