Abstract
In the adult mouse, distinct morphological and transcriptional differences separate stomach from intestinal epithelium. Remarkably, the epithelial boundary between these two organs is literally one cell thick. This discrete junction is established suddenly and precisely at embryonic day (E) 16.5, by sharpening a previously diffuse intermediate zone. In the present study, we define the dynamic transcriptome of stomach, pylorus and intestinal tissues between E14.5 and E16.5. We show that establishment of this boundary is concomitant with the induction of over a thousand genes in intestinal epithelium, and these gene products provide intestinal character. Hence, we call this process intestinalization. We identify specific transcription factors (Hnf4γ, Creb3l3 and Tcfec) and examine signaling pathways (Hedgehog and Wnt) that may play a role in this process. Finally, we define a unique expression domain at the pylorus itself and detect novel pylorus-specific patterns for the transcription factor Gata3 and the secreted protein nephrocan.
Introduction
The vertebrate gastrointestinal (GI) tract consists of a series of connected organs (esophagus, stomach, small intestine, large intestine), each with a highly specialized epithelial surface that enables it to perform a distinct function during digestion. In adults, the epithelial boundaries between some of these adjacent organs are remarkably sharp. At the pylorus, for example, gastric and intestinal cells lie directly next to one another, without a transitional cell type (i.e., the epithelial pyloric border) (Braunstein et al., 2002).
Such discrete organ boundaries have fetal origins. In the embryo, the gut tube is molded from endoderm, along with its associated splanchnic mesoderm (Wells and Melton, 1999). Anteroposterior patterning of the GI tract begins even before tube formation is complete; by embryonic day (E) 10, the developing gut tube has a clear Hox code that marks out the major organ domains and future sphincter locations (Kawazoe et al., 2002). Expression patterns of other gut transcription factors are also established early, including: the HMG-box protein Sox2 in early endoderm of the stomach domain (Sherwood et al., 2009), the caudal-related parahox factor Cdx2 in presumptive intestinal endoderm (Silberg et al., 2000), and the homeodomain protein Nkx2-5 in a thin band of mesenchymal cells at the site of the future pylorus (Smith et al., 2000b). Despite this pattern, the epithelial surface exhibits few obvious morphological differences from stomach to intestine, even as late as E14.5.
While examining the expression pattern of villin, an intestine-specific actin bundling protein, we previously found that, at E14.5, it is expressed in a diminishing gradient (posterior to anterior) at the pyloric border. Two days later however, at E16.5, a sharp anterior expression boundary resolves: villin is detected at high levels in intestinal cells, while neighboring gastric cells exhibit little or no expression (Braunstein et al., 2002). We speculated that the formation of this boundary may reflect an important epithelial compartmentalization event in the GI tract. If so, it is remarkable for its late timing, more than five days after the initial establishment of the broad territorial domains that specify the location of stomach and intestine.
In the present study, we sought to determine whether compartmentalization of villin expression is accompanied by the formation of similar dramatic expression boundaries for other genes. Microdissection and microarray analysis was used to examine gene expression patterns at and around the pylorus at E14.5 and E16.5. Our data reveal that at E14.5, the transcriptomes of stomach, pylorus and intestine are only subtly different. At E16.5, however, hundreds of genes are coordinately upregulated in intestine. Remarkably, this transcriptional burst is seen in the intestinal epithelium but not the mesenchyme, and the batteries of activated genes are involved in the prototypical intestinal functions of absorption and metabolism. Interestingly, a similar large scale burst of gene induction does not occur in the stomach; the transcriptome of this organ changes little between E14.5 and E16.5. We identify several upregulated transcription factors (Hnf4γ, Tcfec and Creb3l3), which, similar to villin, exhibit dramatic pyloric expression boundaries at E16.5. We also investigate signaling pathways (Hedgehog and Wnt) that may be modulated during this compartmentalization event. Finally, we uncover novel genes with expression patterns that are restricted to the pyloric region itself and could participate in this patterning event. These include the zinc finger transcription factor Gata3 and the secreted TGF-β modulator nephrocan.
Results
Diffuse pyloric expression boundaries at E14.5
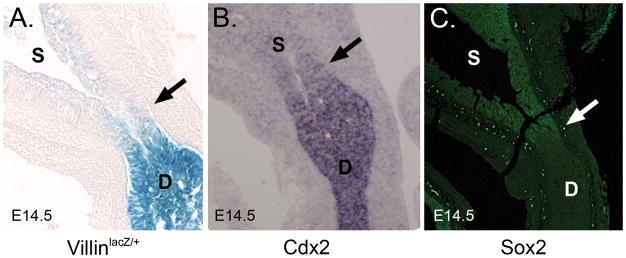
Expression of Cdx2 (an intestinal marker) and Sox2 (a stomach marker) in early foregut endoderm was previously examined using whole mount confocal immunofluorescence (Sherwood et al., 2009). By E9.5, the staining domains of these two proteins at the pylorus appear to be essentially distinct, prompting The Authors of the study to propose that Cdx2 repression of Sox2 establishes this boundary. Indeed, loss of Cdx2 in the intestinal domain leads to increased Sox2 expression and conversion of the epithelium to an esophageal morphology (Gao et al., 2009). If Cdx2 suppression of Sox2 is responsible for formation of the pyloric border and if the expression of these two proteins is compartmentalized across that border by E9.5, as suggested by Sherwood et al., then the compartmentalization of villin expression, which occurs seven days later at E16.5 (Braunstein et al, 2002), likely requires additional cues.
To investigate this more carefully, we examined pyloric Cdx2 and Sox2 expression at E14.5 in sectioned material, and compared these patterns to that of villin. As shown in Figures 1A–C, the boundaries of the expression domains of Cdx2 and Sox2 are, like that of villin, diffuse at the pyloric region. Thus, it appears that while a regional pattern of endoderm identity (Sox2 in stomach and Cdx2 in intestine) is established by E14.5, the precise boundaries of this pattern at the pylorus are not yet mature at the cellular level. The discrepancy between these results and those of Sherwood et al. may be explained by the use of sectioned material and the much higher magnification used here.
Figure 1. The epithelial pyloric boundary is diffuse at E14.5.
A) X-gal staining for β-gal expression in villinlacZ/+ mice (Braunstein et al., 2002). B) In situ hybridization for Cdx2. C) Immunofluorescence (green) staining for Sox2. Staining was performed on sectioned E14.5 material. The presumptive pyloric border is indicated by the arrow. D = duodenum and S = stomach.
Gastric and duodenal transcriptomes are similar at E14.5 but distinct at E16.5
To learn more about the process of epithelial pyloric border formation, we analyzed global gene expression patterns at and around the developing pylorus. Using C57Bl6/J mice, small (1–2mm) tissue fragments were microdissected from the pyloric region itself (easily recognized grossly by the muscular constriction) and from adjacent stomach and duodenum both before (E14.5) and after (E16.5) border formation (Supplemental Figure 1). Triplicate samples of extracted RNA were processed for microarray.
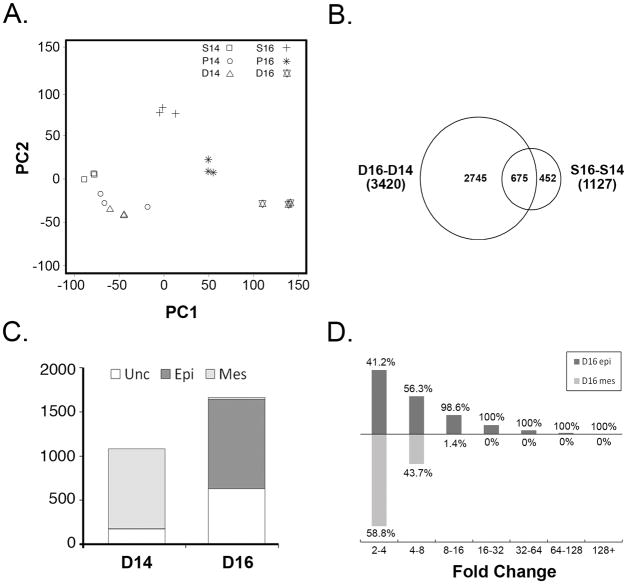
To assess the similarities and differences among the various tissue samples, and to determine the reproducibility of the replicate microarray chips, Principal Component Analysis (PCA) was applied, as described in the legend to Figure 2. The clustering of replicate chip samples seen in Figure 2A demonstrates that the collection process was reproducible. Importantly, the three E14.5 tissue samples (stomach, pylorus and duodenum) are tightly grouped, indicating that the transcriptomes of these tissues are quite similar to one another. In contrast, the three E16.5 groups are clearly different from one another and different from the E14.5 groups. Of the three E16.5 groups, the duodenum shows the most change from E14.5 to E16.5, as measured by the degree of separation between the points along the axis of Principal Component 1 (PC1). Principal Component 1, by definition, contains the majority of the variation in the data (Ringner, 2008). In contrast, the stomach samples exhibit much less change along the PC1 coordinate.
Figure 2. Dramatic upregulation of gene expression in E16.5 duodenal epithelium.
A) Principal components analysis (PCA) of individual microarray chips. The first two principal components (PC1 and PC2), which together represent the majority of the sample variance, are plotted. Note grouping of E14.5 tissues. B) Venn diagram of temporal changes for each tissue (D16-D14 vs. S16-S14). Overlap between these groups indicates probesets that change in both tissues from E14.5 to E16.5. C) Epithelial (Epi) or mesenchymal (Mes) compartmentalization of enriched or depleted probesets in E16.5 duodenum. Probesets with low expression or expression in both compartments are considered unclassified (Unc). D) Histogram of duodenal (D16-D14) fold changes for E16.5 upregulated epithelial probesets (D16 epi) and downregulated mesenchymal probesets (D16 mes). The height of the bar is proportional to the absolute number of probesets. Percentages indicate the relative proportion of D16 epi or D16 mes probesets represented by each bar for each fold change category.
A dramatic change in the duodenal transcriptome at E16.5
To identify gene expression changes underlying the emergence of distinct gastric and duodenal transcriptomes at E16.5, we assembled a list of all significant pairwise probeset enrichments, including comparisons of: a) each tissue at E14.5 to the same tissue at E16.5 (time axis; three comparisons); and, b) stomach, pylorus and duodenum to one another at either E14.5 or E16.5 (tissue axis; six comparisons). A total of 10,499 unique differentially expressed probesets were identified for all comparisons and these are presented in Supplemental Table 1. Of these, 9,137 showed significant changes in the time comparisons, 5,909 in the tissue comparisons, and 4,547 in both. We selected the group of probesets that are dynamic in both time and tissue dimensions for further analysis.
Although some genes were unique to the pyloric region (addressed below), the results of the PCA analysis (Figure 2A) shows that the pyloric area generally (with some exceptions) exhibits probeset intensity values that are essentially the average of stomach and intestine values. Thus, for initial analysis, we compared temporally dynamic probesets in stomach and intestine to catch patterning differences that were emerging between these two tissues at E16.5. Of the 4,547 time and tissue dynamic probesets, 86% (3,917) showed enrichment in either stomach or duodenum at E16.5. Plotting these results in a Venn diagram (Figure 2B) reveals that from E14.5 to E16.5, probesets exhibiting transcriptional change in the duodenum far outnumber the probesets changing in stomach. In the stomach, 1,127 probesets show temporal change, but only 40% of these (452) are specific to the stomach. (675 probesets were changed in both stomach and duodenum.) In contrast, the robust temporal transcriptional change in the duodenum encompasses differences in 3,420 probesets, 80% of which (2,745) change only in the duodenum. These temporally dynamic duodenal probesets could be further annotated as enriched at E16.5 (i.e., depleted at E14.5) or depleted at E16.5 (i.e., enriched at E14.5). Tallying these groups, labeled D16 enriched or D16 depleted, respectively (Supplemental Table 2), reveals that D16 enriched probesets accounts for 61% (1,664 probesets) of the duodenum-specific temporal change.
Upregulated genes in E16.5 duodenum are primarily epithelial, while downregulated genes are mesenchymal
We were next interested to determine whether the dramatic transcriptional change seen in duodenum at E16.5 was primarily due to changes in genes that are expressed in epithelium or mesenchyme. In an earlier study, we separated intestinal tissue using non-enzymatic methods and profiled gene expression in freshly isolated epithelium and mesenchyme to create a catalog of epithelial and mesenchymal genes (Li et al., 2007). Though the earlier study was done using E18.5 intestine, we reasoned that the epithelial/mesenchymal compartmentalization of genes would be largely similar at E16.5 and E18.5. Thus, using the earlier data, we tagged all D16 enriched and D16 depleted probesets as epithelial or mesenchymal. Some probesets could not be classified this way due to low expression or expression in both compartments. For classified D16 enriched probesets, the vast majority (98.3%) were epithelial. These striking results predict that the formation of the distinct epithelial boundary between stomach and intestine is concomitant with a massive transcriptional inductive event in duodenal epithelium. Interestingly, and in contrast, 99.6% of compartment classified probesets that were depleted at E16.5 were mesenchymal (Figure 2C).
Dramatic tissue rearrangement accompanies the emergence of intestinal villi between E14.5 and E16.5. Thus, we were concerned that the transcriptional changes observed might merely reflect an alteration in the ratio of epithelium to mesenchyme. We therefore examined the distribution of fold changes seen among D16 epithelial upregulated probesets or among D16 depleted mesenchymal probesets (Figure 2D). This analysis revealed that 42% of the probesets in the D16 epithelial group are upregulated at levels greater than five-fold; in fact, 20% were upregulated over ten-fold. (Maximum upregulation of over 100-fold was seen in the case of four probesets.) However, only 8% of the D16 mesenchymal probesets are downregulated by five-fold or more and none are downregulated over ten-fold. In fact, nearly 75% of all D16 mesenchymal probesets show less than a four-fold change. Thus, we conclude that tissue rearrangement may account for some of the low level transcriptional change seen among the downregulated (and primarily mesenchymal) probesets. However, the dramatic inductive change observed in the epithelial compartment cannot be accounted for by this mechanism.
Duodenal gene expression changes at E16.5 correlate with functional differentiation of the small intestine
We used functional annotation clustering methods (DAVID; david.abcc.ncifcrf.gov) to classify D16 probesets (Dennis et al., 2003). Since there were only four probesets downregulated in epithelium (Foxa1, Gcg, Mreg and Serpina1c) we could not use DAVID analysis for this group. However, for the eighteen probesets upregulated in mesenchyme, the most enriched annotation terms were apoptosis, immune response and GTP binding. Conversely, among the 73 mesenchymal probesets that are downregulated more than five-fold, we found that cell differentiation, neuronal development and cell migration are the most frequent terms represented (data not shown). Importantly, for the group of 431 epithelial upregulated probesets, genes involved in metabolic processes and cell transport are statistically overrepresented (Supplemental Table 3). This finding suggests that, between E14.5 and E16.5, there is a compartmentalized, functional switch in duodenal gene expression. The mesenchyme downregulates genes involved in development and morphogenesis, while the large burst of gene expression in the epithelium appears to prepare the intestine for its major role in absorption and metabolism. Thus, we can think of this significant epithelial change as a process of intestinalization.
Upregulated duodenal transcription factors display a sharp epithelial boundary at the pylorus
It is of interest to identify which transcription factors show significant change during intestinalization of the epithelium. Table 1 lists the most enriched transcription factors in the D16 epithelial group. Among these genes, Tcfec, Creb3l3, Nr2e3, Mafb and Hnf4γ exhibit expression levels that are over ten-fold enriched in duodenum relative to stomach. Hnf4γ is the lesser studied paralog of Hnf4α, a protein critical for epithelial function in pancreas, liver and colon (Li et al., 2000; Garrison et al., 2006; Miura et al., 2006). Hnf4α is also highly expressed in intestinal epithelium, but is only two- to three-fold enriched in intestine relative to stomach at both E14.5 and E16.5 (Supplementary Table 1). The bZip factor Mafb is mutated in kreisler mice, which exhibit morphogenic defects in hindbrain and inner ear (Cordes and Barsh, 1994; McKay et al., 1994). Interestingly, a recent genome-wide screen links Mafb to polygenic dyslipidemia (Kathiresan et al., 2009). Nr2e3 (also known as PNR or photoreceptor-specific nuclear receptor) was initially implicated in eye development (Chen et al., 2005), but a recent study demonstrated that it is highly induced in intestinal epithelium during midgestation (Choi et al., 2006). Neither Tcfec nor Creb3l3 has previously been associated with intestinal gene regulation.
Table 1.
Summary of transcription factor gene expression changes in E16.5 duodenal epithelium
| Probeset ID | Symbol | Description | D16-S16 | D16-D14 | Epi-Mes |
|---|---|---|---|---|---|
| 1419537_at | Tcfec | transcription factor EC | D16 (41.65) | D16 (5.45) | Epi (18.27) |
| 1424688_at | Creb3l3 | cAMP responsive element binding protein 3-like 3 | D16 (18.95) | D16 (25.92) | Epi (11.26) |
| 1423631_at | Nr2e3 | nuclear receptor subfamily 2, group E, member 3 | D16 (18.68) | D16 (23.80) | Epi (4.60) |
| 1451716_at | Mafb | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) | D16 (14.49) | D16 (14.49) | Epi (2.93) |
| 1460127_at | Hnf4g | hepatocyte nuclear factor 4, gamma | D16 (13.19) | D16 (3.66) | Epi (49.82) |
| 1449051_at | Ppara | peroxisome proliferator activated receptor alpha | D16 (9.09) | D16 (5.04) | Epi (6.58) |
| 1425392_a_at | Nr1i3 | nuclear receptor subfamily 1, group I, member 3 | D16 (8.46) | D16 (8.22) | Epi (6.08) |
| 1419185_a_at | Mlxipl | MLX interacting protein-like | D16 (7.87) | D16 (2.93) | Epi (2.40) |
| 1437473_at | Maf | avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog | D16 (6.89) | D16 (5.38) | Epi (2.41) |
| 1417244_a_at | Irf7 | interferon regulatory factor 7 | D16 (6.77) | D16 (8.09) | Epi (2.18) |
| 1425528_at | Prrx1 | paired related homeobox 1 | D16 (5.35) | D16 (9.57) | Epi (2.73) |
| 1434416_a_at | Solh | Small optic lobes homolog (Drosophila) | D16 (4.34) | D16 (2.64) | Epi (2.84) |
| 1417519_at | Plagl2 | pleiomorphic adenoma gene-like 2 | D16 (3.89) | D16 (2.38) | Epi (7.06) |
| 1440831_at | Bach1 | BTB and CNC homology 1 | D16 (3.84) | D16 (2.32) | Epi (2.67) |
| 1449854_at | Nr0b2 | nuclear receptor subfamily 0, group B, member 2 | D16 (3.31) | D16 (5.04) | Epi (2.55) |
| 1420808_at | Ncoa4 | nuclear receptor coactivator 4 /// predicted gene, EG627557 | D16 (2.98) | D16 (2.53) | Epi (2.72) |
| 1435991_at | Nr3c2 | nuclear receptor subfamily 3, group C, member 2 | D16 (2.55) | D16 (2.57) | Epi (2.36) |
| 1443100_at | Thrb | Thyroid hormone receptor beta | D16 (2.27) | D16 (2.20) | Epi (3.34) |
| 1440870_at | Prdm16 | PR domain containing 16 | D16 (2.20) | D16 (3.13) | Epi (5.24) |
| 1426690_a_at | Srebf1 | sterol regulatory element binding factor 1 | D16 (2.10) | D16 (2.54) | Epi (2.63) |
| 1418437_a_at | Mlx | MAX-like protein X | D16 (2.09) | D16 (2.33) | Epi (2.05) |
| 1419052_at | Ovol1 | OVO homolog-like 1 (Drosophila) | D16 (2.04) | D16 (2.14) | Epi (2.14) |
Note: For each comparison, the label (e.g., D14, S14, D16, S16, Epi, Mes) refers to the time and/or tissue of maximum expression and the number in parentheses is the fold change.
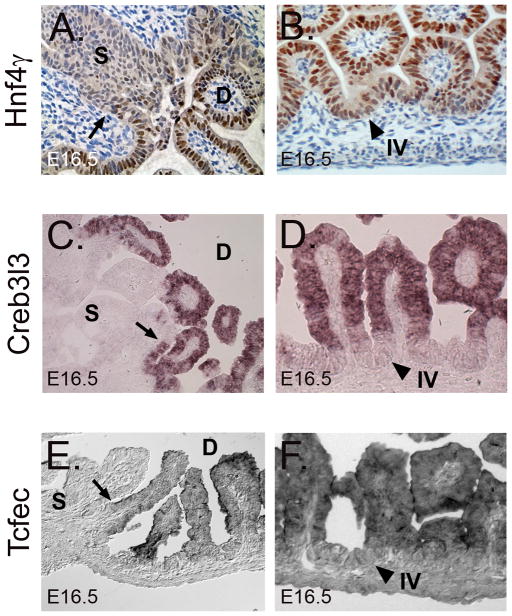
We looked carefully at the expression of three of these highly enriched D16 transcription factors to determine whether, similar to villin, they display a sharp, cell-specific anterior boundary at E16.5. Immunohistochemical staining for Hnf4γ shows both nuclear and cytoplasmic staining in the epithelium. At E16.5, cytoplasmic staining extends into the stomach, but strong nuclear staining is sharply demarcated at the pylorus (Figure 3A). In situ hybridization reveals a sharp anterior expression boundary for Creb3l3 (Figure 3C) and Tcfec (Figure 3E) at E16.5. Additionally, for both Creb3l3 (Figure 3D) and Tcfec (Figure 3F), expression is primarily localized to the differentiated cells of the villus tips. Hnf4γ nuclear staining also appears to display a diminishing villus-crypt expression gradient (Figure 3B). Thus, activation of these three transcription factors (and likely others in Table 1) in E16.5 duodenal epithelium appears to be concomitant with the maturation of the crypt/villus axis.
Figure 3. The duodenum epithelial transcription factors Hnf4γ, Creb3l3 and Tcfec have sharp anterior expression boundaries at E16.5.
A) Immunohistochemical staining for Hnf4γ reveals strong nuclear staining in the intestine, but only weak nuclear staining in the stomach. B) A gradient of nuclear Hnf4γ staining from intervillus (weak) to villus tip (strong) epithelium is present. C) In situ hybridization for Creb3l3 shows a dramatic boundary at the stomach-intestine border at E16.5. Creb3l3 is not expressed at E14.5 (data not shown). D) Creb3l3 is restricted to villus epithelium at E16.5. E) In situ hybridization for Tcfec reveals a discrete boundary of expression at E16.5. F) Tcfec is expressed predominately in differentiated duodenal villus epithelium and not intervillus epithelium. Arrows denote stomach-intestine expression boundary. Arrowheads indicate intervillus (IV) epithelium. D = duodenum and S = stomach.
Modulation of developmental signaling pathways accompanies formation of the epithelial pyloric border
Soluble signaling factors play major roles in gut patterning, and previous data suggest that, in particular, Hedgehog (Zhang et al., 2001) and Wnt (Okubo and Hogan, 2004; Kim et al., 2005; Kim et al., 2007) signaling are critical in the context of intestinal differentiation. Thus, we examined the array results for expression of key elements of these pathways. We also directly tested, using transcriptional reporter readouts, whether there is differential activation of either of these two pathways across the pylorus at E14.5 or E16.5. The results are summarized below and presented in Table 2 (Hedgehog) and Supplemental Table 4 (Wnt).
Table 2.
Summary of gene expression changes in Hedgehog signaling pathway components
| Probeset ID | Symbol | Description | D14-S14 | D16-S16 | S16-S14 | D16-D14 |
|---|---|---|---|---|---|---|
| 1426869_at | Boc | biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein | NC (1.05) | S16 (4.09) | NC (1.25) | D14 (4.86) |
| 1434957_at | Cdon | cell adhesion molecule-related/down-regulated by oncogenes | NC (1.15) | S16 (2.18) | NC (1.40) | D14 (2.64) |
| 1448494_at | Gas1 | growth arrest specific 1 | NC (1.11) | S16 (2.36) | NC (1.67) | D14 (4.36) |
| 1449058_at | Gli1 | GLI-Kruppel family member GLI1 | NC (1.04) | S16 (3.06) | NC (1.14) | D14 (3.65) |
| 1459211_at | Gli2 | GLI-Kruppel family member GLI2 | NC (1.04) | S16 (2.12) | NC (1.99) | D14 (4.40) |
| 1455154_at | Gli3 | GLI-Kruppel family member GLI3 | NC (1.26) | S16 (3.07) | S14 (2.70) | D14 (6.56) |
| 1450704_at | Ihh | Indian hedgehog | NC (1.80) | S16 (2.19) | NC (1.60) | NC (1.32) |
| 1427133_s_at | Lrp2 | low density lipoprotein receptor-related protein 2 | NC (1.45) | D16 (7.29) | NC (1.24) | D16 (4.04) |
| 1439663_at | Ptch1 | Patched homolog 1 | NC (1.31) | NC (1.63) | NC (1.89) | D14 (2.35) |
| 1454876_at | Rab23 | RAB23, member RAS oncogene family | NC (1.02) | S16 (2.28) | NC (1.65) | D14 (3.69) |
| 1436869_at | Shh | sonic hedgehog | NC (1.75) | S16 (6.66) | NC (1.30) | D14 (8.94) |
| 1427049_s_at | Smo | smoothened homolog (Drosophila) | NC (1.03) | S16 (2.21) | NC (1.61) | D14 (3.65) |
| 1434733_at | Stk36 | serine/threonine kinase 36 (fused homolog, Drosophila) | NC (1.12) | NC (1.35) | S14 (2.15) | D14 (2.58) |
Note: NC means no significant change (significant change is FC≥2 and p<0.05). For each comparison, the label (e.g., D14, D16, S14, S16) refers to the time and/or tissue of maximum expression and the number in parentheses is the fold change.
Hedgehog pathway
At E14.5, the Hedgehog ligands Shh and Ihh were expressed similarly in stomach and duodenum. However, by E16.5, Shh, which is expressed in the epithelium and signals in a paracrine manner to the mesenchyme (Madison et al., 2005; Kolterud et al., 2009), was specifically downregulated in the duodenum (6.7 fold relative to stomach). In accordance with the drop in Shh, all three Gli factors (Gli1, Gli2 and Gli3) were reduced in the duodenum, as were the co-receptors Ptch1 and Smo and the mouse Fused homolog Stk36. A number of recently identified pathway modulators (Gas1, Boc, Cdon and Rab23) were also reduced in duodenum (Eggenschwiler et al., 2001; Tenzen et al., 2006; Allen et al., 2007). Only megalin (Lrp2), an endocytic receptor for Hedgehog ligands (McCarthy et al., 2002), is upregulated in epithelium of E16.5 duodenum. However, megalin is not dedicated to the transport of Hedgehog ligands and is known to transport cholesterol and vitamins among the over 50 molecules that it can bind (Kozyraki and Gofflot, 2007).
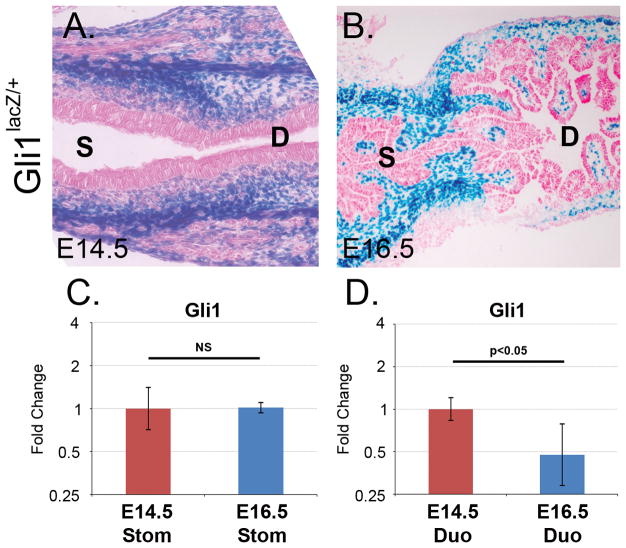
We examined Hedgehog signaling across the pylorus at E14.5 and E16.5 using Gli1lacZ/+ reporter mice as a direct readout of pathway activity (Park et al., 2000). Figure 4A shows that, at E14.5, Gli1lacZ/+ staining is similar in stomach and intestine. However, at E16.5, Gli1 activity is apparently reduced on the duodenal side of the pylorus, but maintained in the stomach (Figure 4B), in agreement with the array results and with another recent study (Kolterud et al., 2009). To validate this observation in a more quantitative manner, we assayed Gli1 mRNA expression using real-time quantitative RT-PCR (qPCR). When normalized to the housekeeping gene Hprt and compared to levels in E14.5 intestine, Gli1 expression was significantly decreased in E16.5 intestine (95% confidence interval: 0.8 to 0.3 fold change; Student’s t-test: p<0.05; Figure 4D). In contrast, Gli1 expression in stomach did not vary significantly between E14.5 and E16.5 (Figure 4C).
Figure 4. Downregulation of Hedgehog signaling in duodenum at E16.5.
A) X-gal staining for β-gal expression in Gli1lacZ/+ mice at E14.5 reveals similar levels of Hedgehog signaling activity across the pyloric border. B) At E16.5, X-gal staining shows an apparent difference in stomach and intestinal Hedgehog signaling. C–D) Histogram of fold change values for normalized Gli1 qPCR data. Compared to the same tissue at E14.5, Gli1 expression was significantly decreased in E16.5 intestine but not in stomach. NS = not statistically significant.
Wnt pathway
The expression of several Wnt ligands is modulated during pyloric border formation. At E16.5, Wnt4, Wnt5a and Wnt11 show significant enrichment in stomach relative to duodenum. The Wnt receptors Fzd1, Fzd2, Fzd6 also show significant enrichment in stomach at this time, as do a number of Wnt pathway modulators (Sfrp1, Sfrp2, Sfrp4, Dkk2 and Dkk3). However, the most robust tissue-specific expression pattern of all Wnt pathway components is that of the secreted modulator Sfrp5, which is highly duodenum-specific at both E14.5 (32 fold relative to stomach) and E16.5 (10.7 fold relative to stomach). Taken together, these findings predict that Wnt signaling activity, like that of Hedgehog, is decreased in intestine at E16.5. To investigate this further, we explored some of these expression patterns by in situ hybridization.
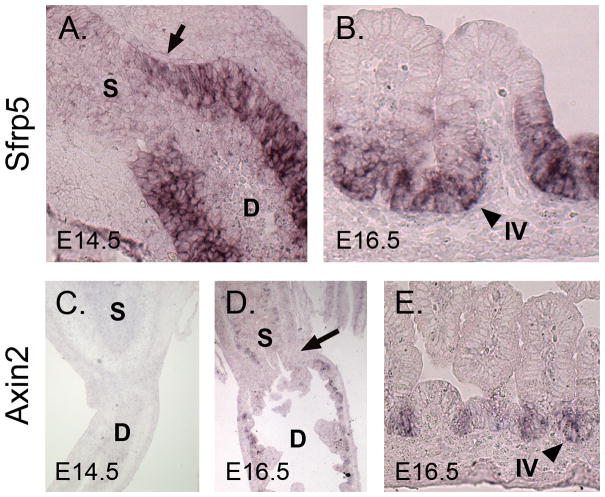
We first examined Sfrp5 expression. By E14.5, Sfrp5 is robustly expressed in duodenal epithelium, with a soft anterior boundary of expression that extends a short distance into the stomach (Figure 5A). By E16.5, when villus formation has begun, this expression domain resolves dramatically; Sfrp5 expression is excluded from the villus tips and present only in cells of the proliferative intervillus region (Figure 5B). The localized expression of this Wnt modulator (thought to be a Wnt inhibitor) in intervillus cells is consistent with the recent study of Kim et al., which concludes that, between E16.5 and birth, canonical Wnt signaling is excluded from intervillus regions and restricted to villus epithelium (Kim et al., 2007).
Figure 5. Canonical Wnt signaling is active across the pylorus at E16.5 and restricted to intervillus epithelium in the duodenum.
A) In situ hybridization for Sfrp5 at E14.5. Note diffuse boundary at pyloric border (arrow). B) Sfrp5 expression in the E16.5 intestine reveals that expression is strong in intervillus but not villus tip epithelium. C) In situ hybridization for Axin2 at E14.5; little or no signal is seen. D) Axin2 expression at 16.5 shows faint staining in both the stomach and duodenal epithelium, with little difference between these tissues. The presumptive pyloric border is indicated by the arrow. E) Higher magnification of Axin2 intestinal staining at E16.5 reveals that staining is confined to the intervillus epithelium. Arrowheads indicate intervillus (IV) epithelium. D = duodenum and S = stomach.
To clearly define the domain of active canonical Wnt signaling and establish whether a gradient of Wnt signaling indeed exists across the pylorus at either E14.5 or E16.5, we examined the expression of Axin2, a Wnt target gene and a commonly accepted read-out of canonical signaling activity (Yan et al., 2001). Previous studies using an Axin2 β-galactosidase (β-gal) reporter mouse suggested that Axin2 is expressed mainly on villus tip cells and excluded from intervillus cells (Kim et al., 2007). Because the detection of β-gal reporter activity in intestinal epithelium can be complicated by the presence of an endogenous β-gal activity in this compartment (indeed, the endogenous β-galactosidase gene, Glb1, is more than six-fold upregulated, specifically in the duodenum at E16.5), we used in situ hybridization with an antisense Axin2 riboprobe to directly examine this question. We found that at E14.5, Axin2 expression is very low in both stomach and duodenum, indicating that the level of canonical Wnt pathway activity is low at this time (Figure 5C). At E16.5, Axin2 expression and therefore, canonical Wnt pathway activity increases considerably, but no visible gradient of canonical signaling activity can be detected across the pyloric region (Figure 5D). Interestingly, in the duodenum, Axin2 staining is exclusively seen in the intervillus region of E16.5 intestine (Figure 5E); no signaling activity is detectable in villus tip epithelium.
A specific domain of gene expression at the pylorus
Using the group of tissue enriched probesets described earlier, we selected probesets that showed a pyloric expression pattern that was significantly different from both the stomach and duodenum at either E14.5 or E16.5. Of these probesets with pylorus-specific expression patterns, fifteen were detected at E14.5 and 79 were detected at E16.5. Known transcription factors and signaling pathway components among these pylorus-specific probesets are shown in Table 3 and the remaining pyloric probesets are listed in Supplemental Table 5.
Table 3.
Summary of pyloric transcription factors and signaling molecules
| Probeset ID | Symbol | Description | P14-S14 | P14-D14 | P16-S16 | P16-D16 |
|---|---|---|---|---|---|---|
| A. Transcription factors | ||||||
| 1447500_at | Cutl2 | cut-like 2 (Drosophila) | NC (1.67) | NC (−1.23) | P16 (2.09) | P16 (2.44) |
| 1448886_at | Gata3 | GATA binding protein 3 | P14 (6.75) | P14 (10.35) | P16 (2.55) | P16 (3.39) |
| 1449566_at | Nkx2-5 | NK2 transcription factor related, locus 5 (Drosophila) | P14 (5.64) | P14 (6.58) | P16 (2.49) | P16 (2.76) |
| 1431899_at | Nkx6-3 | NK6 transcription factor related, locus 3 (Drosophila) | NC (1.51) | P14 (4.56) | P16 (2.00) | P16 (2.08) |
| 1451569_at | Nr2c2 | nuclear receptor subfamily 2, group C, member 2 | NC (1.48) | NC (1.79) | P16 (2.02) | P16 (2.25) |
| B. Signaling molecules | ||||||
| 1451991_at | Epha7 | Eph receptor A7 | NC (1.16) | NC (1.01) | P16 (2.14) | P16 (2.10) |
| 1424007_at | Gdf10 | growth differentiation factor 10 | NC (1.54) | NC (1.23) | P16 (2.74) | P16 (8.62) |
| 1425357_a_at | Grem1 | gremlin 1 | P14 (7.13) | P14 (2.26) | P16 (4.90) | P16 (2.69) |
| 1419065_at | Nepn | Nephrocan | NC (1.10) | P14 (6.31) | P16 (17.06) | P16 (16.73) |
| 1426561_a_at | Npnt | Nephronectin | NC (1.33) | NC (1.13) | S16 (2.85) | D16 (2.05) |
| 1422553_at | Pten | phosphatase and tensin homolog | NC (1.98) | NC (1.94) | P16 (3.36) | P16 (2.02) |
| 1442067_at | Ror1 | Receptor tyrosine kinase-like orphan receptor 1 | NC (1.20) | NC (1.27) | P16 (2.29) | P16 (3.33) |
| 1436892_at | Spred2 | sprouty-related, EVH1 domain containing 2 | NC (1.54) | NC (1.79) | P16 (2.87) | P16 (3.22) |
Note: NC means no significant change (significant change is FC≥2.0 and p<0.05). For each comparison, the label (e.g., P14, P16, D14, D16, S14, S16) refers to the time and/or tissue of maximum expression and the number in parentheses is the fold change.
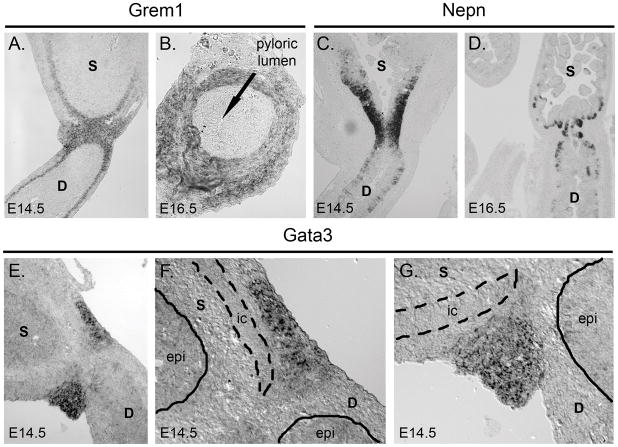
Pylorus-specific genes detectable at both E14.5 and E16.5 include the transcription factor Nkx2-5 and the secreted BMP antagonist gremlin (Grem1), both of which were previously shown to exhibit pyloric expression in chick (Smith et al., 2000a; Moniot et al., 2004). However, the most pylorus-enriched gene detected at E14.5 was Gata3, a transcription factor not previously associated with pylorus patterning or function. Furthermore, at E16.5, the most enriched gene at the pylorus was nephrocan (Nepn), a secreted regulator of TGF-β pathway activity (Mochida et al., 2006). We confirmed these novel findings by in situ hybridization.
Nephrocan and Gremlin
Gremlin and nephrocan are both secreted modulators of TGF-β superfamily signaling and thus it was of interest to compare their expression patterns. At E14.5 and E16.5, gremlin is mesenchymal and expressed in a broad band at the pylorus (Figures 6A–B). Interestingly, nephrocan is expressed in E14.5 pyloric epithelium (Figure 6C); its expression is slightly asymmetric with respect to the pylorus, with a broader expression domain on the stomach side. At E16.5, nephrocan becomes restricted to cells to the intervillus base of the intestine and cells at the base of developing antral glands. Interestingly, despite its epithelial localization, there is no clear boundary of expression across the pylorus for nephrocan at E16.5 (Figure 6D).
Figure 6. Pylorus-specific expression of Gata3, gremlin and nephrocan.
A) In situ hybridization for gremlin (Grem1) at E14.5. Mesenchymal expression is strong at the pylorus and continues in the inner circular muscle of the intestine and more weakly in the inner circular muscle of the stomach. B) Gremlin expression is also mesenchymal at E16.5, as seen in a cross-section through the pylorus. Arrow shows the pyloric lumen. C) In situ hybridization for nephrocan (Nepn) at E14.5 reveals epithelial expression at the pylorus. Expression is more robust towards the stomach, and is primarily localized to epithelial cells closer to the basement membrane. D) Nephrocan expression at E16.5 is restricted to the base of the developing epithelial glands in stomach and developing villi in intestine. E) In situ hybridization for Gata3 at E14.5 reveals expression in a tight band of cells at the pylorus. F–G) Higher magnification of Gata3 pyloric staining reveals expression in mesenchyme but not epithelium. Epithelium (epi) is demarcated by the solid line, while the inner circular (ic) smooth muscle is outlined by the dashed line. D = duodenum and S = stomach.
Gata3
Gata3 expression is detectable in a narrow band at the pylorus at E14.5 (Figure 6E). Staining is restricted to the mesenchyme and appears confined to cells outside of the thick inner circular smooth muscle of the distal stomach (Figure 5F–G). Previous studies of Nkx2-5 showed a similar well-demarcated mesenchymal pattern at the pylorus (Smith and Tabin, 1999). It will be interesting to further compare the extent of possible overlap in both expression and function between these two transcription factors.
Discussion
In this study, we have investigated dynamic gene expression patterns at and around the developing pylorus. Although clear regional patterning of the stomach and intestine occurs prior to E14.5 (e.g., for Sox2, Cdx2, among other genes), this pattern does not play itself out in terms of the global transcriptomes of E14.5 stomach, duodenum, and pylorus tissues, all of which are surprisingly similar. In contrast, at E16.5, a dramatic burst of transcriptional induction occurs in duodenal epithelium and this event generates a distinct compartmentalization of gene expression on the duodenal side of the border. This genetic induction event coordinately activates hundreds of genes involved in absorption and metabolism. As a result, for the first time, epithelial cells of the intestine express genes that unambiguously distinguish their function during digestion from that of the stomach. We call this compartmentalized patterning step intestinalization and note that it occurs strikingly late in fetal development.
Though the vast majority of genes activated in the intestine are epithelial, a few mesenchymal genes are also upregulated at E16.5, several of which are involved in immune response function. This is interesting in light of recent parallel evidence from our laboratory showing that, in adult intestine, decreased Hedgehog signal transduction increases inflammatory signaling (Lees et al., 2008). Whether there is a direct connection between the downregulation of the Hedgehog pathway that we observe in duodenal mesenchyme at E16.5 (see Figures 4B,D) and activation of these mesenchymal inflammatory genes requires further investigation.
An unexpected finding from our array results is that only four genes (Foxa1, Gcg, Mreg and Serpina1c) are specifically downregulated in intestinal epithelium at E16.5 (Supplemental Table 1). Interestingly, Foxa1 has been shown to be required for Shh expression in the developing lung, another foregut endoderm-derived organ (Wan et al., 2005). Because of its concomitant downregulation with Shh in intestinal epithelium, it is tempting to postulate that the attenuation of Foxa1 expression is responsible for reduced Shh expression during intestinalization. If so, it will be important to understand the transcriptional regulation of Foxa1 expression and determine whether downregulation of this gene is required for intestinalization.
Given the ample published evidence of a role for Wnt signaling during foregut specification (Okubo and Hogan, 2004; Kim et al., 2005; Kim et al., 2007), we were surprised to find that the Axin2 expression pattern suggests little or no difference in canonical Wnt signaling across the pylorus, either before or after pyloric border formation. In fact, at E14.5, very little canonical Wnt activity is detectable at all in either stomach or intestine, though we observed clear Axin2 activity in the forestomach at this time (data not shown). The lack of canonical Wnt signal in E14.5 duodenum is concordant with the phenotype of Tcf7l2 (Tcf4)-null mice, which exhibit no apparent defect in E14.5 intestine (Korinek et al., 1998). At E16.5, we see that intestinal Axin2 expression is confined to intervillus regions (see Figure 5E). This is in accordance with the finding that these proliferative cells are dependent upon canonical Wnt signals, as shown by the loss of this proliferative population in the face of either Tcf4 deficiency (Korinek et al., 1998) or Dkk1 overexpression (Pinto et al., 2003; Kuhnert et al., 2004). Together, these findings and the results of our analysis do not support the idea (Kim et al., 2007) that a canonical Wnt signaling compartment exists on villus tips at E16.5.
In this regard, the expression pattern of Sfrp5 at E16.5 is interesting. Recent work indicates that Sfrp5 can modulate either canonical or non-canonical Wnt signals in the Xenopus foregut (Li et al., 2008). In that system, Sfrp5 was able to bind the ortholog of the non-canonical protein Wnt5a. Intriguingly, both overexpression of Sfrp5 (in Xenopus) and deficiency of Wnt5a (in mouse) result in shortened hindgut (Li et al., 2008; Cervantes et al., 2009). Data presented above show that, at E16.5, Sfrp5 expression is restricted to the intervillus region (see Figure 5B), the same area that is positive for Axin2 staining (and, hence, canonical pathway activity). Thus, it will be of interest to test whether Sfrp5 functions in the intervillus zone to modulate canonical signals, non-canonical signals, or both.
Several epithelial transcription factors are dramatically upregulated during intestinalization and may participate directly in large-scale induction of absorptive and metabolic activity in the intestine. The Hnf4γ paralog Hnf4α has been previously implicated in a similarly late developmental maturation event in the liver. In that system, Hnf4α upregulates a large number of structural genes and is thought to be important for the re-epithelialization of hepatic cells following their migration out of the gut tube proper and into the septum transversum (Parviz et al., 2003). It is tempting to speculate that intestinalization is a similar event. Even though intestinal epithelial cells never leave the confines of the epithelial sheet, as developing hepatoblasts do, the epithelium itself is drastically reorganized during the process of villus formation. Perhaps Hnf4α and Hnf4γ are critical in the final stabilization of this remodeled state. Certainly, binding sites for these factors are highly enriched in the promoters of intestine-specific and epithelial-specific genes (Li et al., 2007).
Two previously unstudied factors were among the most upregulated D16 epithelial transcription factors. Creb3l3 (also known as Creb-H), a member of the bZip family of transcription factors, is involved in the endoplasmic reticulum (ER) stress response (Zhang et al., 2006), and interestingly, its expression in the developing liver is dependent upon Hnf4α (Luebke-Wheeler et al., 2008). Since intestinalization involves transcriptional activation of hundreds of genes, several of which are expressed at tremendously high levels, the ER of intestinal epithelial cells may abruptly require a much higher degree of organization and efficiency to deal with the translational onslaught that follows. Indeed, we show that Creb3l3 is expressed in epithelial cells of the villi, exactly the population in which differentiated gene expression is induced. The idea that ER stress might accompany cell differentiation and might activate mediators of the response pathway in order to coordinate protein biosynthesis remains functionally untested here, but has been well documented for a number of secretory cell types (Wu and Kaufman, 2006).
Another transcription factor that is highly induced in E16.5 intestinal epithelium is Tcfec. This bHLH-Zip factor is a member of the MiT family (with Mitf, Tcfeb and Tcfe3), several of which are expressed in a highly tissue- and cell-specific manner. Often these proteins are responsible for the expression of signature proteins that are critical to organ or tissue development and function. For example, MiT family members regulate tartrate-resistant alkaline phosphatase in osteoclasts (Partington et al., 2004), melanin in pigment cells (Tachibana, 2000) and proteases in mast cells (Nechushtan and Razin, 2002). The MiT proteins form both homo- and hetero-dimers, a fact that may explain why mouse knockout models of several family members show no phenotypes, despite the apparent transcriptional importance of these genes (Steingrimsson et al., 2002). According to our microarray data, the related family member Tcfeb is also expressed in the intestine and is upregulated during intestinalization, though not as dramatically as Tcfec.
Intestinalization occurs concomitantly with formation of a sharp boundary of epithelial gene expression at the pylorus. For genes like villin, Cdx2 and Sox2, a broad domain of expression with a diffuse boundary is detectable early and reflects the regional divisions of organ territory in the developing gut tube. But at E16.5, the boundary of expression sharpens exquisitely to allow differentiated intestinal cells to lie directly next to future stomach cells. An interesting question for further analysis is whether boundary formation and intestinalization actually constitute separate events. The process of intestinalization might reflect maturation of the vertical axis of the villus; differentiating cells exiting the proliferative compartment of the last villus next to the stomach may travel only a set distance from that crypt, coming to rest immediately next to a less differentiated neighbor derived from the stomach progenitor compartment. Alternatively, the pyloric border region itself could propagate a signal that promotes intestinalization, similar to the function of a classical organizer (Meinhardt, 2008).
Our data confirm and extend earlier studies (Smith et al., 2000b) that reveal a characteristic domain of gene expression at the pylorus. We show that this domain is present both prior to and after intestinalization. We report here the novel finding that, similar to Nkx2-5, Gata3 is expressed in a narrow band at the pylorus (see Figure 5E). Given that Nkx and Gata family members are known to interact in other developmental systems, these factors may collaboratively regulate pyloric patterning and organogenesis (Charron and Nemer, 1999; Peterkin et al., 2003; Zhang et al., 2007). In addition, we describe the pyloric expression patterns of gremlin (mesenchyme; see Figures 6A–B) and nephrocan (epithelium; see Figures 6C–D), two secreted modulators of TGF-β superfamily signaling. To our knowledge, nephrocan is the first secreted signaling protein to be identified in pyloric epithelium. In this regard, pyloric border patterning might be similar to the boundary patterning events observed at the midbrain-hindbrain border in the developing brainstem and the atrioventricular boundary of the heart. These events involve formation of straight, sharp expression boundaries (Joyner et al., 2000; Canning et al., 2007; Chi et al., 2008), and in both cases, the border region itself has signaling activity that influences neighboring tissues (Bai et al., 2002; Chi et al., 2008).
The intestinalization event that we document occurs without a similar maturation process in the stomach, where only limited transcriptional change occurs. The fact that the intestine, a more posterior tissue, matures prior to the stomach is somewhat surprising, given the tendency of embryonic development to progress in an anterior to posterior direction. Indeed, it is possible that this finding has evolutionary roots, as the stomach is believed to be an added character that first appeared in primitive fish (Smith et al., 2000a). It is possible then, that the stomach epithelium at E16.5 is governed by a program designed to represses the emerging intestinal state in order to preserve the primitive stomach epithelium for the later reception of instructive signals to differentiate as stomach. Though this notion is entirely speculative, it has interesting implications for intestinal metaplasia, a pathological lesion in which patches of epithelium with intestinal character emerge in the stomach. The possibility that active repression of intestinal differentiation in stomach exists during pyloric border formation (and persists throughout adult life) will become amenable to further investigation now that the transcriptomes of stomach and intestine are available during this important developmental event.
Methods
Tissue collection
Embryos from C57Bl/6J mice were collected from timed pregnant females, with the day of vaginal plug detection considered day 0.5. Intestine and stomach were removed and three contiguous segments were collected from antrum (stomach), pylorus, and duodenum (Supplemental Figure 1). For the microarray experiment, a total of 174 E14.5 embryos and 95 E16.5 embryos were dissected, and for each time/tissue group (e.g., E14.5 duodenum; six groups total), collected tissue was randomly pooled into one of three samples for replicate analysis. Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA) and purified using the RNeasy kit (Qiagen, Valencia, CA), per the manufacturers’ instructions. Separate collections were performed for RT-PCR and qPCR validation of microarray results (see Supplemental Methods).
Microarray processing
RNA samples were hybridized to MOE 430.2 microarrays (eighteen total chips: six time/tissue groups, three replicate chips for each group; Affymetrix, Santa Clara, CA) by the University of Michigan Cancer Center MicroarrayCore Facility. Microarrays were scanned and processed using GCOS software (Affymetrix) and the resulting. CEL files were analyzed using RMA (Robust Multiarray Average; affy package in BioConductor; www.bioconductor.org), which does background adjustment, normalization and conversion of probeset intensity data to log2 expression values (Irizarry et al., 2003; Gentleman et al., 2004). Probeset expression values were imported into MeV (Multi-experiment Viewer; www.tm4.org) for evaluation of statistical significance using the Student’s t-test (Saeed et al., 2003). Replicate probeset expression values were averaged and log fold change (LFC) was determined by calculating the difference between any two groups [e.g., LFC = log2(E14.5 duodenum)avg − log2(E14.5 stomach)avg], which was then converted to numerical fold change (FC) [if LFC > 0, FC = 2(LFC); else, FC = −(2(−LFC))]. Probesets with a p-value less than 0.05 and |FC| greater than 2 were selected for further analysis. To provide independent validation for the array results, we evaluated gene expression by RT-PCR. These results, shown in Supplemental Figure 2, reveal concordance with the array findings. Functional Annotation Clustering in DAVID, using the default options and “Medium” Classification Stringency, was performed with the following sets of Gene Ontology terms: GOTERM_BP_ALL, GOTERM_MF_ALL, PANTHER_BP_ALL and PANTHER_MF_ALL.
In situ hybridization, immunostaining, X-gal staining, qPCR, and RT-PCR
Detailed techniques are provided in Supplemental Methods.
Supplementary Material
A photomicrograph showing E14.5 foregut. The pylorus was identified grossly by its muscular constriction (arrows). Small, contiguous pieces of tissue were dissected from the pylorus and surrounding stomach and duodenum. St = stomach, Py = pylorus and D = duodenum.
Supplemental Table 3. DAVID analysis of D16 enriched epithelial genes
Supplemental Table 4. Summary of gene expression changes in Wnt signaling pathway components
Supplemental Table 5. Summary of pyloric genes
Supplemental Table 6. Summary of primer sequences and optimized conditions
Semi-quantitative RT-PCR was carried out as described in Supplemental Methods. Hprt is used as a control; it does not vary with time and tissue. Tissue specific regulation of Cdx1, Cdx2, Isx, Barx1, Sox2 and Sfrp5 is set by E14.5 and does not vary with time. Mafb and Hnf4γ are preferentially, but not specifically, expressed in the intestine. Tcfec and Creb3l3 are greatly induced in E16.5 duodenum.
Acknowledgments
The authors thank Dr. Alex Joyner for providing Gli1lacZ/+ mice. D.G. acknowledges support from R01 DK065850. A.U. is grateful for support from F30 DK082144. This work utilized the Cancer Center Microarray Core through the Michigan Diabetes Research and Training Center funded by P60 DK20572 from the National Institute of Diabetes & Digestive & Kidney Diseases. We also acknowledge support from the Microscopy and Image Analysis Laboratory of the Department of Cell and Developmental Biology, as well as the Morphology Core of the Center for Organogenesis.
Grand sponsor: The National Institute of Diabetes and Digestive and Kidney Diseases
Grant number(s): R01 DK065850 and F30 DK082144
References
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Braunstein EM, Qiao XT, Madison B, Pinson K, Dunbar L, Gumucio DL. Villin: A marker for development of the epithelial pyloric border. Dev Dyn. 2002;224:90–102. doi: 10.1002/dvdy.10091. [DOI] [PubMed] [Google Scholar]
- Canning CA, Lee L, Irving C, Mason I, Jones CM. Sustained interactive Wnt and FGF signaling is required to maintain isthmic identity. Dev Biol. 2007;305:276–286. doi: 10.1016/j.ydbio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MY, Romer AI, Hu M, Lepourcelet M, Mechoor A, Yesilaltay A, Krieger M, Gray PA, Shivdasani RA. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr Opin Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe Y, Sekimoto T, Araki M, Takagi K, Araki K, Yamamura K. Region-specific gastrointestinal Hox code during murine embryonal gut development. Dev Growth Differ. 2002;44:77–84. doi: 10.1046/j.1440-169x.2002.00623.x. [DOI] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;8:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Kim BM, Mao J, Taketo MM, Shivdasani RA. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 2007;133:529–538. doi: 10.1053/j.gastro.2007.04.072. [DOI] [PubMed] [Google Scholar]
- Kolterud Å, Grosse AS, Zacharias WJ, Walton KD, Kretovich KD, Madison BB, Waghray M, Ferris JE, Hu C, Merchant JL, Dlugosz A, Kottman AH, Gumucio DL. Paracrine Hedgehog signaling in stomach and intestine: new roles for Hedgehog in gastrointestinal patterning. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.05.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kozyraki R, Gofflot F. Multiligand endocytosis and congenital defects: roles of cubilin, megalin and amnionless. Curr Pharm Des. 2007;13:3038–3046. doi: 10.2174/138161207782110507. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees CW, Zacharias WJ, Tremelling M, Noble CL, Nimmo ER, Tenesa A, Cornelius J, Torkvist L, Kao J, Farrington S, Drummond HE, Ho GT, Arnott ID, Appelman HD, Diehl L, Campbell H, Dunlop MG, Parkes M, Howie SE, Gumucio DL, Satsangi J. Analysis of germline GLI1 variation implicates hedgehog signalling in the regulation of intestinal inflammatory pathways. PLoS Med. 2008;5:e239. doi: 10.1371/journal.pmed.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Li X, Madison BB, Zacharias W, Kolterud A, States D, Gumucio DL. Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol Genomics. 2007;29:290–301. doi: 10.1152/physiolgenomics.00269.2006. [DOI] [PubMed] [Google Scholar]
- Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 2008;22:3050–3063. doi: 10.1101/gad.1687308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke-Wheeler J, Zhang K, Battle M, Si-Tayeb K, Garrison W, Chhinder S, Li J, Kaufman RJ, Duncan SA. Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology. 2008;48:1242–1250. doi: 10.1002/hep.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RA, Barth JL, Chintalapudi MR, Knaak C, Argraves WS. Megalin functions as an endocytic sonic hedgehog receptor. J Biol Chem. 2002;277:25660–25667. doi: 10.1074/jbc.M201933200. [DOI] [PubMed] [Google Scholar]
- McKay IJ, Muchamore I, Krumlauf R, Maden M, Lumsden A, Lewis J. The kreisler mouse: a hindbrain segmentation mutant that lacks two rhombomeres. Development. 1994;120:2199–2211. doi: 10.1242/dev.120.8.2199. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- Miura A, Yamagata K, Kakei M, Hatakeyama H, Takahashi N, Fukui K, Nammo T, Yoneda K, Inoue Y, Sladek FM, Magnuson MA, Kasai H, Miyagawa J, Gonzalez FJ, Shimomura I. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem. 2006;281:5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]
- Mochida Y, Parisuthiman D, Kaku M, Hanai J, Sukhatme VP, Yamauchi M. Nephrocan, a novel member of the small leucine-rich repeat protein family, is an inhibitor of transforming growth factor-beta signaling. J Biol Chem. 2006;281:36044–36051. doi: 10.1074/jbc.M604787200. [DOI] [PubMed] [Google Scholar]
- Moniot B, Biau S, Faure S, Nielsen CM, Berta P, Roberts DJ, de Santa Barbara P. SOX9 specifies the pyloric sphincter epithelium through mesenchymal-epithelial signals. Development. 2004;131:3795–3804. doi: 10.1242/dev.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan H, Razin E. The function of MITF and associated proteins in mast cells. Mol Immunol. 2002;38:1177–1180. doi: 10.1016/s0161-5890(02)00059-7. [DOI] [PubMed] [Google Scholar]
- Okubo T, Hogan BL. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Partington GA, Fuller K, Chambers TJ, Pondel M. Mitf-PU.1 interactions with the tartrate-resistant acid phosphatase gene promoter during osteoclast differentiation. Bone. 2004;34:237–245. doi: 10.1016/j.bone.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. Embo J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- Smith DM, Grasty RC, Theodosiou NA, Tabin CJ, Nascone-Yoder NM. Evolutionary relationships between the amphibian, avian, and mammalian stomachs. Evol Dev. 2000a;2:348–359. doi: 10.1046/j.1525-142x.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- Smith DM, Nielsen C, Tabin CJ, Roberts DJ. Roles of BMP signaling and Nkx2.5 in patterning at the chick midgut-foregut boundary. Development. 2000b;127:3671–3681. doi: 10.1242/dev.127.17.3671. [DOI] [PubMed] [Google Scholar]
- Smith DM, Tabin CJ. BMP signalling specifies the pyloric sphincter. Nature. 1999;402:748–749. doi: 10.1038/45439. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M. MITF: a stream flowing for pigment cells. Pigment Cell Res. 2000;13:230–240. doi: 10.1034/j.1600-0749.2000.130404.x. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Vertebrate endoderm development. Annu Rev Cell Dev Biol. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, Garcia PD, Randazzo FM, Escobedo J, Fantl WJ, Williams LT. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenthal A, de Sauvage FJ, Shivdasani RA. Downregulation of Hedgehog signaling is required for organogenesis of the small intestine in Xenopus. Dev Biol. 2001;229:188–202. doi: 10.1006/dbio.2000.9953. [DOI] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, Atochina-Vasserman E, Lu MM, Beers MF, Morrisey EE. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134:189–198. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A photomicrograph showing E14.5 foregut. The pylorus was identified grossly by its muscular constriction (arrows). Small, contiguous pieces of tissue were dissected from the pylorus and surrounding stomach and duodenum. St = stomach, Py = pylorus and D = duodenum.
Supplemental Table 3. DAVID analysis of D16 enriched epithelial genes
Supplemental Table 4. Summary of gene expression changes in Wnt signaling pathway components
Supplemental Table 5. Summary of pyloric genes
Supplemental Table 6. Summary of primer sequences and optimized conditions
Semi-quantitative RT-PCR was carried out as described in Supplemental Methods. Hprt is used as a control; it does not vary with time and tissue. Tissue specific regulation of Cdx1, Cdx2, Isx, Barx1, Sox2 and Sfrp5 is set by E14.5 and does not vary with time. Mafb and Hnf4γ are preferentially, but not specifically, expressed in the intestine. Tcfec and Creb3l3 are greatly induced in E16.5 duodenum.