Abstract
The yeast nonchromosomal gene [URE3] is due to a prion form of the nitrogen regulatory protein Ure2p. It is a negative regulator of nitrogen catabolism and acts by inhibiting the transcription factor Gln3p. Ure2p residues 1–80 are necessary for prion generation and propagation. The C-terminal fragment retains nitrogen regulatory activity, albeit somewhat less efficiently than the full-length protein, and it also lowers the frequency of prion generation. The crystal structure of this C-terminal fragment, Ure2p(97–354), at 2.3 Å resolution is described here. It adopts the same fold as the glutathione S-transferase superfamily, consistent with their sequence similarity. However, Ure2p(97–354) lacks a properly positioned catalytic residue that is required for S-transferase activity. Residues within this regulatory fragment that have been indicated by mutational studies to influence prion generation have been mapped onto the three-dimensional structure, and possible implications for prion activity are discussed.
The yeast nonchromosomal genes [URE3] (1) and [PSI] (2) are due to infectious protein forms (prions) of the Ure2 and Sup35 proteins, respectively (ref. 3; reviewed in ref. 4). Ure2p is part of the signal transduction cascade that regulates nitrogen catabolism in yeast, repressing genes for proteins involved in using poor nitrogen sources when a rich nitrogen source is available (5). Sup35p is a subunit of the translation termination factor. The prion forms of Ure2p and Sup35p have lost their normal functions, but have acquired the ability to transmit this abnormality to other molecules of their respective normal forms. Thus, [URE3] prion-carrying cells are derepressed for nitrogen catabolism despite the presence of a good nitrogen source. [PSI+] cells terminate translation inefficiently and thus read through translation termination codons at increased frequency.
Ure2p has an N-terminal asparagine-rich “prion domain” (residues 1–80) that can efficiently induce the de novo generation of the [URE3] prion, and is required in cis for a Ure2p molecule to be inactivated by the [URE3] prion (6–8). The C-terminal part of Ure2p (residues 81–354) is sufficient to carry out the nitrogen regulation function of Ure2p and has significant homology to glutathione S-transferases (GSTs) of bacteria, plants, and animals (9).
In cells with the [URE3] prion, Ure2p is aggregated (10) and partially protease resistant (6), but it is evenly distributed in the cytoplasm and protease sensitive in normal cells. The ability of Ure2p to form amyloid in vitro, the dependence of this amyloid formation on the prion domain of Ure2p, and the similarity of protease-resistant fragments from Ure2p amyloid formed in vitro to those of Ure2p in extracts of [URE3] cells suggest that self-propagating amyloid is the basis for [URE3] prion propagation (11).
Ure2p directly interacts with the DNA binding protein Gln3p, preventing its entry into the nucleus to promote transcription of an array of genes involved in assimilation of poor nitrogen sources (12–15). Ure2p is itself regulated by Mks1p, mediating nitrogen catabolite repression (16), and by the TOR system, a global regulator that transmits nutritional information to many cellular components (14, 17, 18). Inhibiting the TOR system dephosphorylates Ure2p and thereby inactivates it.
Deletions of the Ure2p C-terminal nitrogen regulation domain dramatically elevate the efficiency with which the overexpressed protein induces the de novo appearance of the [URE3] prion (6). Because the activity of the nitrogen catabolite repression pathway does not in itself affect the frequency with which [URE3] arises (7), it has been suggested that the Ure2p C-terminal domain normally interacts with the N-terminal prion domain, thereby stabilizing it. Indeed, by using the yeast two-hybrid system, it has been shown that the C-terminal residues 153–354 interact with the N-terminal residues 1–151 (19). Arguing against interaction of N-terminal and C-terminal domains are the results of denaturation studies. The free energy of denaturation, measured as the urea concentration necessary for half change of the intrinsic fluorescence of the protein is the same for the full-length Ure2p and for a mutant molecule lacking the prion domain (20). However, it is possible that interactions between the N- and C-termini in the full-length Ure2p are replaced by interactions within (or between subunits of) the C-terminal domain in the deleted molecule. Full-length Ure2p purified from Saccharomyces cerevisiae or Escherichia coli is a soluble dimer (11, 20). Recombinant Ure2p 90–354, lacking the prion domain, also has a dimeric structure (20).
Attempts to crystallize the intact Ure2p were frustrated by proteolytic cleavage at residue 95. Here, we report the crystal structure of the nitrogen regulation fragment of Ure2p, Ure2p(97–354). This structure provides a framework to interpret mutation data, identifying residues within this fragment that influence the frequency of prion generation. A structure-based explanation of the observed absence of GST activity for Ure2p is also proposed.
Materials and Methods
Overexpression and Purification of His6-Ure2 Protein.
The His6-Ure2 fusion plasmid (pKT41-1) was constructed by the PCR by using p576 as template and primers 5′-GGAACTCATATGCATCACCATCACCATCACATGTATCCACGTGGGAATATGATGAATAACAACGGC-3′ and 5′-GGAACTGTCGACGAATTCTGTGGTTGGGGTAAC-3′. These primers contain an N-terminal His6 tag, a thrombin cleavage site, a 3′ termination site, and flanking NdeI and SalI restriction sites. In the same PCR, primers 5′-CGGATGAGGTTCGTCGTGTTTACGGTGTAG-3′ and 5′-CTACACCGTAAACACGACGAACCTCATCCG-3′ were included to replace codons 253 and 254 of URE2 with CGT (21). The NdeI-SalI fragment from pKT41-1 was cloned into the pFLAG vector (Kodak). The ExSite Mutagenesis system (Stratagene) was then used to create a plasmid (pTU2-B) coding for Ure2p(97–354) with an N-terminal His6 tag and a thrombin cleavage site.
For selenomethionine incorporation (22), the E. coli methionine auxotroph B834(DE3)pLysS (Stratagene) carrying pTU2-B was grown to log phase in 4 liters of LeMaster Medium containing 55 mg/liter DL-selenomethionine, 100 μg/ml ampicillin, and 50 μg/ml chloramphenicol at 37°C. Overexpression of the C-terminal fusion was initiated by the addition of isopropyl β-d-thiogalactoside (IPTG) to a concentration of 0.4 mM, followed by incubation for 3 h at 30°C. Purification was based on that previously published (11). Briefly, cells (18 g) were resuspended in 50 ml of binding buffer (50 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/5 mM imidazole/1 mM β-mercaptoethanol/1 mM PMSF) and disrupted with a French press. After centrifugation, the supernatant was loaded onto a 2-ml Ni-NTA Superflow (Qiagen, Chatsworth, CA) column, washed with 50 ml binding buffer, and eluted with a 0–500 mM imidazole gradient in binding buffer. Fractions containing pure protein were dialyzed against 20 mM Tris⋅HCl, pH 8.0, with 0.3 M NaCl. The His6 tag was cleaved with thrombin (10 units/mg protein, 6 h), which was subsequently removed by a 2-ml benzamidine Sepharose 2B column (Pharmacia). This column was washed with 5 ml of 20 mM Tris⋅HCl, pH 8.0, containing 0.3 M NaCl, and the protein eluates were combined, concentrated to 3.25 mg/ml, and dialyzed against 20 mM Tris⋅HCl, pH 8.0, with 0.3 M NaCl.
Crystallization.
Ure2p(97–354) was crystallized by vapor diffusion. Droplets containing equal parts (5 μl to 30 μl total volume) of protein and crystallization buffer [reservoir solution with 10−2–10−3 mg/ml of α-chymotrypsin (Bovine pancreas; Sigma)] were equilibrated against reservoir solutions of 2.5 M-3.0 M NaCl and 100 mM bicine, pH 8.8. The growth of the crystal form of Ure2p(97–354) reported here required proteolytic clipping of the protein. Initially, crystallization sporadically occurred, and usually only after a lengthy period, perhaps because of the trace presence of an unknown protease. It was later found that similar crystals could be reproducibly grown after limited proteolysis with chymotrypsin. This treatment clipped Ure2p(97–354) after M272 and before F295. Large well-formed crystals (0.6 mm maximum) grew within several days.
Data Collection.
Crystals were transferred into a final cryoprotectant solution of 21.75% (wt/vol) glycerol, 2.5 M–3.0 M NaCl, and 100 mM bicine, pH 8.8, and were flash frozen with liquid propane. Multiwavelength anomalous diffraction data at three separate wavelengths (Table 1) were collected at beamline X-9B of the National Synchrotron Light Source (NSLS) at 100 K by using an Area Detector Systems Corporation Quantum 4 charge-coupled device (CCD) detector. The crystal belonged to space group P212121, with cell dimensions a = 64.3 Å, b = 69.2 Å, and c = 150.0 Å. The Matthews' coefficient (23) is 3.02 Å3 Da−1, with two monomers per asymmetric unit.
Table 1.
Data collection and refinement statistics
| Data set
|
|||
|---|---|---|---|
| λ1 | λ2 | λ3 | |
| Wavelength, Å | 0.9791 | 0.9789 | 0.9686 |
| Resolution, Å | 20.0–2.3 | 20.0–2.3 | 20.0–2.3 |
| No. of observations | 184,409 | 180,695 | 168,715 |
| No. of unique reflections | 27,098 | 30,005 | 29,499 |
| Completeness (%)* | 89.1 (92.6) | 98.6 (99.8) | 97.6 (99.5) |
| Rsym (%)*† | 5.8 (37.5) | 5.7 (32.9) | 4.8 (32.0) |
| Phasing (F/σ(F) ≥ 1.0) | |||
| Resolution, Å | 2.7 | 2.4 | 2.3 |
| Phasing Power‡ | 1.62 | 2.54 | — |
| Figure of merit | 0.49 for 23,123 phased reflections | ||
| 0.79 post-solvent flattening | |||
| Refinement statistics (λ3 data set) | |||
| No. of reflections (F/σ(F) ≥ 2.0) | |||
| Working | 25,681 | ||
| Test | 1,343 | ||
| R (Rfree)§ | 22.1% (27.5%) | ||
| No. of protein atoms | 3,666 | ||
| No. of waters | 204 | ||
| 〈B-factor〉, Å2 | |||
| Monomer A | 55.0 | ||
| Monomer B | 54.1 | ||
| Water | 57.1 | ||
| r.m.s.d. from ideality | |||
| Bond lengths, Å3 | 0.0118 | ||
| Angles, deg | 1.62 | ||
Values in parentheses refer to statistics for data in the 2.38–2.30 Å resolution shell.
Rsym = ∑|I − 〈I〉|/∑〈I〉.
Phasing power: FHC/E for the isomorphous case and 2FHC"/E for the anomalous case, where FHC is the calculated heavy atom structure factor and E is the rms lack of closure.
R = ∑|FO − FC|∑FO; Rfree computed using randomly selected 5% of the data that were excluded throughout the refinement.
Structure Determination and Refinement.
X-ray data were processed by using HKL (24). Nine of the twelve selenium atoms in the asymmetric unit were located by using solve (25) and SnB (26). The three undetected sites were later revealed to be located on the protein surface and subject to high thermal vibrations. The selenium positions were refined by using maximum-likelihood phase refinement, and the resultant phases were modified by using solvent flattening. These pseudo-MIRAS calculations (Table 1) were undertaken in phases-95 (27). The protein model was built into a good quality map into the density by using o (28), and was refined by using cns version 1.0 (29). Bulk solvent correction and an anisotropic temperature factor correction were used. Before refinement, 5% of the data were randomly selected as a test data set, to be used for calculating Rfree, whereas the model was refined against the remaining 95% of the data. Rfree was used throughout the refinement to optimize the refinement scheme and to prevent over-fitting of the data. The final model included the use of weak noncrystallographic symmetry (ncs) restraints (ncs-related main chain atoms, 50 kcal/mol/Å2; ncs-related side chain atoms, 25 kcal/mol/Å2; weakly ncs-related main chain atoms, 25 kcal/mol/Å2; weakly ncs-related side chain atoms, 15 kcal/mol/Å2; and no restraints on side chain atoms judged not related by ncs) and restrained individual temperature factors. Water molecules were added to the model at the later stages of refinement. The final protein model was evaluated by using procheck (30). Figures were created by using ribbons (31).
Results and Discussion
Overall Structure.
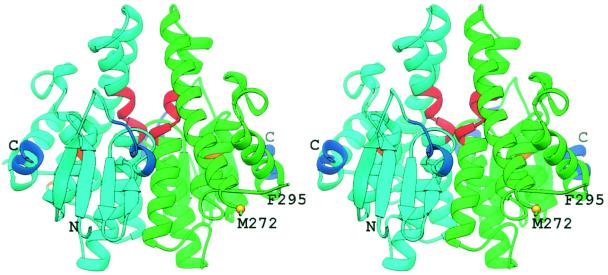
The structure of Ure2p(97–354) displays a fold similar to that of the GST superfamily (Fig. 1), as suspected from the sequence similarity between these proteins (9, 32–34). However, it should be noted that no GST activity has been observed in Ure2p (9, 32). The Ure2p(97–354) monomer has two domains and dimensions of approximately 57 Å × 55 Å × 36 Å. The N-terminal domain, residues Glu-112 to Gly-197, contains a single four-stranded β-sheet and three α-helices in a βαβαββα motif (Fig. 2). The core of this domain is the mixed β-sheet, in which β1 and β2 are parallel, and β1 and β3, and β3 and β4 are anti-parallel. Helix α2 is present in a highly solvent exposed loop structure. Residues Asn-198 to Asp-204 form a linker between the first and second domains. The N-terminal residues Val-97 to Phe-105 of Monomer A, and Val-97 to Gln-109 of Monomer B were excluded from the model, because they were not observed in the electron density maps, and are presumably disordered.
Figure 1.
Stereoview of the Ure2p(97–354). Monomer A is green and Monomer B is cyan. Prion-inhibiting regions (His-151 to Ser-158 and Val-347 to Glu-354) are indicated in blue, and the prion-promoting region (Ser-221 to Ile-227) is indicated in red. Gold labels the position of two additional residues implicated in affecting prion-induction, K127 and V271.
Figure 2.
Sequence alignment of Ure2p and structurally similar GSTs [PDB code 1A0F: E. coli GST (37); PDB code 1GNW: A. thaliana theta class GST (38)]. Secondary structural elements identified in the Ure2p(97–354) crystal structure are indicated above the sequence. Residues conserved in all sequences are indicated by an asterisk below the sequence. Catalytic residues are cyan. G-site (GSH-binding) residues are green. H-site (hydrophobic electrophile-binding) residues are violet. Underlined residues indicate residues contributed to a binding site by the second monomer of the dimer. The indicated residues in Ure2p have been selected by analogy to these similar GSTs.
The C-terminal domain, residues Asp-205 to Glu-354, contains six α-helices and a single turn of 310 helix. This domain contains the chymotrypsin cleavage sites, and the clipped region (Glu-273 to Phe-294) is a solvent exposed loop structure that extends from the core of the domain. In sequence alignments between Ure2p and GSTs, there is a high tendency for this clipped region in Ure2p to be absent in GST (Fig. 2).
The domain interface within the monomer is formed by the helices α1, α3, α4, α6, and α9, and has a combination of hydrophobic and hydrogen bonding interactions, together with a single salt bridge between Glu-134 and Arg-344. The total buried surface area at this interface is approximately 2100 Å2.
Ure2p(97–354)'s dimer assembly is consistent with that observed within the theta class GST family, but with some difference in the relative orientation of the two interacting monomers. The theta class GSTs exhibit increased variability at the dimer interface (35) compared with other classes of GST. The rms deviation between the two Ure2p(97–354) monomers is 0.38 Å and 1.21 Å, calculated over Cα and all atoms, respectively. The dimer interface is composed of the extended loop region between β2 and β3, strand β4, and helices α3, α4, and α5. A total surface area of approximately 3600 Å2 is buried at the dimer interface.
Crystal Contacts.
The crystal form of Ure2p(97–354) reported here has a higher than average solvent content of approximately 60%. The crystal packing contacts involve solely like monomers. That is, the only contacts between Monomer A and Monomer B exist within the intact dimer, which constitutes the asymmetric unit. The two types of crystal packing interactions, those involving only Monomer As and those involving only Monomer Bs, are similar but not identical. Contacts of the first type are more extensive, with a total buried surface area of 1750 Å2, opposed to only 1150 Å2 buried in contacts of the second type. The phenyl group of Phe-295 in the Monomer B contacts makes fewer hydrophobic interactions and is more solvent exposed than the corresponding group in the Monomer A contacts.
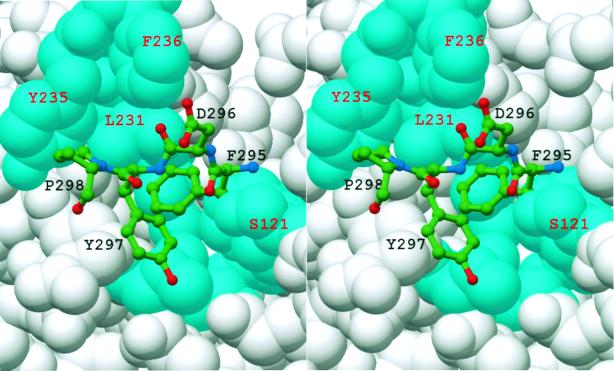
A further point of interest regarding the crystal contacts concerns the residues involved. At the 2-fold screw axis, the residues near the proteolytic clip sites (i.e., Met-272 and Phe-295) of one monomer protrude into a cleft formed between the two domains of a symmetry related monomer (Fig. 3). Phe-295, Asp-296, and Tyr-297 make contact by being inserted into the cleft, and residues Arg-267 to Met-272 interact with the cleft's outer edge. More specifically, this cleft is defined by the loop between β1 and α1 and the loop between β2 and β3 in the first domain, and by α4, α6, the 310 helix immediately after α6, and α9 on the second domain. In the structurally similar GSTs, the analogous cleft contains the catalytic active site, including the G- (glutathione) and the H- (hydrophobic electrophile) substrate binding sites (36–38). The Ure2p(97–354) residues within the cleft that participate in crystal contacts are in a region equivalent to the GST H-site. Thus, there is a possibility that the crystal packing interactions are mimicking biologically relevant interactions formed between Ure2p and an endogenous binding partner.
Figure 3.
Space-filling model of Ure2p(97–354) Monomer A about the region comparable to the GST H-site (cyan). Residues from a symmetry related molecule are represented as balls-and-sticks. Additional residues from the neighboring molecule, which also forms crystal contacts in this region, have been excluded for clarity.
Structural Comparisons with GSTs.
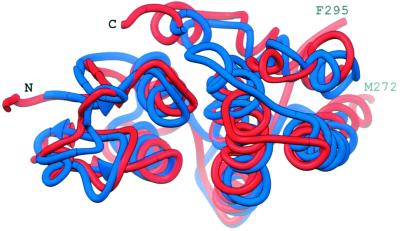
The structure of monomeric Ure2p(97–354) was compared with the entries in the PDB by using dali (39) and vast (40), and all of the closest neighbors belonged to the GST superfamily (reviewed in refs. 36, 41, and 42). A GST from E. coli (PDB code 1A0F) (43) had the lowest rms deviation (2.3 Å) from Ure2p(97–354) over Cα atoms from 187 residues. The sequence identity was 20%. The plant theta class GST from Arabidopsis thaliana (PDB code 1GNW) (38) has a rms deviation of 2.6 Å over 199 residues (23% identity) when compared with the Ure2p(97–354) monomer (Fig. 4). Ure2p(97–354)'s deep cleft has a strong structural resemblance to the regions identified as the G- and H-sites in the GSTs. This similarity is particularly true for the comparison with the theta class GST from A. thaliana.
Figure 4.
The superposition of Monomer A of Ure2p(97–354) (red) and a monomer of A. thaliana GST (38) (blue). This representation is viewed into the cleft between the two domains, which in GST contains the G- and H-sites.
Despite the broad sequence variation in GSTs, there are several residues and motifs that are highly conserved, and these tend to also be present in Ure2p. Koonin et al. (33) identified two conserved sequence motifs within the GST superfamily. Ure2p(97–354) contains motif I between Asn-162 and Asn-190, and contains motif II between Trp-300 and Asn-317. Two structurally important residues belonging to the two motifs are also found in Ure2p(97–354) as cis Pro-166 and an aspartate with its sidechain buried (Asp-310). The cis proline participates in the proper formation of the G-site, and the buried aspartate has been implicated as playing a role in the initial folding steps in GSTs.
A consensus sequence pattern has been generated for the theta class GSTs (34). This consensus pattern contains restrictions on 35 sequence positions within the N-terminal domain. The corresponding residues in Ure2p conform to this pattern in all but two positions. One of these disagreements is at position 71 of the consensus pattern (a tyrosine), which is His-187 in Ure2p. This residue is in α3 of Ure2p(97–354) and interacts with the sidechain of Asp-170, which is in β3. In the A. thaliana GST crystal structure (38), which conserves this consensus tyrosine, a similar interaction is observed with an aspartate on β3. Thus, Ure2p's violation of the consensus pattern at this position would appear to be minor.
Ure2p's second deviation from the GST theta class consensus sequence is more significant. Position 9 of the consensus sequence is a serine, but the corresponding position is Ala-122 in Ure2p. In the known structures of theta class GSTs (38, 44, 45), this serine's Oγ atom is either positioned, or may be positioned through a simple change of rotamer, to form a hydrogen bond with the sulfhydryl group of bound glutathione (GSH). This interaction has been proposed to stabilize the GSH as a thiolate anion, and thus activate GSH for the nucleophilic addition of the GSH sulfur atom to electrophilic groups of hydrophobic substrates (43). Mutation of this catalytic serine, Ser-11, to alanine in the A. thaliana theta class GST not only dramatically reduced activity to less than 0.5% the wild-type enzyme, but also significantly reduced the mutant's affinity for immobilized GSH (45). In the other GST classes, the hydroxyl group of a conserved tyrosine serves to stabilize the thiolate anion. Ure2p also lacks this corresponding tyrosine.
The lack of observed catalytic activity could be due to the insertion of alanine for serine at residue 122 of Ure2p(97–354). The cleft in Ure2p(97–354), comparable to the GSH binding site in GSTs, was analyzed for other residues that might stabilize an activated GSH thiolate anion. Adjacent to Ala-122 is Ser-121, and this initially appeared to be a likely suspect. However, the sidechain of this residue is directed away from what is comparable to the G-site, and is ill positioned, even with a change of rotamer, to interact with GSH, much less to stabilize the thiolate anion. On superposition of the monomers of both structures, the distance separating the Cβ atoms of Ser-121 and the catalytic Ser-11 of A. thaliana GST (38) is 6.3 Å, and the distance between the respective Cα atoms is 4.3 Å. Moreover, the directions of the Cα-Cβ bond of the two respective serines differ by greater than 90° in the superimposed structures. No other residues within Ure2p(97–354)'s cleft appear to be positioned to serve this catalytic role without the protein undergoing significant conformational changes.
In the E. coli GST, neither the conserved tyrosine nor serine catalytic residues are present. Instead, the mainchain amide group of Cys-10 and the sidechain of His-106 of this GST are proposed to be the catalytically important groups (37). An analogous arrangement is absent in the Ure2p(97–354) structure. Yet, the possibility that Ure2p contains a novel set of catalytic residues cannot be excluded.
Several alternative conclusions may be drawn regarding Ure2p and its lack of observed GST activity. First, it may lack activity because of the absence of a key catalytic residue leading to the protein's inability to stabilize the active form of GSH. Second, Ure2p may indeed possess GST activity, but requires an as yet untested and possibly unique substrate or set of reaction conditions. For example, theta class GSTs exhibit substrate specificity, having a tendency toward low specific activity when assayed with the small substrates 1-chloro-2,4-dinitrobenzene (CDBN), 1,2-dibromomethane, and 1,2-epoxy-3-(p-nitrophenoxy)-propane (38, 46). There are examples of GST and GST-like proteins that possess functionality that is independent of GST activity. A human pi class GST has been reported as serving as a regulator of signal transduction (47). Both the wild-type pi class GST and an enzymatically inactivate mutant were shown to inhibit activation of the nuclear transcriptional activating protein jun by jun kinase (JNK), with the GST's C-terminal residues (194 to 201) being implicated as important for this inhibitory activity. The S-crystallins of cephalopods, which perform a refractive function in the eye lens, have evolved from a sigma class GST (48, 49).
Implications for Prion Formation.
The isolated nitrogen regulation fragment of Ure2p lacks both prion-inducing and prion propagation activity (6, 7). However, there are data that this fragment of the protein influences the prion activity of Ure2p's prion domain (residues 1–80). First, the prion-inducing activity of the prion domain alone is much greater than that of the intact protein (6), and the nitrogen regulatory activity of the nitrogen regulation fragment alone is also less efficient than that of intact Ure2p (6). Several deletion mutants within the nitrogen regulation fragment alter the prion-inducing activity of the otherwise intact Ure2p (8). Deletion of residues His-151 to Ser-158, which reside in the loop between β2 and β3, or the C-terminal residues Val-347 to Glu-354, increases prion-inducing activity approximately 100-fold while also eliminating the nitrogen regulatory activity (Fig. 1). These prion-inhibiting regions lie on the same face of the protein as does the N terminus of the Ure2p(97–354) fragment, consistent with the hypothesis that the prion-inhibiting property of these residues is due to direct interaction between them and residues within the prion domain. Residues from His-151 to Ser-158 of Ure2p(97–354) correspond to a region in GSTs that participates in forming the G-site.
Deletion of residue Ser-221 to Ile-227 eliminates the prion-inducing activity of the otherwise intact Ure2p, while not affecting the nitrogen-regulating activity. These residues have been termed prion promoting (8), but no explanation for this activity is readily forthcoming from the structure. These residues are located in the middle of the long helix α4, and include the kink within this helix. Although these residues are near the dimer interface, only Ser-221 actually forms a contact across the interface. In fact, many of these residues are solvent accessible because α4 forms a wall that separates the cleft in one monomer from its mate in the dimer. Two possible models regarding the role of these residues in prion generation are suggested. In the first model, the deletion of these residues eliminates prion induction simply by the fortuitous creation of stabilizing interactions between the prion domain and the regulation fragment, while not altering the region responsible for the nitrogen regulatory activity. In the second model, Ser-221 to Ile-227 are necessary for interacting with a factor that enhances prion generation. One candidate for this factor is the protein Mks1p, which has been demonstrated to be necessary for prion generation (16, 50).
Two double mutants of Ure2p have been identified that enhance prion-induction by approximately 10-fold (51). Each mutant includes a single mutation in the prion domain and a single mutation in the nitrogen regulation fragment, neither of which alone results in any change in prion generation. The first such double mutant is Ure2p(S10L/V271E), which retains regulatory activity. Residue V271 is located at the C-terminal end of helix α5 (Fig. 1). It is distant from the N terminus of Ure2p(97–354) of the same monomer, but, in the context of the dimer, it lies near the N terminus of the second monomer. The second Ure2p double mutant is Ure2p(R17C/K127E). This mutant has lost nitrogen regulatory activity, as has the single K127E point mutation. Lys-127 is located on helix α1, and its sidechain amino group is located across the cleft from the prion-promoting region in α4 (Ser-221 to Ile-227), with the Nζ atom of Lys-127 6.8 Å from the Cβ atom of Ala-224. Whereas Lys-127 and the prion-promoting region map to the same structural region, the R17C/K127E double mutant and the Ser-221 to Ile-227 deletion mutant result in opposite affects on both prion induction and nitrogen regulatory activities.
Concluding Remarks.
The mechanisms through which Ure2p participates in nitrogen catabolite repression and may be transformed into a yeast prion are still not understood. Nitrogen catabolite repression involves an interplay between Ure2p, Gln3p, the TOR system, Mks1p, and the phosphorylation states of Ure2p and Gln3p. Mks1p is also a necessary factor for de novo [URE3] prion generation. However, because the frequency of prion generation is not a function of the available nitrogen source (7), Mks1p probably has two distinct interactions with Ure2p, one dependent and one independent of the available nitrogen source.
The crystal structure of Ure2p(97–354) demonstrates that this fragment is structurally similar to the GST superfamily. However, the absence within Ure2p(97–354) of catalytic residues conserved within GSTs may be the reason why no GST activity has been observed for Ure2p. In the least, it indicates that perhaps Ure2p is a novel GST for which the appropriate enzymatic assay has not yet been applied. Additionally, this work allows for the location of residues implicated by mutational analysis as influencing prion inducement within the context of the three-dimensional structure. Perhaps more intriguing and harder to interpret are those residues that are located within the cleft between the two domains. This cleft is structurally comparable to the GST active site. One possibility is that, through evolution, Ure2p has lost GST activity and instead this cleft is used for the binding of some or all of the Ure2p's partners in nitrogen regulation and perhaps also factors that influence prion formation. The crystal contacts observed within and near the cleft may mimic these binding interactions.
Acknowledgments
We thank Fred Dyda and K. R. Rajashankar for help in data collection. We thank Xinhua Ji and Dan Masison for the careful reading of this manuscript and their thoughtful comments.
Abbreviations
- GST
glutathione S-transferase
- GSH
reduced glutathione
- ncs
noncrystallographic symmetry
- G-site
glutathione-binding site
- H-site
hydrophobic electrophile-binding site
Note Added in Proof.
A paper describing a similar construct of Ure2p, although in different crystal forms, has just appeared (52).
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1HQO).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041607898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041607898
References
- 1.Lacroute F. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox B S, Tuite M F, McLaughlin C S. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 3.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 4.Wickner R B, Taylor K L, Edskes H K, Maddelein M-L, Moriyama H, Roberts B T. Microbiol Mol Biol Rev. 1999;63:844–861. doi: 10.1128/mmbr.63.4.844-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 283–317. [Google Scholar]
- 6.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 7.Masison D C, Maddelein M-L, Wickner R B. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maddelein M L, Wickner R B. Mol Cell Biol. 1999;19:4516–4524. doi: 10.1128/mcb.19.6.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coschigano P W, Magasanik B. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edskes H K, Gray V T, Wickner R B. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor K L, Cheng N, Williams R W, Steven A C, Wickner R B. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 12.Courchesne W E, Magasanik B. J Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blinder D, Coschigano P W, Magasanik B. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck T, Hall M N. Nature (London) 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 15.Cox K H, Rai R, Distler M, Daugherty J R, Coffman J A, Cooper T G. J Biol Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edskes H K, Hanover J A, Wickner R B. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Bellot E, Guillemet E, Baudin-Baillieu A, Gaumer S, Komar A A, Cullin C. Biochem J. 1999;338:403–407. [PMC free article] [PubMed] [Google Scholar]
- 20.Perrett S, Freeman S J, Butler P J G, Fersht A R. J Mol Biol. 1999;290:331–345. doi: 10.1006/jmbi.1999.2872. [DOI] [PubMed] [Google Scholar]
- 21.Komar A A, Guillemet E, Reiss C, Cullin C. Biol Chem. 1998;379:1295–1300. [PubMed] [Google Scholar]
- 22.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews B W. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. In: in Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. Orlando, FL: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger T C, Berendzen J. Acta Cryst. 1997;D53:571–579. doi: 10.1107/S0907444997005398. [DOI] [PubMed] [Google Scholar]
- 26.Weeks C M, Miller R. J Appl Cryst. 1999;32:120–124. [Google Scholar]
- 27.Furey W, Swaminathan S. In: in Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 277. Orlando, FL: Academic; 1997. pp. 590–620. [DOI] [PubMed] [Google Scholar]
- 28.Jones T A, Cowan S, Zou J-Y, Kjeldgaard M. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, Rice L M, Simonson T, Warren G L. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 31.Carson M. J Appl Cryst. 1991;24:958–961. [Google Scholar]
- 32.Choi J H, Lou W, Vancura A. J Biol Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 33.Koonin E V, Mushegian A R, Tatusov R L, Altschul S F, Bryant S H, Bork P, Valencia A. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossjohn J, Board P G, Parker M W, Wilce M C. Protein Eng. 1996;9:327–332. doi: 10.1093/protein/9.4.327. [DOI] [PubMed] [Google Scholar]
- 35.Prade L, Hof P, Bieseler B. Biol Chem. 1997;378:317–320. doi: 10.1515/bchm.1997.378.3-4.317. [DOI] [PubMed] [Google Scholar]
- 36.Wilce M C, Parker M W. Biochim Biophys Acta. 1994;1205:1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 37.Nishida M, Harada S, Noguchi S, Satow Y, Inoue H, Takahashi K. J Mol Biol. 1998;281:135–147. doi: 10.1006/jmbi.1998.1927. [DOI] [PubMed] [Google Scholar]
- 38.Reinemer P, Prade L, Hof P, Neuefeind T, Huber R, Zettl R, Palme K, Schell J, Koelln I, Bartunik H D, Bieseler B. J Mol Biol. 1996;255:289–309. doi: 10.1006/jmbi.1996.0024. [DOI] [PubMed] [Google Scholar]
- 39.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 40.Gibrat J F, Madej T, Bryant S H. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 41.Dirr H, Reinemer P, Huber R. Eur J Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 42.Salinas A E, Wong M G. Curr Med Chem. 1999;6:279–309. [PubMed] [Google Scholar]
- 43.Board P G, Coggan M, Wilce M C, Parker M W. Biochem J. 1995;311:247–250. doi: 10.1042/bj3110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilce M C, Board P G, Feil S C, Parker M W. EMBO J. 1995;14:2133–2143. doi: 10.1002/j.1460-2075.1995.tb07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossjohn J, McKinstry W J, Oakley A J, Verger D, Flanagan J, Chelvanayagam G, Tan K L, Board P G, Parker M W. Structure. 1998;6:309–322. doi: 10.1016/s0969-2126(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 46.Ploemen J P, Wormhoudt L W, Haenen G R, Oudshoorn M J, Commandeur J N, Vermeulen N P, de Waziers I, Beaune P H, Watabe T, van Bladeren P J. Toxicol Appl Pharmacol. 1997;143:56–69. doi: 10.1006/taap.1996.8004. [DOI] [PubMed] [Google Scholar]
- 47.Adler V, Yin Z, Fuchs S Y, Benezra M, Rosario L, Tew K D, Pincus M R, Sardana M, Henderson C J, Wolf C R, Davis R J, Ronai Z. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomarev S I, Zinovieva R D. Nature (London) 1988;336:86–88. doi: 10.1038/336086a0. [DOI] [PubMed] [Google Scholar]
- 49.Ji X, von Rosenvinge E C, Johnson W W, Tomarev S I, Piatigorsky J, Armstrong R N, Gilliland G L. Biochemistry. 1995;34:5317–5328. doi: 10.1021/bi00016a003. [DOI] [PubMed] [Google Scholar]
- 50.Edskes H K, Wickner R B. Proc Natl Acad Sci USA. 2000;97:6625–6629. doi: 10.1073/pnas.120168697. . (First Published May 23, 2000; 10.1073/pnas@12016897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez-Bellot E, Guillemet E, Cullin C. EMBO J. 2000;19:3215–3222. doi: 10.1093/emboj/19.13.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bousset L, Belrhali H, Janin J, Melki R, Morera S. Structure Fold Des. 2001;9:39–46. doi: 10.1016/s0969-2126(00)00553-0. [DOI] [PubMed] [Google Scholar]