Abstract
The main function of the mitotic spindle is to accurately segregate replicated chromosomes during cell division. This dynamic, microtubule-based structure is assembled by a dividing cell and facilitates the orchestrated movement of chromosomes that is the hallmark of mitosis. Steady-state spindle size and morphology are relatively constant for cells of a specified type but vary considerably from one cell type to the next. Despite these differences, all eukaryotic spindles share basic architectural similarities, perhaps the most important of which is bipolar symmetry. At its core, assembling a bipolar spindle is a mechanical process that requires dynamic microtubules be moved and arranged to realize some ultimate functional form. These movements are the result of forces generated either by microtubule polymer dynamics or molecular motors. In this review we focus specifically on the motor-dependent mechanisms that shape the spindle and defer a more comprehensive treatment of spindle assembly and other motor functions during mitosis to others [1].
Keywords: microtubule motors, spindle assembly, chromosome segregation, spindle poles, length control
Introduction
The main function of the mitotic spindle is to accurately segregate replicated chromosomes during cell division. This dynamic, microtubule-based structure is assembled by a dividing cell and facilitates the orchestrated movement of chromosomes that is the hallmark of mitosis. Steady-state spindle size and morphology are relatively constant for cells of a specified type but vary considerably from one cell type to the next. Despite these differences, all eukaryotic spindles share basic architectural similarities, perhaps the most important of which is bipolar symmetry. At its core, assembling a bipolar spindle is a mechanical process that requires dynamic microtubules be moved and arranged to realize some ultimate functional form. These movements are the result of forces generated either by microtubule polymer dynamics or molecular motors. In this review we focus specifically on the motor-dependent mechanisms that shape the spindle and defer a more comprehensive treatment of spindle assembly and other motor functions during mitosis to others [1].
Overview of Spindle Assembly
To ensure assembly of a functional spindle, a cell must build a single bipolar microtubule array which typically requires integration of microtubules generated at multiple, spatially distinct sites. In higher eukaryotes, the spindle microtubules are nucleated by microtubule organizing centers (MTOCs or centrosomes) or acentrosomally through chromatin-mediated pathways. In budding and fission yeast, spindle pole bodies (SPBs) imbedded in the nuclear envelop nucleate spindle microtubules. In cells with centrosomes, the initial stages of spindle assembly are marked by motor-dependent separation and movement of centrosomes to opposite sides of the prophase nucleus (Fig. 1A, Video 1). When only two centrosomes are present, this migration establishes spindle bipolarity with the trajectory between the two centrosomes ultimately becoming the interpolar axis of the spindle (the presence of more than two centrosomes or spurious MTOCs requires additional mechanisms to ensure bipolarity). Subsequent breakdown of the nuclear envelope permits centrosome-nucleated microtubules to enter the nuclear space. The growing microtubule (plus) ends exhibit dynamic instability characterized by abrupt transitions between periods of growth and shortening [2]. This characteristic allows them to efficiently search space and establish attachments with chromosomes, the cell cortex, and other microtubules [3–5]. Observations of mitosis in animal cells suggests that the chromosome-directed pathway contributes to spindle assembly in centrosome-containing cells as well [6, 7]. The sum of these processes results in the establishment of a single spindle.
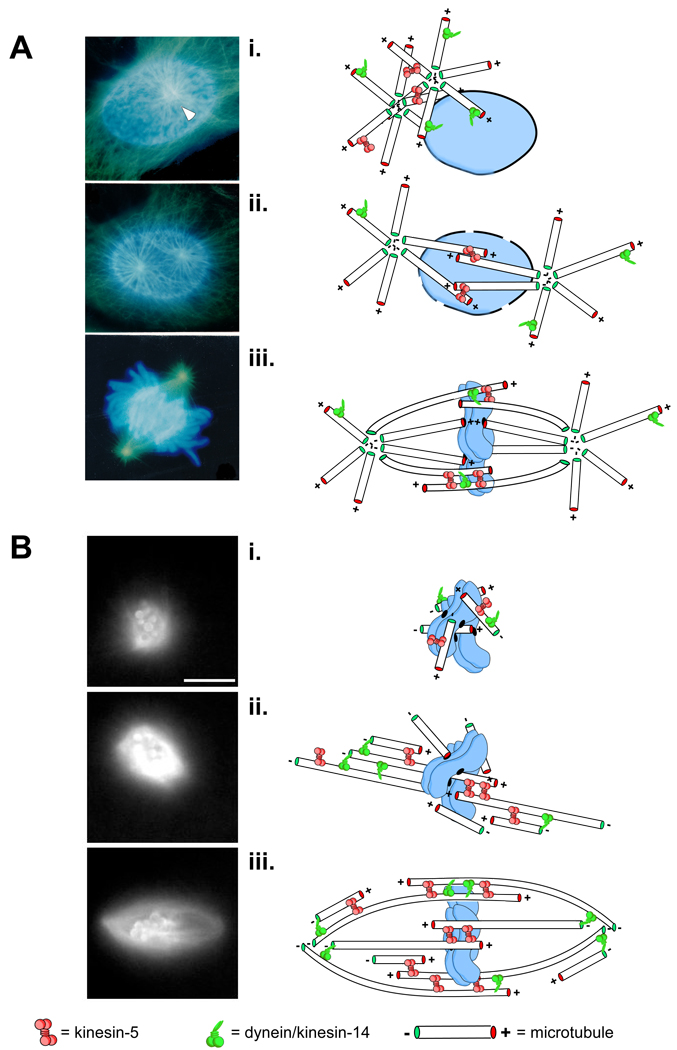
Figure 1. Eukaryotic spindle assembly pathways.
Pathways to spindle assembly are typically defined by the source of the microtubules used to construct the spindle. Some mitotic systems rely more heavily on one pathway or the other but spindle assembly likely involves some combination of both centrosome- and chromosome-nucleated microtubules. A, spindle assembly in cells from newt lungs is dominated centrosome-derived microtubules. Images are taken from fixed samples at different times during spindle assembly. In early prophase, astral microtubules (green) emanate from a pair of centrosomes (white arrowheads) clustered at a single locus on one side of the intact nucleus which bears condensed chromosomes, labeled in blue (i). The centrosomes are then forced apart by motor-dependent microtubule-microtubule sliding (e.g. kinesin-5 at sites of antiparallel overlap) as well as motors anchored on the nuclear envelope and at the cell cortex (ii). After nuclear envelop breakdown, centrosomally derived microtubules can search for and capture targets such as kinetochores and other microtubules ultimately adopting a spindle-like shape (iii). Acentrosomal spindle assembly, shown in B, is characterized by a burst of microtubule nucleation around chromatin (or chromatin-coated beads) that requires localized signals from the Ran-GTP and CPC pathways (see text; i). Monochromatic images were taken from a time-lapse recording of spindle assembly around chromatin-beads in Xenopus egg extracts doped with fluorescently labeled tubulin. Newly nucleated microtubules are arranged by sliding filament mechanisms eventually forming two prominent loci of focused minus ends, precursors to the spindle poles (ii). Eventually, microtubule polymer density equilibrates and the motors continue to shape the microtubule arrays until it achieves the bipolar, fusiform shape of a typical spindle (iii).
In contrast to cells with centrosomes, cells without rely exclusively on chromosome-directed pathways in which microtubules are nucleated and stabilized near chromosomes and kinetochores by the Ran GTPase [8, 9] and the chromosomal passenger complex (CPC) respectively [10, 11]. In animal cells, centrosomal spindle assembly begins after dissolution of the nuclear membrane with a burst of microtubules polymerization near chromosomes. These microtubules extend outward from the clustered chromosomes in all directions (Fig. 1B, Video 2). Compared to centrosome-directed spindle assembly, it is less clear how this initial asymmetry is broken and bipolarity established. However, over time these randomly oriented microtubules are sorted and bundled by microtubule motor and non-motor proteins into symmetric, antiparallel arrays with microtubule minus-ends facing away from the chromosomes [12, 13]. The entire assembly eventually adopts a fusiform spindle shape with interdigitated plus ends near chromosomes and minus ends focused at the poles [14, 15].
Construction of structures with dynamic building blocks presents a unique challenge, particularly if its lifetime is to be longer than its component parts. Such is the case for the spindle and the inherently dynamic microtubules that comprise it. This requires that once a bipolar spindle has been established, its form must be maintained. How this stability is achieved with unstable polymers is a fundamental question in the spindle mechanics field. Although we are a long way from a full conceptual understanding, it is clear that microtubule motors play a critically important role.
Sliding-filament mechanisms
Microtubule motors are protein machines that convert the energy released by ATP hydrolysis into step-wise movement along microtubules. A given type of motor typically exhibits a characteristic maximum speed (when unloaded) and a preferred direction of travel along the microtubule, i.e. it moves predominantly towards the fast growing plus end (plus end-directed) or towards the relatively stable minus end (minus end-directed) of the inherently polar polymer. In this way, motors direct the transport of associated cargoes from one location in the cell to another. Many different types of cargoes are transported along microtubules, including other microtubules. Thus, motors can act as dynamic cross-links, moving microtubules relative to each other. This is the basis of sliding-filament mechanisms of spindle assembly [16].
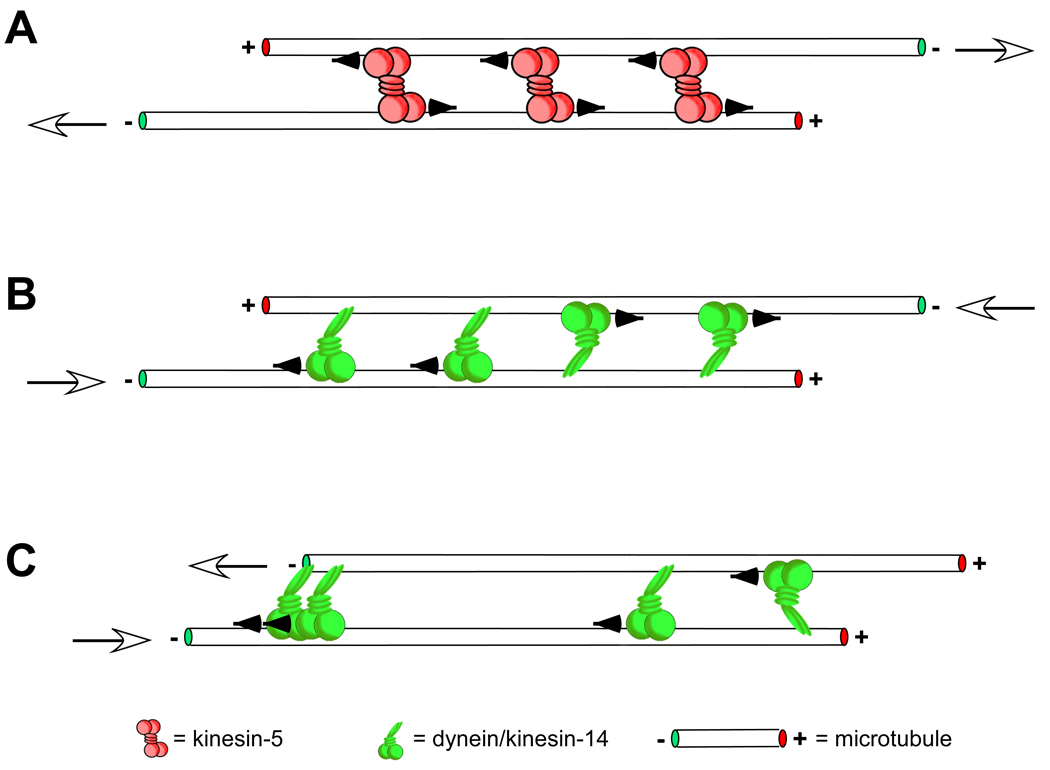
The most fundamental sliding “unit” of an interconnected microtubule array like the spindle is a pair of microtubules cross-linked by a single type of motor (Fig. 2). In this simple system, the preferred directionality of the motor and the orientation of the microtubules dictate the final spatial arrangement. Consider two microtubules of equivalent length juxtaposed and initially oriented in an anti-parallel configuration, i.e. with the plus end of one microtubule aligned with the minus end of the other (Fig. 2A; see also [17]). The action of a plus end-directed motor (e.g. a kinesin-5), would work to align the plus ends, pushing the minus ends apart. Conversely, a minus-end directed motor (e.g. dynein or a kinesin-14 family member) would slide the minus ends closer to one another and push the plus ends apart (Fig. 2B). If the same microtubules were initially aligned end-to-end but with a parallel orientation (Fig. 2C), no relative sliding would occur, regardless of the directionality of the motor involved. However, parallel sliding, revealed by dynein-dependent transport of polarity-marked microtubule seeds to spindle poles [9], has been observed within the spindle, likely stemming from interactions between parallel microtubules with unaligned ends. This implicitly suggests that motor binding kinetics at the microtubule end differ from those along the microtubule length.
Figure 2. Fundamental sliding filament mechanisms.
The orientation of cross-linked microtubules and the directional preference of the cross-linking motor dictate how sliding filament mechanisms contribute to spindle assembly and maintenance. In A, microtubules overlapped at their plus ends with an antiparallel orientation, which commonly occurs near the midzone of the spindle (see [20, 84, 85]), are pushed outward, toward the poles, by plus end-directed kinesin-5 motors. White arrows show the resulting direction of microtubule sliding. Conversely, minus end-directed motors such as dynein or kinesin-14 family members would act to pull the minus ends of the same microtubules together, sliding the microtubules inward, toward the spindle midzone ([20, 37]; B). A balance between these opposing forces is thought to contribute to achieving a steady-state spindle length. The other extreme of microtubule orientations is parallel alignment (C). In this geometry the effect of the motor on microtubule sliding depends on whether the motor “hangs on” once it reaches a microtubule end. A motor with bipolar symmetry like kinesin-5 would bind to and move processively along both the microtubules it cross-links without producing any relative sliding. In this way the motor may act to better align parallel microtubules along their lengths by “zippering” them up without sliding them [56]). For an asymmetric motor like dynein, which binds to microtubules via a static non-motor and steps along the other, the effect of stochastic binding of its non-motor end to either microtubule results in a net force of zero (e.g. the two motors on the right-hand side of the cross-linked microtubules have equal and opposite effects on microtubule sliding, producing no net sliding or force). However, a higher motor binding affinity to microtubule minus ends would allow for sliding and end alignment (for an excellent treatment of these mechanisms please see [17]).
Poleward microtubule flux
One of the two most evident manifestations of sliding-filament mechanisms within the spindle is microtubule flux. Flux, the slow (2–4 µm/min) poleward movement of microtubules, is driven predominantly by members of the kinesin-5 family of microtubule motors (Eg5 in Xenopus [18], Klp61F in drosophila [19, 20]). Kinesin-5 motors form homotetrameric complexes with a central rod-like domain flanked on either side by a pair of globular, microtubule binding domains [21]. These motor complexes cross-link anti-parallel microtubules with overlapped plus-ends and are capable of moving along each cross-linked microtubule at the same time, pushing their minus ends apart [22]. The activity of kinesin-5 motors in sliding anti-parallel microtubules is causally linked to both the establishment of bipolarity and the regulation steady-state spindle length. Perturbation of motor function prevents centrosome separation, in cells that have them, resulting in the formation of abnormal microtubule asters with single poles (monopoles; [23, 24]). Kinesin-5 function is important in establishing bipolar symmetry in acentrosomal systems as well, where inhibition of the motor during spindle assembly results in the formation of monopoles [25, 26]. The motor also plays a critical role in maintaining spindle morphology once it is established: motor inhibition following spindle assembly causes spindle shortening and, if sufficiently strong, complete spindle collapse into monopoles [27].
Pole formation by minus end focusing
The other prominent sliding-filament mechanism within the spindle is pole formation by alignment and focusing of microtubule minus-ends [9, 15, 28–30]. This process is mediated by the minus-end directed motors of the kinesin-14 family (HSET in humans [31], XCTK2 in Xenopus [29], non-claret disjunction (Ncd) in Drosophila [32]) and also by cytoplasmic dynein (dynein; [9, 33]). Unlike kinesin-5, the structures of these motors are not bipolar or tetrameric and instead typically contain only two microtubule binding domains actively used to “walk” along a single microtubule. The valency required for microtubule cross-linking and sliding is achieved by additional microtubule binding domains within the motor protein itself [34] or by accessory proteins (such as dynactin [35] or NuMA in the case of dynein [33]). It is unclear if pole-focusing mechanisms rely exclusively on parallel microtubule sliding or if these motors can also act in regions of antiparallel overlap as well [36–38]. However, as mentioned above, parallel microtubule-microtubule sliding by minus end-directed motors requires differential binding kinetics at microtubule ends, specifically at the relatively stable minus ends [9, 17]. The molecular basis for this difference remains a mystery due in part to the lack of a bona fide molecular marker for microtubule minus ends which are thought to be capped by microtubule nucleating γ-TuRComplexes [39] and may also associate with members of the recently characterized augmin complex which plays a critical role in spindle-mediated nucleation of microtubules [40–42]. Thus, we do not yet understand how minus end-directed motor proteins, either through their own domains or those of accessory proteins, interact with microtubule minus ends.
Motor antagonism and spindle length
There is substantial experimental evidence from a variety of eukaryotic organisms to support motor antagonism as a means to regulate steady-state spindle length. In general, kinesin-5 function is thought to promote increased spindle length (outward directed forces) whereas minus end motors such as dynein and kinesin-14 function to promote spindle shortening (inward directed force)i [27, 43–45]. The mechanistic basis for this antagonism, however, is poorly understood. Some sliding-filament models posit that motors of opposite polarity antagonize each other directly at sites of antiparallel overlap [37, 38, 43]. Competitive in vitro motility assays have been used to investigate how the actions of two motors with opposite polarities might affect microtubule sliding [46, 47]. In the work by Scholey and colleagues [46], kinesin-5 (Klp61F) and Ncd were deposited on glass coverslips and the amount of each motor relative to the other was varied. Over a broad range of molar ratios, sliding velocity and directionality were dominated by one motor. As the relative amount of the less abundant opposing motor was increased the velocity of sliding slowed, consistent with the idea of opposing motors acting as molecular “brakes” [48]. Only over a very small range of molar ratios does the sliding velocity approach and fluctuate around zero. In light of these results, it is interesting that microtubules in Xenopus extract spindles can flux at rates similar to those observed for the unloaded motor in vitro, at least near the spindle midzone [22, 49], suggesting that anti-parallel antagonism by minus end directed motors has a minimal effect on sliding speedii or that other mechanisms contribute to generating force required for sliding.
Other models for spindle length suggest a different sort of sliding filament mechanism to explain motor antagonism, where pole-focusing forces on microtubule minus ends oppose flux-forces produced by kinesin-5 at sites of plus end antiparallel overlap [54]. These “slide and cluster” models are based on observations in Xenopus extract spindles and require microtubule minus ends to be distributed throughout the spindle [55], a condition which may be specific to the architecture of meiotic spindles which rely heavily on chromatin-directed assembly pathways. Here kinesin-5 forces dominate near the spindle midzone where the arrangement of microtubules provides the greatest opportunity for anti-parallel overlap of microtubule plus ends. Closer to the poles, where parallel microtubule orientations are more prevalent, dynein-mediated minus end clustering forces dominate. The authors argue that this explains regional differences in flux rates, which do slow down near the poles. However, the model requires that flux velocity slows to zero near the pole and there is some discrepancy in the literature as to whether this is the case (compare flux rates measured in [49, 54]). Slower flux rates near the pole may also result from regional differences in motor function due to changes in microtubule orientation. Dynein has been show to transport Eg5 to the poles [56] where the motor has been hypothesized to bundle parallel microtubules, perhaps exerting forces on unaligned microtubule plus ends in a manner analogous to those at the minus ends, i.e. by hanging onto the end briefly while sliding along another parallel microtubule [22]. Alternatively, slower flux rates near the poles might result from spatial regulation of motor activities within the spindle [57, 58].
Motor-dependent regulation of microtubule dynamics
Microtubule-based motors can also regulate microtubule dynamics and in turn dictate spindle assembly and length regulation. Indeed, polymer dynamics models of spindle length regulation are based on the idea that spindle and microtubule lengths scale. Thus, mechanisms that control the length of microtubules tend to influence spindle size as well (e.g. [44]), though there are exceptions to this idea (discussed below). Motor-based regulation of microtubule dynamics is also important in establishing spindle bipolarity and in chromosome congression mechanisms, where the control of kinetochore-fiber dynamics contributes to alignment of chromosomes on the metaphase plate [59, 60].
Molecular basis for microtubule dynamics
In vitro, the plus ends of microtubules exhibit abrupt changes between periods of growth and shortening termed “dynamic instability” [2] and it is generally assumed, although not experimentally verified, that microtubules within the spindle undergo the same dynamics. The switch from growth to shortening is commonly referred to as a “catastrophe” while that from shortening to growth is called a “rescue”. This unique property of microtubules is explained in part by their composition and structure. Microtubules are filamentous polymers comprised of individual subunits of α- and β- tubulin heterodimers. These dimers are arranged end-to-end in one of the thirteen protofilaments that make up a typical microtubule. Newly added dimers are bound to GTP which is hydrolyzed shortly after incorporation into the microtubule lattice at the plus end. The delay in hydrolysis is thought to create a stable GTP “cap” at the growing plus end (for recent review see [61]). For free tubulin dimers, the GTP-bound form maintains a straight conformation while hydrolysis to the GDP-bound induces a bend in the dimer. When hydrolysis occurs within the lattice, this conformational change is prevented by stabilizing lateral bonds between GDP-tubulin dimers of adjacent protofilaments so much of the free energy liberated by hydrolysis is actually stored as strain in the microtubule (although pockets of GTP-tubulin seem to remain in the lattice [62]). Removal of the GTP cap is thought to cause rapid depolymerization of the plus end with individual protofilaments peeling and splaying outward as the tubule shortens [63, 64].
Motor-dependent microtubule length regulation during mitosis
Many proteins are known to influence microtubule dynamics and thereby affect change in microtubule length (e.g. op18/stathmin, ch-TOG/xMAP215, EB1, mast/orbit/CLASP], but the focus of this review limits our discussion to motor-based mechanisms. These mechanisms typically limit microtubule length, i.e. result in shorter microtubules, perhaps explaining the bounded and relatively narrow microtubule length distributions observed during eukaryotic mitosis (for discussion see [65]). Though the precise mechanism of action is unknown, it is thought that motor binding to the GTP-cap at the end of a growing microtubule, and perhaps hydrolysis of motor-bound ATP, induces a conformational strain on protofilaments which destabilizes the end and elicits catastrophe [66–68]. Thus, at the plus end, motor-depolymerases reduce the average microtubule length by increasing the catastrophe frequency, not by increasing the rate at which microtubules undergoing a catastrophe shorten. Some microtubule depolymerases can remove tubulin dimers from microtubule minus ends as well, but the mechanistic basis for this depolymerization has not been elucidated and, due to the stability of the minus end, must be independent of catastrophe.
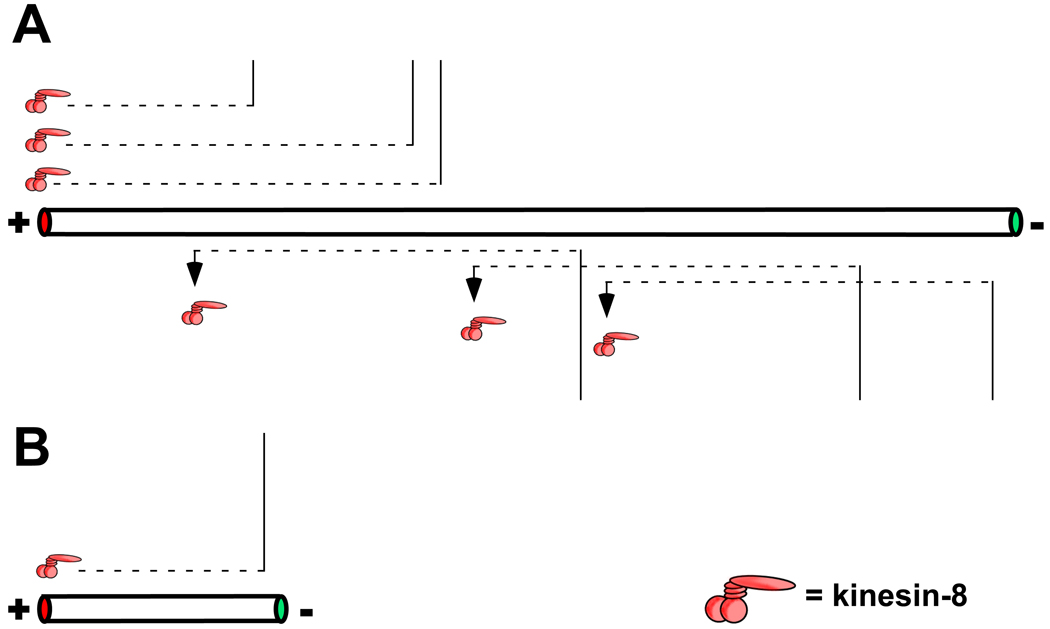
To shorten a microtubule, microtubule-depolymerases need to get to the microtubule ends. This can be achieved through intrinsic means in which the motor gets itself to the end or by indirect targeting that requires additional proteins/motors. Intrinsic mechanisms of end localization occur by microtubule lattice-based diffusion, as is the case for the kinesin-13 MCAK [67, 69], or by processive motor movement as has been observed for the kinesin-8 motor Kip3p [70, 71]. Lattice-based diffusion is potentially advantageous as it is unbiased, allowing the motor to target either microtubule end and explaining in vitro observations that MCAK can depolymerize both microtubule ends [69]. Furthermore, by constraining motor movement to one axis, lattice-based diffusion achieves more rapid targeting to microtubule ends than does simple diffusion from the surrounding cytoplasm/medium. In contrast, kinesin-8 is a processive, plus end-directed motor. Thus, a typical kinesin-8 motor will travel a longer distance compared to a nonprocessive motor like MCAK and only affects plus end polymerization dynamics. The processive nature of kinesin-8 travel on the microtubule confers an interesting functional consequence: length-dependent microtubule length regulation (Fig. 3; [71]). In essence the microtubule acts as an antenna, collecting motors as a function of its length. Motors that bind to the microtubule close enough to reach the plus end during a processive run concentrate there whereas motors that bind further away never reach it. Thus, longer microtubules have a greater chance of having more motor-depolymerases at their plus ends compared to microtubules that are shorter than the run-length of the motor. This hypothesis may explain observations of length-dependent regulation of microtubule dynamics in Xenopus egg extracts where longer microtubules undergo catastrophe more frequently than shorter ones [72].
Figure 3. Microtubule length-dependent regulation of microtubule length by motor-depolymerases.
The ability of a motor-depolymerases to affect microtubule length in a length-dependent manner is derived from the distance the motor can travel during its association with the microtubule track. For a processive motor, this distance is proportional the velocity of the motor and the amount of time it spends attached to the microtubule [71]. For a motor that relies on lattice based diffusion, such as MCAK, this is likely dependent on diffusion length which is proportional to the amount of time the motor dwells on the microtubule and its diffusion coefficient [69]. The diffusion length for MCAK is less than 1 µm while that for the processive kinesin-8 is ~ 8 µm. Microtubules longer than the motor’s typical travel length will collect the most motors at their tips, increasing the chance of a catastrophe (A). Motors that bind the microtubule too far from the ends will fall off before reaching them and not influence catastrophe frequency. In B, shorter microtubules will not collect as many motors and therefore will have fewer motors at their tips reducing the chance of catastrophe.
There is evidence that depolymerases are also targeted to microtubule ends indirectly by hitchhiking on other motors and proteins. Dynein has been implicated in targeting the kinesin-13 Kif2A to microtubule minus ends [73] where it may contribute to microtubule flux [24] and the flux component of chromosome segregation during anaphase-A in drosophila embryos (Klp10A; [74]). MCAK has been shown to target to growing microtubule plus ends via binding to +TIP proteins [75–77]. The kinesin-5 motors in yeast (Cin8, Kip1) also accumulate at microtubule-plus ends in the spindle of budding yeast [59, 78], where they promote (directly or indirectly) plus-end disassembly. Their contribution to the spindle outward force may therefore reflect distinct functions. First, as discussed above is the ability of these motors to cross-link and slide anti-parallel microtubules, and a second, disassembly of longer kinetochore microtubules. While, there is no evidence that kinesin-5 motors promote microtubule shortening in vitro, these studies reveal interesting and important differences and challenges in reconciling in vitro with in vivo approaches. Toward this end, Gardner et al were able to shift Cin8 to the cytoplasm and observe shortening of individual astral microtubules. Cin8’s ability to regulate microtubule length is therefore not restricted to the specialized structure of a mitotic spindle. These results highlight the complexities that we face when evaluating motor function in the confines of living cells.
Motor-depolymerases can localize to similar places within the spindle yet have distinct functions in the context of spindle assembly and maintenance. For example, there are three genes for kinesin-13 motor-depolymerases in humans, MCAK, Kif2A, and Kif2B. Both MCAK and Kif2B localize to centromeres/kinetochores, whereas Kif2A localizes predominantly to spindle poles. Cells depleted of MCAK assemble relatively normal spindles during metaphase while depletion of either Kif2A or Kif2B yields mostly monopolar mitotic structures [79]. Rogers et al. demonstrated that the Drosophila kinesin-13s have distinct roles in regulating chromosome to pole movement during anaphase-A [74]. Although Klp10A localizes to both centromeres and spindle poles, the motor is thought to contribute to chromosome segregation by disassembly of kinetochore-fiber microtubules only at their minus ends, reeling in the attached chromosomes. On the other hand, Klp59C is localized exclusively near centrosomes/kinetochores and contributes to chromosome segregation by depolymerizing the same microtubules at their plus ends (i.e. the “Pac-man” mechanism). How the function of motor-depolymerases translates to changes in spindle morphology is poorly understood, but it is clear that depolymerizing microtubules can exert forces on the cellular structures they are attached to [80]. Thus, motor-depolymerases might function not only to promote microtubule disassembly but also to transmit the force derived from that depolymerization to whatever they are attached to, e.g. spindle poles [24, 73, 74], hence shaping the spindle.
Concluding remarks
Mitosis has fascinated cell biologists since Flemming’s first observations of cell division in the late eighteenth century [Flemming, 1882 #529]. Yet despite much progress that has been made toward a better collective understanding of spindle assembly mechanisms, the question of how a dividing cell co-opts its interphase microtubule cytoskeleton to form a steady-state spindle remains only partially answered. For example, we are just now beginning to understand the spatiotemporal regulation of microtubule motors and how post-translational modification might affect their function (e.g. [82, 83]). We also know little about the behavior of complex motor assemblies within the spindle, which are likely comprised of motors with different sizes, speeds, processivities, and directional preferences. Additionally, elucidation of the structure of the microtubule minus end and the proteins that associate with it should facilitate a more complete picture of how sliding filament mechanisms shape the spindle and also how motor-depolymerases might function there. Within the mitosis field, the low-hanging fruit has been plucked from the tree. To reach ever higher branches and resolve these issues will require continued integration of ideas and experimental approaches from diverse research fields. If the present is any indication of what the future might bring, it looks bright.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Minus end motors, specifically cytoplasmic dynein, can also generate forces at sites where astral microtubules interact with the cell cortex. These forces would act to pull the poles apart and thus differ from intrinsic dynein forces within the spindle which act to pull the poles together. The cortical dynein force may be larger than the intrinsic force, perhaps explaining why dynein and kinesin-5 promote increased spindle length in drosophila [43].
Motor velocity and load are often modeled as inversely related with maximum velocity of the motor occurring in the absence of a load and motor stalling occurring at some maximum load (or opposing force; 50,51]. However, this relationship does not appear to be generally applicable to all motors [52–53].
References
- 1.Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 4.Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci U S A. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the 'search-and-capture' process during mitotic-spindle assembly. Curr Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 7.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 9.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 10.Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle Assembly in the Absence of a RanGTP Gradient Requires Localized CPC Activity. Curr Biol. 2009 doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walczak CE. CLASP fluxes its mitotic muscles. Nat Cell Biol. 2005;7:5–7. doi: 10.1038/ncb0105-5. [DOI] [PubMed] [Google Scholar]
- 13.Kwok BH, Yang JG, Kapoor TM. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol. 2004;14:1783–1788. doi: 10.1016/j.cub.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 14.Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcintosh JR, Hepler PK, Van Wie DG. Model for Mitosis. Nature. 1969;224:659–663. [Google Scholar]
- 17.Nedelec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J Cell Biol. 2002;158:1005–1015. doi: 10.1083/jcb.200202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto DT, Perlman ZE, Burbank KS, Groen AC, Mitchison TJ. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholey JM. Kinesin-5 in Drosophila embryo mitosis: Sliding filament or spindle matrix mechanism? Cell Motil Cytoskeleton. 2009 doi: 10.1002/cm.20349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- 21.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 23.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 24.Cameron LA, Yang G, Cimini D, Canman JC, Kisurina-Evgenieva O, Khodjakov A, Danuser G, Salmon ED. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J Cell Biol. 2006;173:173–179. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 26.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- 30.Merdes A, Cleveland DW. Pathways of spindle pole formation: different mechanisms; conserved components. J Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 34.Furuta K, Toyoshima YY. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol. 2008;18:152–157. doi: 10.1016/j.cub.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 35.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 36.Faruki S, Cole RW, Rieder CL. Separating centrosomes interact in the absence of associated chromosomes during mitosis in cultured vertebrate cells. Cell Motil Cytoskeleton. 2002;52:107–121. doi: 10.1002/cm.10036. [DOI] [PubMed] [Google Scholar]
- 37.Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, Mitchison TJ, Salmon ED. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Curr Biol. 2009;19:287–296. doi: 10.1016/j.cub.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. Embo J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 40.Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 41.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 43.Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 45.Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 47.Vale RD, Malik F, Brown D. Directional instability of microtubule transport in the presence of kinesin and dynein, two opposite polarity motor proteins. J Cell Biol. 1992;119:1589–1596. doi: 10.1083/jcb.119.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tawada K, Sekimoto K. Protein friction exerted by motor enzymes through a weak-binding interaction. J Theor Biol. 1991;150:193–200. doi: 10.1016/s0022-5193(05)80331-5. [DOI] [PubMed] [Google Scholar]
- 49.Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2:718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 51.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 53.Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 55.Burbank KS, Groen AC, Perlman ZE, Fisher DS, Mitchison TJ. A new method reveals microtubule minus ends throughout the meiotic spindle. J Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uteng M, Hentrich C, Miura K, Bieling P, Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- 59.Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O'Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED, Bloom K, Odde DJ. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 62.Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Pous C, Perez F. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 2008;322:1353–1356. doi: 10.1126/science.1165401. [DOI] [PubMed] [Google Scholar]
- 63.Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 67.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 69.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 70.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 71.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 72.Dogterom M, Felix MA, Guet CC, Leibler S. Influence of M-phase chromatin on the anisotropy of microtubule asters. J Cell Biol. 1996;133:125–140. doi: 10.1083/jcb.133.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 75.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, Jelesarov I, Winkler FK, Wuthrich K, Akhmanova A, Steinmetz MO. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 76.Jiang K, Wang J, Liu J, Ward T, Wordeman L, Davidson A, Wang F, Yao X. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009;10:857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee T, Langford KJ, Askham JM, Bruning-Richardson A, Morrison EE. MCAK associates with EB1. Oncogene. 2008;27:2494–2500. doi: 10.1038/sj.onc.1210867. [DOI] [PubMed] [Google Scholar]
- 78.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 81.Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig: F.C.W. Vogel; 1882. [Google Scholar]
- 82.Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A, Sosa HJ, Sharp DJ. Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J Cell Biol. 2009 doi: 10.1083/jcb.200902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawin KE, Mitchison TJ. Microtubule flux in mitosis is independent of chromosomes, centrosomes, and antiparallel microtubules. Mol Biol Cell. 1994;5:217–226. doi: 10.1091/mbc.5.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maddox P, Desai A, Salmon ED, Mitchison TJ, Oogema K, Kapoor T, Matsumoto B, Inoue S. Dynamic confocal imaging of mitochondria in swimming Tetrahymena and of microtubule poleward flux in Xenopus extract spindles. Biol Bull. 1999;197:263–265. doi: 10.2307/1542641. [DOI] [PubMed] [Google Scholar]