Abstract
Galectin-3 is expressed in a cell-type specific manner in human pituitary tumors and may have a role in pituitary tumor development. In this study, we hypothesized that Galectin-3 is regulated by RUNX proteins in pituitary tumors. Transcription factor prediction programs revealed several putative binding sites in the LGALS3 (Galectin-3 gene) promoter region. A human pituitary cell line HP75 was used as a model to study LGALS3 and RUNX interactions using Chromatin immunoprecipitation assay and electrophoresis mobility shift assay. Two binding sites for RUNX1 and one binding site for RUNX2 were identified in the LGALS3 promoter region. LGALS3 promoter was further cloned into a luciferase reporter, and the experiments showed that both RUNX1 and RUNX2 upregulated LGALS3. Knock-down of either RUNX1 or RUNX2 by siRNA resulted in a significant downregulation of Galectin-3 expression and decreased cell proliferation in the HP 75 cell line. Immunohistochemistry showed a close correlation between Galectin-3 expression and RUNX1/RUNX2 level in pituitary tumors. These results demonstrate a novel binding target for RUNX1 and RUNX2 proteins and suggest that Galectin-3 is regulated by RUNX1 and RUNX2 in human pituitary tumor cells by direct binding to the promoter region of LGALS3 and thus may contribute to pituitary tumor progression.
Keywords: Galectin-3, Pituitary tumor, RUNX1, RUNX2, Gene regulation
Introduction
Galectin-3 (Gal-3) is a β-Galactoside-binding protein, which has important roles in diverse biological events, such as embryogenesis, cell adhesion, proliferation, apoptosis, mRNA splicing, and regulation of the immune system [1–3]. Gal-3 is expressed in various cells and tissues, such as activated macrophages, eosinophils, neutrophils, mast cells, epithelium of the gastrointestinal and respiratory tracts, kidneys and some sensory neurons, and is also involved in tumorigenesis, angiogenesis, and tumor metastasis [4, 5]. Gal-3 expression is increased in many tumors, including those of the pancreas, colon, and thyroid [6, 7]. Our previous studies showed that Gal-3 had an important role in promoting pituitary cell proliferation and tumor progression [8, 9]. However, the mechanism of Gal-3 regulation in pituitary tumors is not well understood.
Recent studies in mouse skeletal system showed that LGALS3 was regulated by RUNX2 through the binding to LGALS3 promoter [10]. Another study using HOBIT, a human osteoblast-like cell line, showed that, after treatment with nucleotides (ATP and UTP), RUNX2 was highly expressed, and the Gal-3 protein level was also increased [11]. We hypothesized that LGALS3 was regulated by RUNX proteins in human pituitary tumors.
RUNX protein family includes RUNX1, RUNX2, and RUNX3 which are all transcription factors containing a highly conserved region of 128 amino acids designated as the “runt homology domain (RHD),” and bind to a consensus sequence 5′-ACCPuCPu-3′ through the RHD, so RUNX1 and RUNX3 could also regulate LGALS3 in pituitary tumors. After screening with transcription factor prediction programs, we found two putative binding sites for RUNX1 and one site for RUNX2, but none for RUNX3 in the human LGALS3 promoter. We designed a series of overlapping PCR primers which covered the entire region of the LGALS3 promoter. Serial Chromatin Immunoprecipitation Assay (ChIP) performed with HP75 cells showed that one specific region in LGALS3 promoter was pulled down by anti-RUNX1 antibody while another specific region was pulled down by anti-RUNX2 antibody. Using Electrophoresis Mobility Shift Assay (EMSA), we found that HP75 nuclear extract specifically bound wild type oligonucleotides but not mutant oligonucleotides derived from the binding region of promoter. Luciferase reporter with the LGALS3 promoter demonstrated that LGALS3 was upregulated by RUNX1 and RUNX2, while siRNA knock-down of RUNX1 or RUNX2 significantly decreased Gal-3 expression and cell proliferation in HP75. These findings indicate that RUNX1 and RUNX2 may play important roles in human pituitary tumor through regulation of LGALS3.
Materials and methods
Cell culture
The human pituitary adenoma cell line HP75 [12] was maintained in DMEM (Invitrogeen, Carlsbad, CA) supplemented with 15% horse serum, 2.5% fetal bovine serum and 1 μg/ml insulin. As previously reported the HP75 cell line was derived from a non-functioning human pituitary adenoma and produces low levels of FSH in culture [12]. HeLa (human cervical carcinoma) and SK-BR-3 (human breast adenocarcinoma) (ATCC, Manassas, VA) were grown in DMEM and McCoy’s 5A (Mediatech, Inc, Herndon, VA), respectively, with 10% fetal bovine serum. All media contain 1% Antibiotic-Antimycotic (100 units/ml penicillin G, 100 μg/ml streptomycin sulfate, and 250 ng/ml amphotericin B; Life Technologies, Inc., Grand Island, NY.). Cells were cultured in a humidified incubator at 37°C with 5% CO2.
PCR primers for ChIP and oligonucleotides for EMSA
Five pairs of PCR primers for ChIP were designed based on the human LGALS3 promoter sequence (Genbank AF031421. Table 1). Originally it was reported that the human LGALS3 promoter length is 977 bp [10], after comparison with the genomic sequence of LGALS 3 on Chromosome 14:54648570–54698569 and our sequencing results, we added 12 bp, so that the intact promoter length was 989 bp. We designed five pairs PCR primers to cover the human LGALS3 promoter from upstream to downstream (called region 1–5), see Table 1 and Fig. 1a.
Table 1.
Primers used in ChIP assay
| Primers | Position (Transcription start as +1) | Sequences |
|---|---|---|
| Primer 1 | −884 | 5′-CAGGCCAGCAGATTTGATGT-3′ |
| −611 | 5′-CATTTTTACTGTCAGGGTGCTA-3′ | |
| Primer 2 | −662 | 5′-AACTTTGGCGGGATATAAACA-3′ |
| −412 | 5′-CCTAGGGAACTGACTACAAATTGA-3′ | |
| Primer 3 | −458 | 5′-GCCTATTGATCTAGAATAAGTAGTC-3′ |
| −247 | 5′-CTGCCACCGTTAGGTTCC-3′ | |
| Primer 4 | −264 | 5′-GGAACCTAACGGTGGCAG-3′ |
| −56 | 5′-GAGCCTCAAATACTCCCAGC-3′ | |
| Primer 5 | −77 | 5′-GGGCTGGGAGTATTTGAGG-3′ |
| +154 | 5′-ACAGGCTGTGGGGCTCTC-3′ |
Fig. 1.
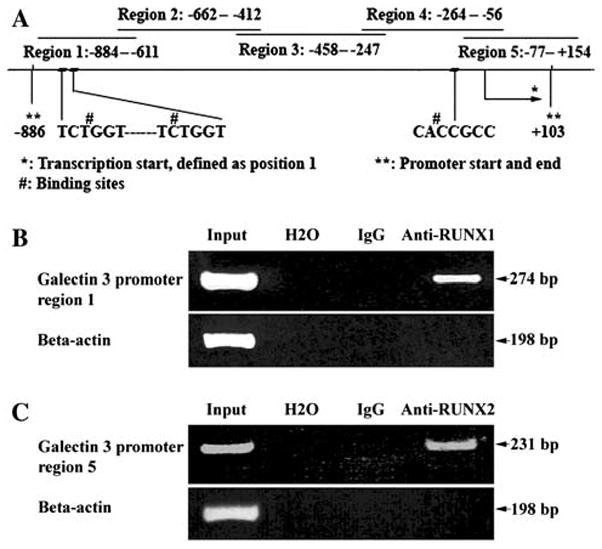
RUNX1 and RUNX2 bound to Gal-3 promoter. a Schematic drawing of LGALS3 promoter, PCR-amplified regions, and putative RUNX1 and RUNX2 binding sites are shown. b ChIP assay. HP 75 cells were sonicated and incubated with anti-RUNX1 antibody. c ChIP assay. HP 75 cells were sonicated and incubated with anti-RUNX2. Immune complex was precipitated by protein A-Sepharose, DNA was extracted by phenol, and PCR amplified with 5 pairs of primers derived from Gal-3 promoter and beta-actin
Beta-actin (Genbank: M10277) was used as input control. Primers are 5′-ACA GAC TCC CCA TCC CAA GAC for sense and 5′-GAG GCG TAC AGG GAT AGC AC for antisense. Searching of DNA sequences for putative transcription factor-binding sites was performed using the web-based prediction programs MatInspector: Transfac, (http://transfac.gbf.de/) [13], TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html), TESS (Transcription Element Search System, http://www.cbil.upenn.edu/tess/), and TFBIND (http://tfbind.hgc.jp) [14]. The MatInspector thresholds for core similarity and matrix similarity were set to 0.85 and 0.90, respectively. The TFSEARCH minimum score was set to 85.0 points. TFBIND minimum score was set to 90.0 points (default cutoff value) [14]. TESS Maximum Allowable String Mismatch % (tmm) was set to 10, Minimum log-likelihood ratio score (ts-a) to 12, Minimum string length (tw) to 10. Putative RUNX-binding sites were identified by searching for sequences matching the published consensus sequence [10, 15–18]. Thus, the sequence was searched for the motif 5′-ACCPuCPu-3′ or its complementary sequence (YGYGGTY); positions 2 and 3 (CC) were considered to be most important. Two putative binding sites for RUNX1 and one putative binding site for RUNX2 were found in the LGALS3 promoter region (Fig. 1a). No binding site was found for RUNX3. The two binding sites for RUNX1 have same sequence (TCTGGT) and they are located in the upstream region of the LGALS3 promoter (−827 and −809, transcription start was defined as +1. This is called region 1) and very close (12 bp apart) to each other. RUNX2 binding sequence (CACCGCC) is located in the downstream region of the LGALS3 promoter (This is called region 5). For EMSA, we designed one oligonucleotide probe to cover the two putative binding sites of RUNX1, and another oligonucleotide to cover the putative binding site of RUNX2. In mutant types of oligonucleotides, the two GG were changed to CC in TCTGGT for RUNX1 or CC to GG in CACCGCC for RUNX2 (Table 2). Single-stranded oligonucleotides were purchased from IDT Inc. (Coralville, IA). Wild type and mutant type of oligonucleotides were labeled with biotin at 5′-end (hot probe); non-labeled wild-type (cold probe) was used as competitors.
Table 2.
Probes used in EMSA (the binding motifs are highlighted by bold letters; mutated nucleotides were in lower case)
| Probes (i.e., Oligos. In cold probe, there was not biotin labeled) | Sequences |
|---|---|
| Wild type for region 1 | |
| Sense | 5′-biotin-TGATGTCTGGTGAGGGCCTGCTTTCTGGTTCACAG |
| Antisense | 5′-biotin-CTGTGAACCAGAAAGCAGGCCCTCACCAGACATCA |
| Mutant 1 for region 1 site 1 | |
| Sense | 5′-biotin-TGATGTCTccTGAGGGCCTGCTTTCTGGTTCACAG |
| Antisense | 5′-biotin-CTGTGAACCAGAAAGCAGGCCCTCAggAGACATCA |
| Mutant 2 for region 1 site 2 | |
| Sense | 5′-biotin-TGATGTCTGGTGAGGGCCTGCTTTCTccTTCACAG |
| Antisense | 5′-biotin-CTGTGAAggAGAAAGCAGGCCCTCACCAGACATCA |
| Wild type for region 5 | |
| Sense | 5′-biotin-GGCTCGGAGCCACCGCCCCGCCGGCGCCC |
| Antisense | 5′-biotin-GGGCGCCGGCGGGGCGGTGGCTCCGAGCC |
| Mutant type for region 5 | |
| Sense | 5′-biotin-GGCTCGGAGCCAggGCCCCGCCGGCGCCC |
| Antisense | 5′-biotin-GGGCGCCGGCGGGGCccTGGCTCCGAGCC |
The complementary oligonucleotides were dissolved in TE buffer (pH 8.0) and combined together, and then annealed in a PCR cycler. All annealed oligonucleotides were stored at −20°C.
ChIP
ChIP assay Kit (Upstate Biotechnology, Lake Placid, NY) was used according to the method recommended by the manufacturer, with minor modifications. In brief, 1 × 106 HP75 cells were plated on a 10 cm dish, and fixed in the presence of 1% formaldehyde for 10 min at 37°C. A soluble chromatin fraction containing fragmented DNA of approximately 200–500 bp was obtained after cell lysis and sonication. The sonication was set as follows with an Ultrasonic Processor GEX130 sonicator (Cole-Parmer Instruments, Vernon Hills, IL): 50% output power, 20 cycles of 10 s sonication, resting 10 s between cycles. Pre-cleared lysates were aliquoted in two parts with equal volume. One aliquot was subject to ChIP by incubating with 5 μg of anti-RUNX1 or anti-RUNX2 or anti-RUNX3 polyclonal antibody overnight (Calbiochem, La Jolla, CA), the other aliquot was immnunoprecipitated using rabbit-IgG as a control with the same procedure. Immnunoprecipitated DNA was extracted by phenol/chloroform/isoamyl alcohol. The DNA was resuspended in TE buffer and stored at −20°C for PCR analysis. PCR reactions were performed using 1 μl of sample or 1 μl from 1:100 dilution of input. Pilot experiments were done to ensure that the number of cycles was within the range of exponential amplification. ChIP assays were repeated at least 3 times for each of anti-RUNX1, anti-RUNX2, and anti-RUNX3 antibodies. Positive controls (input chromatin DNA before immunoprecipitation) and negative controls (mock immunoprecipitation with rabbit-IgG) were run in each experiment. Beta-actin was used as another internal control.
Nuclear protein extraction
Nuclear extracts were prepared using the NucBuster protein extraction kit (Novagen, Inc., Madison, WI.) as recommended by the manufacturer. The protein concentration in nuclear extracts was measured using a WPA Biowave Spectrophotometer (Brinkmann Instruments Inc., Westbury, NY) and the samples were stored in aliquots at −80°C.
EMSA
Binding reactions were performed using the LightShift EMSA kit (Pierce, Rockford, IL) according to the manufacturer’s instructions. About 20 μg of nuclear extract and 20 fmol of biotin-labeled double-stranded oligonucleotide probes were used in the binding reaction. For competition assays, unlabeled double stranded probes were added to the reaction mixture prior to addition of the labeled probes with an excess of 200-fold molar concentrations. For supershift reactions, 1 μg of anti-RUNX1 or anti-RUNX2 antibodies (Biochem, La Jolla, CA.) was added to the EMSA reaction mixture 20 min before the biotin-labeled probes were added. EBNA system was run as a control per instruction in the kit manual. Signal was detected by Biotin-Streptavidin-HRP conjugation method (Chemiluminescent Nuclear Acid Detection Module, Pierce, Rockford, IL). Three to five independent experiments were done for each binding site.
Construction of plasmids and site-directed mutagenesis
Plasmids
The firefly luciferase reporter pGL3-Basic Vector and Renilla were purchased from Promega (Madison, WI). The empty and RUNXs expression vectors, pEF-BOS, pEF-BOS-HA-RUNX1, and pEF-BOS-RUNX2 were kind gifts from Dr. Ito [19, 20]. The entire promoter region of LGALS3 was amplified from human genomic DNA by PCR with the following primers: sense 5′-GCG CCTCGAGTCCAGGCCAGC-3′, antisense 5′-GCGCA AGCTTACCCTCTCCGGAC. pGL3-LGALS3 was constructed by inserting the intact 989 bp of the human LGALS3 promoter region in the pGL 3 basic vector between the XhoI and HindIII sites. Site-directed mutagenesis was performed with a kit from Stratagene (QuickChange II, La Jolla, CA) with the following primers: Primer 1 for Region 1 site 1: Sense 5′-AGGCCAGCA GATTTGATGTCTccTGAGGGCCTGC-3′, Antisense 5′-GCAGGCCCTCAggAGACATCAAATCTGCTGGCCT-3′; Primer 2 for Region 1 site 2: Sense 5′-GAGGGCCTGCTT TCTccTTCACAGAGGGAGCC-3′, Antisense 5′-GGCTCC CTCTGTGAAggAGAAAGCAGGCCCTC-3′; Primer 3 for region 5: Sense 5′-TGAGGCTCGGAGCCAggGCCCCG CC-3′, Antisense 5′-GGCGGGGCccTGGCTCCGAGCCTC A-3′. Mutated nucleotides were in lower case, from gg to cc or vice verca.
Dual luciferase reporter assay
HeLa and SK-BR-3 cells were plated in 6-well plates at 200,000 cells/well with antibiotics-free media. Transient transfection was performed, respectively, with pGL3-LGALS3-Luc and pEF-BOS-HA-RUNX1, pEF-BOS-RUNX2, or pEF-BOS using the TransIt-LT1 reagent (Mi-rus, Madison, WI). Mock-transfected cells were used as control. Simian Virus 40-Renilla luciferase (phRL-SV40; Promega) plasmids were included in the HeLa and SK-BR-3 experiments as internal controls for transfection efficiency. Cells were lysed in passive lysis buffer 48 h post-transfection and tested with a Dual-Glo Luciferase Assay System according to the manufacturer’s protocol (Promega Corporation, Madison, WI) by using a TD-20e luminometer (Turner Designs Inc., MT. View, CA). Luciferase activity was normalized to Renilla activity to control for transfection efficiency variation. To identify the key nucleotides in the binding motifs, mutant types of pGL3-LGALS3 were also transfected and tested with the same procedures.
RNA interference
To determine whether downregulation of RUNX1 or RUNX2 could inhibit LGALS3 expression in pituitary tumor, siRNA of RUNX1 and RUNX2 were transfected into HP75 cells, and the expression of RUNX1, RUNX2, and Gal-3 mRNAs were tested by RT-PCR. Validated siRNA (on-Target plus) duplex oligoribonucleotide against RUNX1, RUNX2, negative control (Non-targeting), and positive control (GAPDH) were synthesized by Dharmacon (Lafayette, Colo). The sequences are as follows: (i) RUNX1 (Genbank NM_001754.3) sense 5′-UGACAACCCUCUCU GCAGAUU-3′, antisense 5′-UCUGCAGAGAGGGUUGU CAUU-3′; (ii) RUNX2 (Genbank NM_001015051) sense 5′-CAAGGACAGAGUCAGAUUAUU-3′, antisense 5′-U AAUCUGACUCUGUCCUUGUU-3′; (iii) GAPDH (NM-002046) 5′-GUCAACGGAUUUGGUCGUA-3′. The duplex oligoribonucleotide were re-suspended in siRNA buffer provided by Dharmacon to make a 20 μM solution and stored at −20°C till further use. siRNAs were used at a final concentration of 100 nM. HP 75 cells were transfected in a 12-well plate at approximately 50% confluent using the DharmaFECT1 transfection reagent (Dharmacon) according to the manufacturer’s protocol. Transfections were carried out for 48 h in serum and antibiotic-free media. Knock-down of RUNX1, RUNX2, and Gal-3 mRNA was confirmed by RT-PCR. All experiments were performed in triplicates and repeated 3 times.
Sequencing
All PCR products in ChIP assays, plasmid constructs, and site-directed mutagenesis products were sequenced for confirmation.
[3H]Thymidine incorporation
HP 75 cells plated in six-well plates were grown for 96 h after transfection with RUNX1, RUNX2 siRNA as described above. The cells were then incubated in fresh medium and [3H] thymidine for 5 h (5 uCi/ml). The cells were harvested, washed three times in PBS, and 1 ml of PBS was added to each pellet for cell counting. Ten thousand cells were placed in each scintillation vial, vortexed, and counted in a scintillation counter (Beckman Instruments Inc., Palo Alto, CA). All experiments and controls were performed in duplicate with three separate experiments.
Tissue microarray and immunohistochemistry
A total of 191 pituitary tumor tissues from patients who had surgery between 1990 and 2007 at Mayo Clinic, Rochester, MN, were used. Institutional review board permission was obtained. These included 39 corticotroph (ACTH), 49 lactotroph (PRL), 47 gonadotroph (GTH), and 56 null cell (NC) tumors. Tissue microarrays were constructed as previously described [21]. Briefly, 0.6 mm cores from selected blocks of formalin-fixed, paraffin-embedded tissue were taken in triplicate and placed in the TMA block. TMA sections cut at 4 μm were used for immunohistochemical staining. Antigen retrieval was performed by microwaving in 0.1 mM citrate buffer at pH 6.0 for 5 min. Antibodies used for TMA slides included Gal-3 (1:500, Vector Laboratories, Burlingame, CA), RUNX1, and RUNX2 (1:20 and 1:50, Calbiochem, La Jolla, CA). Immunostaining was performed with the avidin biotin peroxidase complex method (Vector Corp., Burlingame, CA), and diaminobenzidene was used as the chromogen. Evaluation of immunohistochemical results was performed by grading the intensity of staining on a scale of 0–4 with 0 for negative staining, 1+ for weak staining, 2+ for moderate staining, 3+ for strong staining, 4+ for very strong staining. The TMA triplicates were then averaged. Substitution of the primary antibody with normal serum was used as negative control for immunohistochemical staining. The specificity of each antibody was evaluated by Western blotting.
Results
RUNX1, RUNX2 bind to LGALS3 promoter regions
Because Gal-3 and RUNX1, 2, 3 were all highly expressed in some specific type of pituitary tumors, we designed a series of ChIP assays to determine if RUNX proteins could bind to the promoter region of LGALS3 in the HP75. Five pairs of primers were designed to amplify the LGALS3 promoter DNA. These 5 primers overlapped each other, covering all regions during the ChIP assays to determine if there was binding between RUNXs and LGALS3 promoter. We successfully amplified region 1 from the immunoprecipitated DNA with the anti-RUNX1 antibody as a 274 bp fragment (Fig. 1b). We also successfully amplified region 5 from the immunoprecipitated DNA with the anti-RUNX2 antibody as a 231 bp product (Fig. 1c). Although no binding motif was found in region 4, a 209 bp product (region 4) was also detected in pellets precipitated by anti-RUNX2 antibody (data not shown). All the 3 PCR products were confirmed by sequencing. There was no amplificatory product from the five regions from the immunoprecipitated DNA by anti-RUNX3 antibody. Beta-actin was not amplified in any ChIP assays except in the control input. These results confirmed that the fragments of LGALS3 promoter DNA were specifically pulled down by the anti-RUNX1 or anti-RUNX2 antibodies and not by carryover. The rabbit IgG immunoprecipitated pellets failed to amplify beta-actin or any fragment of LGALS3 promoter, further indicating that there was no non-specific binding.
EMSA supported the findings from ChIP assays. With one oligonucleotide derived from LGALS3 region 1, we found two shifted bands with the HP75 nuclear extract (Fig. 2a), probably from different isoforms (RUNX1 gene has at least 3 isoforms, AMLla 250AA, AML1b 453AA, AML1c 480AA) [22–25]; these two bands were ablated by a 200-fold excess of the cold oligonucleotide, indicating that they were specific binding sites. Adding anti-RUNX1 antibody slowed the migration of one band (super-shift), and confirmed the specific binding between this oligonucleotide and HP75 nuclear extract, but the other band disappeared. Similar to this finding, we also observed a shifted band on the oligonucleotide derived from LGALS3 region 5 (Fig. 2b), this band was ablated by 200-fold excess of its cold oligonucleotide, but we were not able to get a super-shifted band after adding anti-RUNX2 antibody.
Fig. 2.
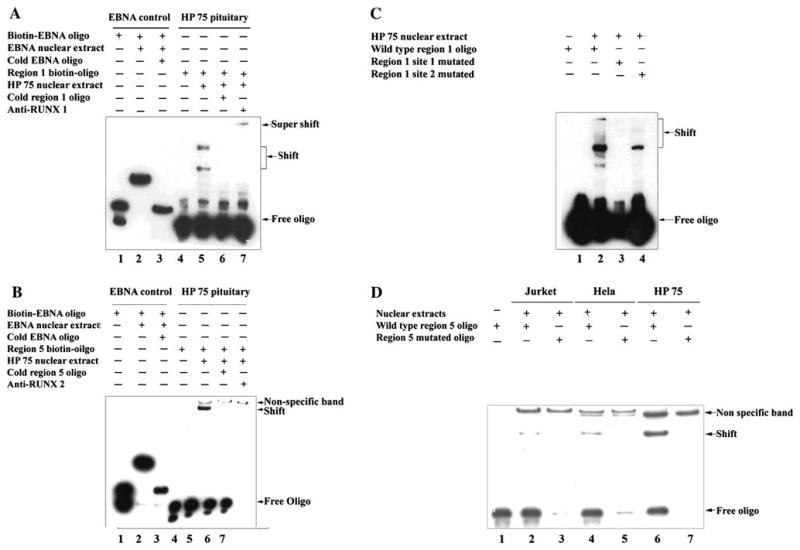
HP75 nuclear extract bound to oligonucleotides (probes) derived from Gal-3 promoter. EMSA was performed. EBNA system from EMSA kit was served as control. a Shift was observed with HP75 nuclear extract, super-shift was observed by adding anti-RUNX1 antibody. b HP75 nuclear extract bound to oligonucleotide (probe) derived from Gal-3 promoter region 5. Anti-RUNX2 antibody disrupted this binding. c Mutations in region 1 oligonucleotide disrupted its binding to HP75 nuclear extract. Same oligonucleotide was used as in a, but key nucleotides in two different binding sites were mutated separately. Site 1 mutation abolished its binding; site 2 mutation significantly decreased its binding to HP75 nuclear extract. d Mutation in region 5 oligonucleotide disrupted its binding to HP75 nuclear extract. Same oligonucleotide was used as in c, but key nucleotides were mutated. Nuclear extracts from Jurket and HeLa cells were served as positive controls
RUNX1 and RUNX2 upregulate LGALS3 by binding specific sequences of its promoter
Based on the ChIP and preliminary EMSA study results, we found that both RUNX1 and RUNX2 bind to the promoter region of LGALS3, but how the bindings affect expression of LGALS3 was uncertain. We then mutated the binding consensus sequence in EMSA probes, and repeated the experiments. We found that mutated region 1 probe in the first binding site lost its binding for LGALS3, while mutated region 1 probe in the second binding site significantly decreased the binding (Fig. 2c). Mutated region 5 probe also lost the ability to bind HP75 nuclear extract (Fig. 2d). These results indicated that these binding sites are sequence specific.
The full length LGALS3 promoter [23] was then cloned into pGL3 Basic vector, a luciferase reporter construct without promoter. This construct showed high luciferase activity, indicating that it was a real promoter. The reporter construct was then co-transfected with pEF-BOS or pEF-BOS-HA-RUNX1 or pEF-BOS-RUNX2 in Hela cells which express low level or lack endogenous RUNX1 activity [24, 26]. There was significantly increased luciferase activity in the cells transfected with RUNX1 and RUNX2 vector compared to the empty vector (Fig. 3a). In co-transfection with the RUNX1 vector, luciferase activity increased ~3.9 times, while in co-transfection with RUNX2 vector, the luciferase activity increased ~2.5 times. With increasing dosages of RUNX1 or RUNX2, luciferase activity was also increased (Fig. 3b). We repeated this co-transfection experiment in SK-BR-3, which does not express endogenous Gal-3, and obtained the same results, luciferase activity increased 4.8 and 6.2 times by RUNX1 and RUNX2 separately (Fig. 3a, b). We chose HeLa and SK-BR-3 rather than HP75 because HP75 expresses high levels of endogenous RUNX1, RUNX2, and Gal-3 which may have higher background to these analyses. These experiments demonstrated that both RUNX1 and RUNX2 can upregulate Gal-3. Since the CC (or GG) binucleotides are the most important ones in the binding consensus sequence [10, 15–18], we further mutated the consensus sequence in Region 1 (TCTGGT → TCTccT) and Region 5 (CACCGCC → CAggGCC). There was a significant decrease of luciferase activity in the co-transfection experiments (Fig. 3c, d), indicating that these short DNA sequences are responsible for the interactions between RUNXs and LGALS3 promoter. These data support the hypothesis that RUNXs specifically regulate LGALS3 expression driven by the regulatory region.
Fig. 3.
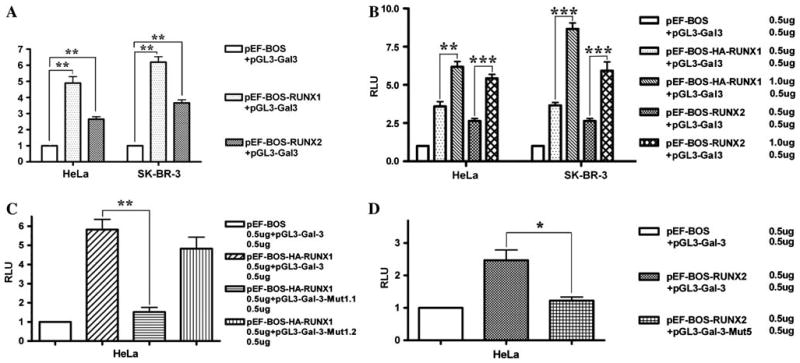
RUNX1 and 2 upregulated LGALS3 promoter activities in a sequence specific manner. a pEF-BOS or pEF-BOS-HA-RUNX1 or pEF-BOS-RUNX2 were co-transfected with pGL3-LGALS3 in either HeLa or SK-BR-3 cells, all vectors were transfected at 0.5 μg/well in six-well plates. About 0.1 μg of Renilla vector was also co-transfected as an internal control. b Different doses of pEF-BOS or pEF-BOS-HA-RUNX1 or pEF-BOS-RUNX2 were co-transfected with pGL3- LGALS3 in either HeLa or SK-BR-3 cells. Renilla vector was co-transfected as in a. c Region 1 mutations disrupted binding of RUNX1 with LGALS3 promoter by luciferase reporter system. pGL3-LGALS3 was mutated by site-directed mutagenesis, site 1 and site 2 binding sites were mutated from TCTGGT to TCTccT separately, and then co-transfected with pEF-BOS or pEF-BOS-HA-RUNX1 or pEF-BOS-RUNX2. Renilla vector was co-transfected as in a. d Region 5 mutation disrupted binding of RUNX2 protein with LGALS3 promoter by luciferase reporter system. pGL3-LGALS3 was mutated by site-directed mutagenesis, binding motif from CACCGCC to CAggGCC, and then co-transfected with pEF-BOS or pEF-BOS-HA-RUNX1 or pEF-BOS-RUNX2. Renilla vector was co-transfected as in a. * P < 0.05; ** P < 0.01; *** P < 0.001; ANOVA test
RUNX1 and RUNX2 both upregulate LGALS3 in pituitary tumor cells
We used the HP75 as a pituitary tumor model, and knocked down RUNX1 and RUNX2 by siRNA. In the RNA interference experiments, there was a significant decrease in Gal-3 expression in HP75 cells after knock-down RUNX1 or RUNX2, indicating that both RUNX1 and RUNX2 upregulated LGALS3 in pituitary tumor cells (Fig. 4a, b).
Fig. 4.
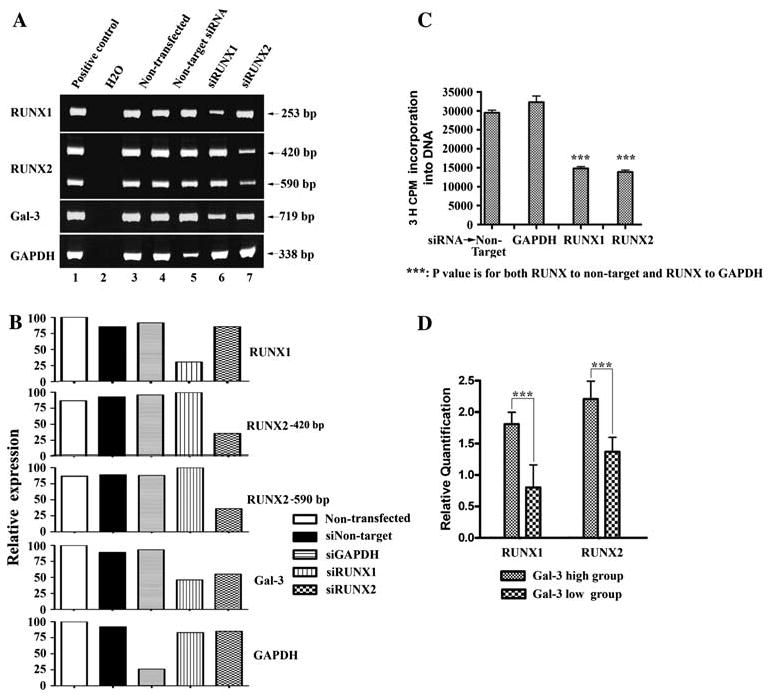
RUNX1 and RUNX2 upregulate Gal-3 in HP75 cells and pituitary tumors, promote HP75 cell growth. a siRNA of RUNX1 and RUNX2 knocked down RUNX1 and RUNX2 expression, Gal-3 was knocked down by either of the siRNA. GAPDH served as positive control; non-transfected and non-target siRNA served as negative controls (RT-PCR) in RNA interference experiments. First left lanes are positive controls for PCR reactions. b Densitometry for a. c [3H] Thymidine incorporation experiments showed that knockdown of RUNX1, RUNX2 slowed HP75 cell growth. d Immunohistochemistry analysis with tissue microarray showing correlation of Gal-3, RUNX1, and RUNX2 expression levels in pituitary tumors. Gal-3 was expressed by PRL and ACTH tumors, but not by GH, FSH, LH, null cell, or TSH tumors. In the normal pituitary only PRL, ACTH, and folliculostellate cells expressed Gal-3.
*** P < 0.001; ANOVA test
RUNX1 and RUNX2 promote pituitary tumor growth in HP75
To determine whether RUNX1 and RUNX2 play a role in pituitary tumor growth, [3H] thymidine incorporation experiments were performed in the HP75 cells. After siR-NA knock-down of RUNX1/RUNX2 for 96 h, HP75 cell proliferation was significantly decreased (Fig. 4c). This showed the importance of RUNX1 and RUNX2 in pituitary tumor growth.
RUNX1 and RUNX2 expression are correlated with Gal-3 level in pituitary tumors
Different subtypes of primary pituitary tumors were divided into two groups according to their Gal-3 expression level. There were 88 tumors in the Gal-3 high-expressing group which consisted of PRL and ACTH tumors, and 103 tumors in the Gal-3 low-expressing group which consisted of GH, FSH, LH, null cell, and TSH tumors. In the normal pituitary Gal-3 was produced only by PRL, ACTH, and folliculostellate cells. RUNX1 and RUNX2 immunostaining was scored in each group and compared between the two groups. There was a significant correlation between Gal-3 and RUNX1/RUNX2 expression levels (Fig. 4d).
Discussion
A large body of evidence has shown roles of Gal-3 in tumor progression and metastasis [1, 27, 28]. Recently, Gal-3 expression has emerged as a potential diagnostic and/or prognostic marker for some cancers, such as thyroid cancers [29]. Previously we reported that Gal-3 was expressed in a subset of normal pituitary cells, including lactotroph (PRL), corticotroph (ACTH), and folliculo-stellate (FS) cells. We also showed that Gal-3 had a role in the development of pituitary tumors [8, 9]. Treatment with transforming growth factor beta 1, which regulates pituitary cell proliferation, reduced Gal-3 as well as p27 expression levels in cultured HP75 pituitary tumor cells [8]. Inhibition of LGALS3 gene expression by RNA interference also decreased HP75 cell proliferation and increased apoptosis [8]. Consistent with this, a study from Roncaroli [30] demonstrated expression of Gal-3 in pituitary tumors derived from folliculostellate cells. These findings suggest that Gal-3 has an important regulatory role in pituitary tumor cell development and progression. The present studies show that RUNX1 and RUNX2 also have regulatory roles on Gal-3 expression in pituitary tumors.
The RUNX family members are important transcription factors which share a common partner, core-binding factor-β (CBF β). They are characterized by a highly conserved 128-amino-acid region homologous to the Drosophila protein Runt [31]. The Runt domain is responsible for DNA binding, protein–protein interaction, ATP binding, and contributes to nuclear localization [32–34]. In mammals, the RUNX family comprises three genes, encoding for RUNX1, RUNX2, and RUNX3. These proteins play crucial roles in regulating distinct developmental pathways. RUNX1 is important for hematopoiesis [35, 36], RUNX2 is a fundamental transcription factor for osteogenesis [37, 38], and RUNX3 is crucial for gastrointestinal organogenesis and neurogenesis [39, 40]. RUNX also has a role in tumor proliferation and progression, but whether they are oncogenes or tumor suppressor genes remains controversial. It is possible that their role as oncogenes and/or tumor suppressor genes is context-dependent. Studies in mouse skeletal system by Stock et al [10] indicated that RUNX2 protein regulates LGALS3 through binding to its promoter region. We questioned whether RUNX2 and possibly its family members, RUNX1 and RUNX3, could regulate LGALS3 and play a role in human pituitary tumor development.
ChIP assays showed that there was binding between RUNX1, RUNX2 proteins and LGALS3 promoter in the human pituitary tumor cell line HP75. RUNX1 binds to an upstream region on LGALS3 promoter which contains the predicted RUNX1 binding consensus sequence “TCTG GT”; RUNX2 binds to a downstream region which contains the predicated RUNX2 binding consensus sequence “CACCGCC.” We also observed that the anti-RUNX2 antibody pulled down the region 4 in the LGALS3 promoter in which there is no binding sequence predicted by the softwares; however, the binding sequence in region 5 is very close to region 4, with only 5 base pair distances. Considering the DNA length after sonication is generally 200–500 bp, and our PCR products are 200–300 bp, it is reasonable to observe this precipitation of region 4 by anti-RUNX2 antibody. These results were supported by EMSA experiments, which showed that the synthetic oligonucleotides containing “TCTGGT” or “CACCGCC” specifically bind to HP 75 nuclear extract. We also identified the key nucleotides in the binding sequences, since mutations from “TCTGGT” to “TCTccT” or “CACCG CC” to “CAggGCC” completely abolished these bindings. Anti-RUNX1 antibody slowed one band in region 1 probe further strengthening the hypothesis of specific binding between RUNX1 and region 1; but anti-RUNX1 failed to slow the other band. In contrast, the antibody eliminated this protein/DNA binding. The reason could be that, anti-RUNX1 antibody binds to the same site of this isoform of RUNX1 protein which also binds the oligonucleotides, so adding antibody interrupted the formation of a DNA/RUNX1 complex. In EMSA with region 5 probe, anti-RUNX2 also failed slowing the band but ablated the protein/DNA binding. The reason is probably the same as in RUNX1 experiment. These experiments provided evidences for a role of RUNX1 and RUNX2 in pituitary tumor development.
Proliferation studies using [3H] thymidine incorporation provided more biological evidence of the regulatory role of RUNX1 and RUNX2 in pituitary tumor, because knockdown of RUNX1 and RUNX2 in HP75 cells significantly decreased the cell growth.
There is increasing evidence that RUNXs are involved in the development of tumors, where they act as either oncogenes or tumor suppressor genes depending on the context. Amplification of RUNX1 has been linked with poor prognosis in childhood B-cell leukemia [41], while RUNX2 expression has been implicated in bone metastasis of breast cancer [42]. One study has also shown that the ability to induce premature senescence in primary murine embryonic fibroblast is a common feature of all three RUNX genes [43]. A more recent study demonstrated that both RUNX1 and RUNX2 but not RUNX3 are highly expressed in human glioma cells; this study also showed that RUNX2 mediates expression of Galectin-3 in glioma [44]. Therefore, elucidating the signaling pathway of the RUNX genes is very important. Here, we showed that LGALS3, which is an oncogene and plays critical roles in many cancers, is a direct target gene of RUNX1 and RUNX2.
In summary, we showed for the first time that LGALS3 is upregulated by RUNX1 and RUNX2 in human pituitary tumors and that different RUNX family member binds different DNA sequences in the LGALS3 promoter region. These studies suggest that RUNX1 and RUNX2 upregulate LGALS3 by direct binding to its promoter region and contribute, in part, to pituitary tumor growth regulation.
Acknowledgments
This research was supported in part by NIH Grant CA90249 and a Grant from the Jarislowsky Foundation to RVL) and NIH Grant R37CA46120-19 (to AR).
Contributor Information
He-Yu Zhang, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA.
Long Jin, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA.
Gail A. Stilling, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA
Katharina H. Ruebel, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA
Kendra Coonse, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA.
Yoshinori Tanizaki, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA.
Avraham Raz, Tumor Progression and Metastasis Program, Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA.
Ricardo V. Lloyd, Email: lloyd.ricardo@mayo.edu, Department of Pathology, Mayo Clinic College of Medicine, 200, 1 Street SW, Rochester, MN 55905, USA
References
- 1.Shekhar MP, Nangia-Makker P, Tait L, Miller F, Raz A. Alterations in galectin-3 expression and distribution correlate with breast cancer progression: functional analysis of galectin-3 in breast epithelial-endothelial interactions. Am J Pathol. 2004;165:1931–1941. doi: 10.1016/S0002-9440(10)63245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong HC, Honjo Y, Nangia-Makker P, Hogan V, Mazurak N, Bresalier RS, Raz A. The NH2 terminus of galectin-3 governs cellular compartmentalization and functions in cancer cells. Cancer Res. 1999;59:6239–6245. [PubMed] [Google Scholar]
- 3.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 4.Yamaoka K, Mishima K, Nagashima Y, Asai A, Sanai Y, Kirino T. Expression of galectin-1 mRNA correlates with the malignant potential of human gliomas and expression of antisense galectin-1 inhibits the growth of 9 glioma cells. J Neurosci Res. 2000;59:722–730. doi: 10.1002/(SICI)1097-4547(20000315)59:6<722::AID-JNR4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Takenaka Y, Inohara H, Yoshii T, Oshima K, Nakahara S, Akahani S, Honjo Y, Yamamoto Y, Raz A, Kubo T. Malignant transformation of thyroid follicular cells by galectin-3. Cancer Lett. 2003;195:111–119. doi: 10.1016/s0304-3835(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 6.Hughes RC. The galectin family of mammalian carbohydrate-binding molecules. Biochem Soc Trans. 1997;25:1194–1198. doi: 10.1042/bst0251194. [DOI] [PubMed] [Google Scholar]
- 7.Hughes RC. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim Biophys Acta. 1999;1473:172–185. doi: 10.1016/s0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- 8.Riss D, Jin L, Qian X, Bayliss J, Scheithauer BW, Young WF, Jr, Vidal S, Kovacs K, Raz A, Lloyd RV. Differential expression of galectin-3 in pituitary tumors. Cancer Res. 2003;63:2251–2255. [PubMed] [Google Scholar]
- 9.Ruebel KH, Jin L, Qian X, Scheithauer BW, Kovacs K, Nakamura N, Zhang H, Raz A, Lloyd RV. Effects of DNA methylation on galectin-3 expression in pituitary tumors. Cancer Res. 2005;65:1136–1140. doi: 10.1158/0008-5472.CAN-04-3578. [DOI] [PubMed] [Google Scholar]
- 10.Stock M, Schafer H, Stricker S, Gross G, Mundlos S, Otto F. Expression of galectin-3 in skeletal tissues is controlled by Runx2. J Biol Chem. 2003;278:17360–17367. doi: 10.1074/jbc.M207631200. [DOI] [PubMed] [Google Scholar]
- 11.Costessi A, Pines A, D’Andrea P, Romanello M, Damante G, Cesaratto L, Quadrifoglio F, Moro L, Tell G. Extracellular nucleotides activate Runx2 in the osteoblast-like HOBIT cell line: a possible molecular link between mechanical stress and osteoblasts’ response. Bone. 2005;36:418–432. doi: 10.1016/j.bone.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Jin L, Kulig E, Qian X, Scheithauer BW, Eberhardt NL, Lloyd RV. A human pituitary adenoma cell line proliferates and maintains some differentiated functions following expression of SV40 large T antigen. Endocr Pathol. 1998;9:169–184. [Google Scholar]
- 13.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 14.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 15.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 16.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnikova IN, Crute BE, Wang S, Speck NA. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaiman AL, Lewis AF, Crute BE, Speck NA, Lenz J. Transcriptional activity of core binding factor-alpha (AML1) and beta subunits on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEB-P2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadrofske MM, Openo KP, Wang JL. The human LGALS3 (galectin-3) gene: determination of the gene structure and functional characterization of the promoter. Arch Biochem Biophys. 1998;349:7–20. doi: 10.1006/abbi.1997.0447. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi Y, Kurokawa M, Imai Y, Izutsu K, Asai T, Ichikawa M, Yamamoto G, Nitta E, Yamagata T, Sasaki K, Mitani K, Ogawa S, Chiba S, Hirai H. AML1 is functionally regulated through p300-mediated acetylation on specific lysine residues. J Biol Chem. 2004;279:15630–15638. doi: 10.1074/jbc.M400355200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YW, Bae SC, Huang G, Fu YX, Lu J, Ahn MY, Kanno Y, Kanno T, Ito Y. A novel transcript encoding an N-terminally truncated AML1/PEBP2 alphaB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol Cell Biol. 1997;17:4133–4145. doi: 10.1128/mcb.17.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howcroft TK, Weissman JD, Gegonne A, Singer DS. A T lymphocyte-specific transcription complex containing RUNX1 activates MHC class I expression. J Immunol. 2005;174:2106–2115. doi: 10.4049/jimmunol.174.4.2106. [DOI] [PubMed] [Google Scholar]
- 27.Fukumori T, Oka N, Takenaka Y, Nangia-Makker P, Elsamman E, Kasai T, Shono M, Kanayama HO, Ellerhorst J, Lotan R, Raz A. Galectin-3 regulates mitochondrial stability and antiapoptotic function in response to anticancer drug in prostate cancer. Cancer Res. 2006;66:3114–3119. doi: 10.1158/0008-5472.CAN-05-3750. [DOI] [PubMed] [Google Scholar]
- 28.Oka N, Nakahara S, Takenaka Y, Fukumori T, Hogan V, Kanayama HO, Yanagawa T, Raz A. Galectin-3 inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by activating Akt in human bladder carcinoma cells. Cancer Res. 2005;65:7546–7553. doi: 10.1158/0008-5472.CAN-05-1197. [DOI] [PubMed] [Google Scholar]
- 29.Volante M, Bozzalla-Cassione F, Orlandi F, Papotti M. Diagnostic role of galectin-3 in follicular thyroid tumors. Virchows Arch. 2004;444:309–312. doi: 10.1007/s00428-004-0993-5. [DOI] [PubMed] [Google Scholar]
- 30.Roncaroli F, Scheithauer BW, Cenacchi G, Horvath E, Kovacs K, Lloyd RV, Abell-Aleff P, Santi M, Yates AJ. ‘Spindle cell oncocytoma’ of the adenohypophysis: a tumor of folliculostellate cells? Am J Surg Pathol. 2002;26:1048–1055. doi: 10.1097/00000478-200208000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 32.Crute BE, Lewis AF, Wu Z, Bushweller JH, Speck NA. Biochemical and biophysical properties of the core-binding factor alpha2 (AML1) DNA-binding domain. J Biol Chem. 1996;271:26251–26260. doi: 10.1074/jbc.271.42.26251. [DOI] [PubMed] [Google Scholar]
- 33.Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 37.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 38.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 39.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, Yamashita N, Itohara S, Kudo N, Ito Y. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 41.Robinson HM, Broadfield ZJ, Cheung KL, Harewood L, Harris RL, Jalali GR, Martineau M, Moorman AV, Taylor KE, Richards S, Mitchell C, Harrison CJ. Amplification of AML1 in acute lymphoblastic leukemia is associated with a poor outcome. Leukemia. 2003;17:2249–2250. doi: 10.1038/sj.leu.2403140. [DOI] [PubMed] [Google Scholar]
- 42.Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64:4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- 43.Kilbey A, Blyth K, Wotton S, Terry A, Jenkins A, Bell M, Hanlon L, Cameron ER, Neil JC. Runx2 disruption promotes immortalization and confers resistance to oncogene-induced senescence in primary murine fibroblasts. Cancer Res. 2007;67:11263–11271. doi: 10.1158/0008-5472.CAN-07-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vladimirova V, Waha A, Luckerath K, Pesheva P, Probstmeier R. Runx2 is expressed in human glioma cells and mediates the expression of galectin-3. J Neurosci Res. 2008;86:2450–2461. doi: 10.1002/jnr.21686. [DOI] [PubMed] [Google Scholar]


