p107, a member of the retinoblastoma susceptibility protein family, influences lipid metabolism in muscle and other tissues by repressing the PPAR-γ co-factor PGC-1α.
Abstract
Mice lacking p107 exhibit a white adipose deficiency yet do not manifest the metabolic changes typical for lipodystrophy, and instead exhibit low levels of serum triglycerides and a normal liver phenotype. When fed a high fat diet, p107-null mice still did not accumulate fat in the liver, and display markedly elevated energy expenditures together with an increased energy preference for lipids. Skeletal muscle was therefore examined, as this is normally the major tissue involved in whole body lipid metabolism. Notably, p107-deficient muscle express increased levels of peroxisome proliferator–activated receptor gamma co-activator-1α (PGC-1α) and contained increased numbers of the pro-oxidative type I and type IIa myofibers. Chromatin immunoprecipitation revealed binding of p107 and E2F4 to the PGC-1α proximal promoter, and this binding repressed promoter activity in transient transcription assays. Ectopic expression of p107 in muscle tissue in vivo results in a pronounced 20% decrease in the numbers of oxidative type IIa myofibers. Lastly, isolated p107-deficient muscle tissue display a threefold increase in lipid metabolism. Therefore, p107 determines the oxidative state of multiple tissues involved in whole body fat metabolism, including skeletal muscle.
Introduction
The retinoblastoma susceptibility protein (Rb) family, including pRb, p107, p130, and their associated proteins, have been implicated in adipogenesis and regulation of energy metabolism (Hallenborg et al., 2009). Recently, we found that mice lacking p107 displayed an extensive replacement of white adipose tissue (WAT) with brown adipose tissue (BAT; Scimè et al., 2005). The p107-deficient WAT depots contain brown-type adipocytes, express peroxisome proliferator–activated receptor gamma co-activator-1α (PGC-1α) and uncoupling protein 1 (UCP-1), and appear poorly differentiated. Deficiencies in adipose tissue, or lipodystrophy, are normally associated with a metabolic syndrome that resembles obesity due to an inability to process fat in the diet (Reue and Phan, 2006). Paradoxically, p107−/− mice exhibit a lean metabolic phenotype in spite of an apparent lipodystrophy (Scimè et al., 2005).
When high levels of lipid cannot be stored in WAT, it is predominately partitioned into skeletal muscle, which can lead to metabolic consequences of impaired insulin responsiveness (insulin resistance) and type II diabetes mellitus (Lelliott and Vidal-Puig, 2004; Kelley, 2005). In humans and animals, increased capacity for lipid oxidation by muscle is correlated to increased insulin sensitivity, reduced adipocyte size, leanness, and aerobic fitness (Luquet et al., 2003; Ryder et al., 2003; Ukropcova et al., 2005).
Skeletal muscle fiber types are generally classified according to four major myosin heavy chain (MyHC) isoforms that display differences with respect to their metabolic potential (Pette and Staron, 2001). They consist of slow contracting type I oxidative, fast contracting type IIa oxidative, and fast contracting glycolytic type IIx and IIb fibers. Type I and IIa muscle fibers are highly oxidative, containing many mitochondria that provide the muscle with the production of a large supply of energy in the form of ATP. Type IIx and IIb fibers rely for the most part on glycolytic metabolism as a major energy source, have few mitochondria, exert quick contractions, and fatigue easily.
Many studies have demonstrated that obesity and type II diabetes are associated with a lower percentage of oxidative type I fibers and a higher percentage of glycolytic type II fibers (Hickey et al., 1995; Nyholm et al., 1997; Tanner et al., 2002). Moreover, type I fibers are more insulin sensitive compared with type IIb fibers (Lillioja et al., 1987; Hickey et al., 1995).
The major site for oxidative metabolism is skeletal muscle, with as much as 90% of the energy needs during rest derived from fatty acids (Dagenais et al., 1976). A leading abnormality in obesity is a reduction in the capacity of muscle for lipid oxidation as measured in the whole body, or from muscle homogenates or strips (Kelley et al., 1999; Kim et al., 2000; Hulver et al., 2003; Thyfault et al., 2004). The fatty acid oxidation impairment is also apparent within primary muscle cells of extremely obese individuals (Hulver et al., 2005; Ukropcova et al., 2005). In addition, the reduced capacity in lipid oxidation is correlated with a propensity toward weight gain and obesity (Zurlo et al., 1990; Marra et al., 2004).
In response to physiological pressures such as exercise and diet, skeletal muscle can remodel to switch fiber types by activating signaling pathways that inevitably control gene expression patterns in progenitor cells and/or muscle fibers (Bassel-Duby and Olson, 2006). Recently, the transcriptional coactivator PGC-1α has been implicated to play an important pro-oxidative role in muscle (Puigserver et al., 1998; Wu et al., 1999). PGC-1α has been shown to regulate a myriad of metabolic activities including enhancing mitochondrial function and biogenesis in skeletal muscle. Fiber-type enrichment of pro-oxidative type I and type IIa fibers is observed when PGC-1α is expressed at physiological levels in transgenic mice under a muscle specific promoter (Lin et al., 2002, 2005).
The lean metabolic phenotype of p107−/− mice suggested the hypothesis that the mice may have an elevation in lipid metabolism in a tissue such as skeletal muscle. Nuclear p107 expression has been well documented in differentiated myotubes and muscle tissue by immunostaining (Baldi et al., 1997; Parakati and DiMario, 2005), which supports a role for p107 in noncycling myofibers. In this study, we find that p107 plays a central role in determining the fiber type character of skeletal muscle. Our findings demonstrate that the p107-ablated animals have higher levels of oxidative MyHC type I and type IIa fibers. This plays a role in the increased efficiency of fatty acid metabolism in p107−/− mice when fed a high fat (HF) diet. Indeed, p107 influences the oxidative nature of skeletal muscle by directly repressing PGC-1α expression in myogenic cells. These results indicate a central role for p107 in the control of whole body fat metabolism. Therefore, our data suggest p107 as a therapeutic target for the treatment of various metabolic and neuromuscular disorders by influencing the oxidative state of muscle.
Results
Mice lacking p107 exhibit a life-long white adipose deficiency
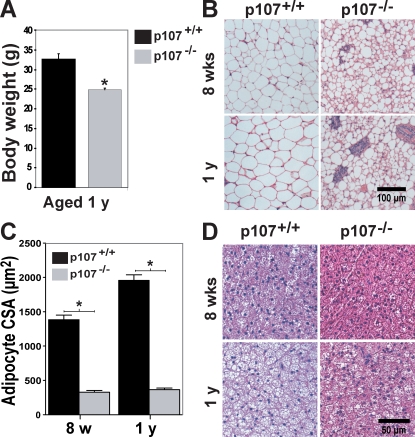
White adipocyte differentiation is deficient in p107−/− BALB/c mice, resulting in a lean phenotype exemplified by a significant reduction in total adiposity (Scimè et al., 2005). We assessed whether the reduced adiposity remained throughout the lifetime of the mutant animals. Both male and female p107−/− mice remained leaner than their wild-type siblings, weighing as much as 30% less after 1 yr of age (P < 0.0003; Fig. 1 A). As the fat content is the main contributor to the leanness in young mice, we determined if lipids had accumulated in adipose tissue of aged p107−/− mice.
Figure 1.
Aged p107−/− mice display underdeveloped WAT depots. (A) Graphical representation of the weight of 1-yr-old wild-type and p107−/− mice (n = 5 for p107+/+ and n = 4 for p107−/−). The asterisk denotes significance (P < 0.0003). (B) H&E staining of inguinal WAT for wild-type and p107−/− mice at 8 wk and 1 yr of age. (C) Graphical representation of the mean cross-sectional area (CSA) of white adipocytes for wild-type and p107−/− mice at 8 wk (w) and 1 yr (yr) of age. The asterisk denotes significance (P < 0.009). (D) H&E staining of interscapular BAT for wild-type and p107−/− mice at 8 wk and 1 yr of age. Note the increased lipid deposition in the wild-type animals after 1 yr. Error bars indicate SD.
The adipocyte paucity was evident, as shown by the reduced lipid levels within WAT and BAT in older aged mice (Fig. 1, B and C). At 1 yr of age, p107−/− WAT is characterized as nonuniform smaller sized adipocytes appearing much the same as in 8 wk of age (Fig. 1 B). By comparison, adipocytes within the WAT of 1-yr-old wild-type mice increased in area by as much as 42.8% (Fig. 1 C). BAT for older p107-deficient mice retained small sized multilocular adipocytes as compared with wild-type mice aged 1 yr that had become engulfed by lipids and infiltrated with unilocular-appearing cells (Fig. 1 D). In addition, 2-yr-old p107−/− mice did not exhibit any significant weight gain and were similar in weight to 3-mo-old mutant mice. Collectively, these results suggest that the p107−/− lean phenotype is retained throughout the lifetime of the animal.
Mice lacking p107 are resistant to weight gain
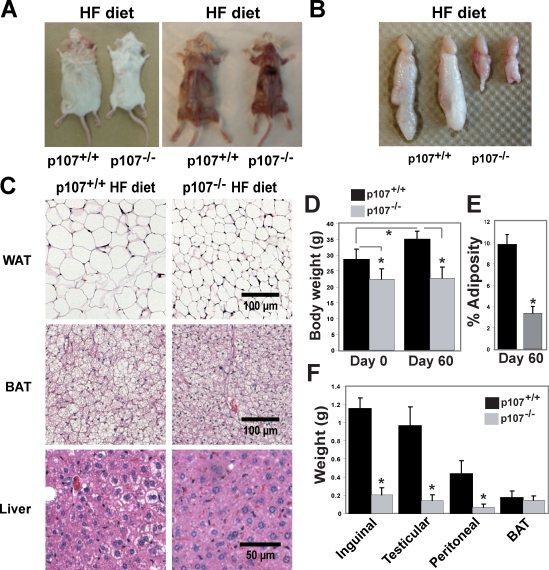
Male mice were challenged with an HF diet, which normally induces obesity, containing 60% (wt/wt) fat. Wild-type mice fed an HF diet for 60 d exhibited an obvious weight gain relative to the maintenance of leanness in the p107−/− animals (Fig. 2 A). This was further illustrated by the testicular fat pads from representative animals on the HF diet that revealed extremely blunted weight gain by the p107−/− mice (Fig. 2 B).
Figure 2.
p107−/− mice are refractory to fat accumulation after HF feeding. (A) Representative dorsal view of wild-type and p107−/− mice after 60 d on an HF diet. Note the lack of WAT pads for the p107−/− mouse. (B) Representative gross comparison of testicular fat depots between adult wild-type and p107−/− mice after 60 d on HF diet. (C) H&E staining of inguinal WAT, interscapular BAT, and livers for representative wild-type and p107−/− mice after 60 d on an HF diet. Note the accumulation of lipids within cells of the various tissues for the wild-type mouse but not the p10−/− mouse. (D) Graphical representation of body weight for wild-type and p107−/− mice before (day 0) and after an HF diet for 60 d (day 60; n = 5 and 4 for p107+/+ and p107−/−, respectively). Asterisks indicate significance between wild-type and p107−/− at day 0 (P < 0.03) and day 60 (P < 0.002) and between day 0 and day 60 for wild-type mice (P < 0.02, n = 5 and 4 for p107+/+ and p107−/−, respectively). (E) Percent adiposity for wild-type and p107−/− mice after 60 d of being fed an HF diet (day 60; n = 5 and 4 for p107+/+ and p107−/−, respectively). Asterisks denote significance (P < 0.000006; n = 3). (F) Weights of various fat depots between p107−/− and wild-type male mice after an HF diet for 60 d. Asterisks denote significance for inguinal (P < 0.002), testicular (P < 0.0005), and peritoneal (P < 0.001; n = 3) samples. Note that p107−/− mice do not gain any appreciable weight at the level of lipid accumulation in WAT after HF feeding. Error bars indicate SD.
The HF diet resulted in considerable lipid accumulation and hypertrophy in both brown and white adipocytes of wild-type animals relative to p107−/− mice (Fig. 2 C). As expected, hematoxylin and eosin (H&E) staining of liver tissue sections also revealed that the wild-type mice were unable to fully accommodate the increased fat load in their diet. The wild-type mice developed fatty liver as demonstrated by the presence of lipids within their hepatocytes (Fig. 2 C). Strikingly, the livers of p107−/− mice did not appreciably manifest any liver anomalies despite ingesting the same diet, and displayed an overall normal appearance.
The total weight and adiposity of the control and experimental animals were also evaluated before and after feeding on the HF diet for 60 d. The p107-deficient mice failed to gain any weight, whereas littermate controls significantly (P < 0.02) gained as much as 27% more weight after completion of the HF diet (Fig. 2 D, compare day 0 versus day 60). Moreover, the body weight for p107−/− mice compared with controls before and after the HF diet was significantly lower (P < 0.03 and P < 0.002, respectively). To test if the weight gain differences observed were caused by fat mass expansion, we assessed the percentage adiposity of the animals (Fig. 2 E). The total percentage of fat in the wild-type animals is at least three times greater after completion of the fat-laden diet as compared with the p107−/− animals (Fig. 2 E). This is corroborated by significantly higher weight in individual fat depots in control mice (Fig. 2 F). Contrarily, the size of the BAT depots between the animals remained relatively similar. Collectively, these results indicate that p107−/− mice remain remarkably lean when challenged with an HF diet.
Mice lacking p107 are more efficient at β oxidation
Typically, lipodystrophic mice exhibit elevated triglycerides and free fatty acids in the serum as well as fatty liver changes, and they do not show an elevation in energy expenditures (Reue and Phan, 2006). However, p107−/− mice do not exhibit these changes despite the increased lipid load (Figs. 2 and S1). Notably, p107−/− mice fed a regular diet have significantly higher metabolic rates than their littermate controls (Scimè et al., 2005). Therefore, we tested if this characteristic was responsible for maintaining the p107 mutant phenotype when challenged with an HF diet.
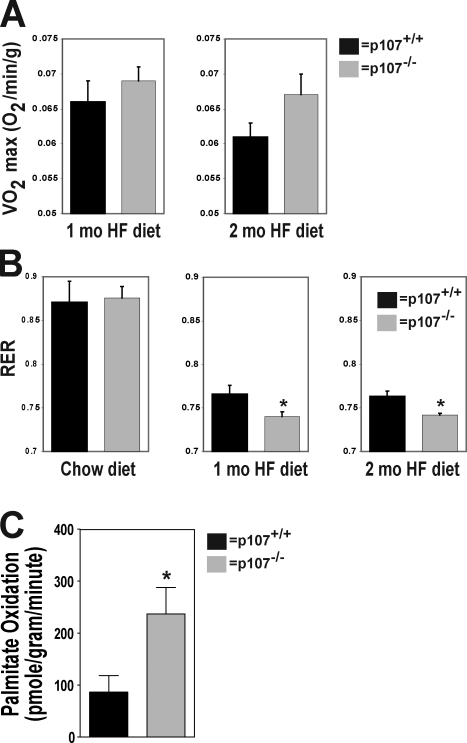
After 60 d on the HF diet, the mutant mice exhibited significantly higher metabolic rates than their wild-type controls. This suggests that the maintenance of a higher metabolic rate leads to an increase in fatty acid metabolism (Fig. 3 A). We therefore evaluated the fuel preference of the mice before and after the HF diet by measuring their respiratory exchange ratios (RERs). A lower RER is an indication of a greater fat metabolism, whereas a higher RER is associated with a greater carbohydrate metabolism. On the normal diet, the RER values for both p107−/− and control mice were not significantly different and reflected the nature of this diet (Fig. 3 B). As expected on the HF diet, because lipids are the predominate source of energy, both p107−/− and wild-type mice had lower RER values than when feeding on the regular diet (Fig. 3 B). However, the p107−/− mice had a significantly lower RER after 1 and 2 mo on the diet (P < 0.05 and P < 0.03, respectively).
Figure 3.
Elevated metabolic rate in p107−/− mice induced by an HF diet. (A) Mean 24 h mass-adjusted total energy expenditure for wild-type and p107−/− mice after 1 mo and 2 mo of HF feeding (n = 4 for both p107+/+ and p107−/−). (B) RER for wild-type and p107−/− mice on a chow diet or at the end of 1 and 2 mo of an HF diet (n = 4 and 5 for p107−/− and p107+/+, respectively). Asterisks denote significance P < 0.05 for 1 mo and P < 0.03 at 2 mo (n = 4 for both p107+/+ and p107−/−). (C) Exogenous palmitate oxidation per gram per minute of gastrocnemius muscle ex vivo over time (n = 12 and 8 for p107−/− and p107+/+, respectively). The asterisk denotes significance (P < 0.02). Error bars indicate SD.
The major site for whole body oxidative metabolism is skeletal muscle, with as much as 90% of the energy needs during rest derived from fatty acids (Dagenais et al., 1976). Therefore, we next asked whether the high-lipid metabolism phenotype of p107−/− mice can be attributed to skeletal muscle. Lipid metabolism in muscle was assessed by measuring the oxidative potential of isolated gastrocnemius using radiolabeled palmitic acid (Fig. 3 C). Indeed, we found a significant threefold increase in β-oxidative potential in p107−/− muscle. This marked increase strongly supports the assertion that the increased capacity in β-oxidative potential in muscle has a significant role in contributing to the lean phenotype of p107 mutant mice.
Muscle in p107−/− mice exhibits enhanced pro-oxidative capacity
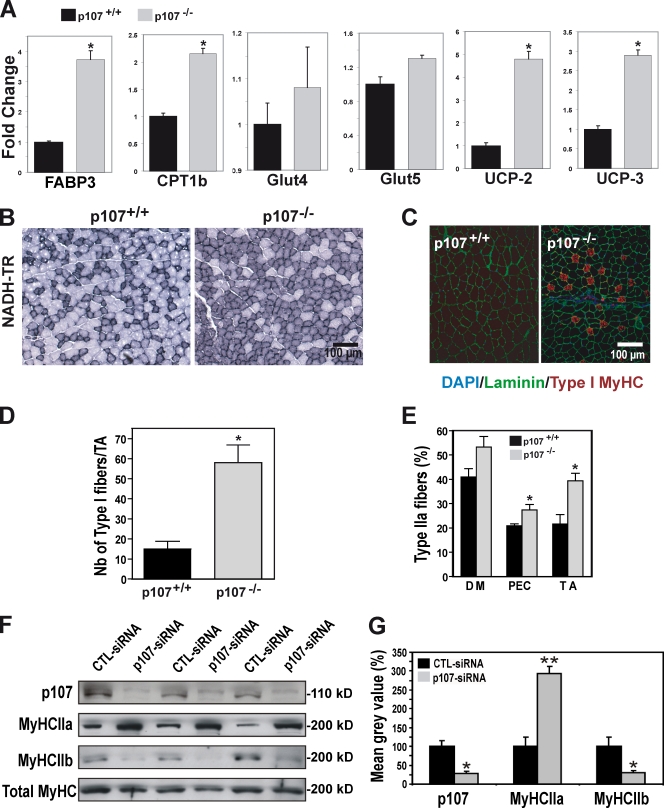
To investigate the basis for the enhanced ability of p107-deficient muscle to metabolize lipids, we analyzed the expression levels of various genes involved in regulating energy metabolism in the tibialis anterior (TA) muscle when fed a normal diet. In adult mice, we first examined the expression of fatty acid binding protein 3 (FABP3), which is involved in the intramyocellular transport of free fatty acid, and carnitine palmitoyltransferase Ib (CPT1b), which is involved in the uptake of fatty acid from the cytoplasm into the mitochondrial matrix. Strikingly, we observed a 3.6-fold increase in FABP3 and a 2.1-fold increase in CPT1b levels by quantitative RT-PCR (qRT-PCR; Fig. 4 A). In contrast, Glut4 and Glut5, which are involved in carbohydrate metabolism, did not display significant changes in expression levels (Fig. 4 A). Lastly, UCP-2 and UCP-3 were significantly up-regulated in the p107−/− muscle relative to wild-type controls (Fig. 4 A). Collectively, these results indicate that the molecular components involved in fatty acid metabolism and β oxidation are markedly increased in p107−/− skeletal muscle.
Figure 4.
Elevated oxidative function of p107−/− skeletal muscle. (A) Graphical representation of the relative fold change expression between wild-type and p107−/− mice for FABP3, CPT1b, Glut4, Glut5, UCP-2, and UCP-3 in TA muscle using qRT-PCR. Asterisks denote significance (P < 0.05, P < 0.004, P < 0.005, and P < 0.005 for FABP3, CPT1b, UCP-2, and UCP-3, respectively). n = 4 for both p107+/+ and p107−/−. (B) Representative NADH reductase staining with NBT (NADH-TR) of TA fibers for wild-type and p107−/− mice. Note the obvious abundance of darkly stained pro-oxidative fibers for the p107−/− TA. (C) Type I MyHC (red), Laminin (green), and DAPI (blue) fluorescence immunostaining of representative TA sections for wild-type and p107−/− mice. Note the increased levels of oxidative fibers for the p107−/− muscle in the mainly mixed type II muscle. (D) Graphical representation of the number of type I fibers per TA for wild-type and p107−/− mice. The asterisk denotes significance (P < 0.02; n = 3 for both p107+/+ and p107−/−). (E) Graphical representation of the number of type IIa fibers per diaphragm (DM), pectoralis (PEC), and TA for wild-type and p107−/− mice. Note that the different muscle groups have pro-oxidative fibers. The asterisks denote significance (P < 0.05 and P < 0.02 for PEC and TA, respectively; n = 3 for both p107+/+ and p107−/−). (F) Immunoblot triplicates of p107-siRNA knockdown myotubes using antibodies directed against the indicated proteins. A nonsilencing siRNA (CTL-siRNA) was used as a control, and equal protein loading was confirmed by blotting for total MyHC. MyHCIIa, MyHC type IIa; MyHCIIb, MyHC type IIb. (G) Background-corrected gray values from F were normalized to the total MyHC levels, and the CTL-siRNA was set to 100%. Compiled data are expressed as mean ± SEM, with the level of significance indicated as: **, P < 0.01; * P < 0.05. Error bars indicate SD.
Type I and IIa muscle fibers are highly oxidative, containing many mitochondria that provide the muscle with the production of a large supply of energy in the form of ATP. Therefore, we examined the distribution of fiber types in p107-deficient muscle. We first assessed the mitochondrial oxidative enzyme activity in the TA by staining with nitro-blue tetrazolium (NBT), a histochemical stain against the pro-oxidative component NADH-reductase (Fig. 4 B). Oxidative muscle fibers are darkly stained in NBT, whereas intermediately oxidative and nonoxidative fibers are stained lighter. Representative tissue sections of NBT staining demonstrated that the p107−/− mice had a dramatically higher content of darkly and intermediate staining fibers within their TAs, compared with the littermate control (Fig. 4 B). Collectively, these data suggested that p107-deficient fibers are much more oxidative.
To investigate whether the increased oxidative capacity was due to increased numbers of pro-oxidative type I and/or type IIa fibers, we immuno-stained cross sections of the TA from p107−/− and wild-type mice for slow type I MyHC fibers (Fig. 4 C). Type I immunostaining revealed that p107−/− TA displayed a fourfold (P < 0.02) increase in type I fibers relative to their wild-type littermate controls (Fig. 4 D). The increase in type I fibers was also evident in the mixed glycolytic and oxidative fiber type gastrocnemius muscle (unpublished data).
We next assessed the percentage of type IIa pro-oxidative fibers within various muscle groups to find if they influence the oxidative nature of p107−/− muscle. We found significantly increased percentages of type IIa fibers within the TA (P < 0.02) and pectoralis (P < 0.05) muscles, and higher overall levels within the diaphragm muscles of p107−/− mice (Fig. 4 E). p107−/− TAs and pectoralis muscles had a mean increase of 82.4% and 31.1% in the numbers of type IIa fibers, respectively, as compared with wild-type littermate controls. Notably, the total number of fibers in the p107−/− TA was similar to wild-type TA, which suggests that the number of IIb fibers must be correspondingly decreased.
We also tested if the observed muscle fiber type phenotype in the p107−/− mice was caused by a myoblast cell autonomous affect. This was accomplished by knocking down p107 expression by transient transfection of siRNA specific for p107 in three different primary myoblast isolates before their differentiation into myotubes (Fig. 4 F). By Western blot analysis, siRNA-mediated inhibition of p107 protein expression revealed increased oxidative type IIa (MyHC IIa) fiber content and a corresponding decrease in type IIb fibers (MyHC IIb) after differentiation (Fig. 4 F). Quantification of the data demonstrated that the levels of MyHC IIa significantly (P < 0.01) increased twofold, and that MyHC IIb significantly decreased (P < 0.05) by as much as 75% when compared with the nonsilencing control siRNA (CTL-siRNA; Fig. 4 G).
Our experiments thus document that p107−/− muscle contains greatly increased numbers of pro-oxidative type fibers that result from a cell-autonomous deficit in p107 during myogenic precursor differentiation. Together, these data suggest that the increased pro-oxidative nature of skeletal muscles in p107−/− mice is responsible for the overall increased lipid utilization.
PGC-1α transcription is negatively regulated by p107
Recent work has implicated PGC-1α in muscle fiber type determination and switching. For example, muscle-specific expression of PGC-1α in transgenic mice induces activation of genes typical of oxidative fibers (Lin et al., 2002). Our recent study demonstrated that p107 negatively regulates PGC-1α and thus UCP-1 in white adipose tissue (Scimè et al., 2005), which suggests the hypothesis that p107 similarly regulates PGC-1α in muscle. Therefore, we evaluated the levels of PGC-1α in the TA muscle by qRT-PCR.
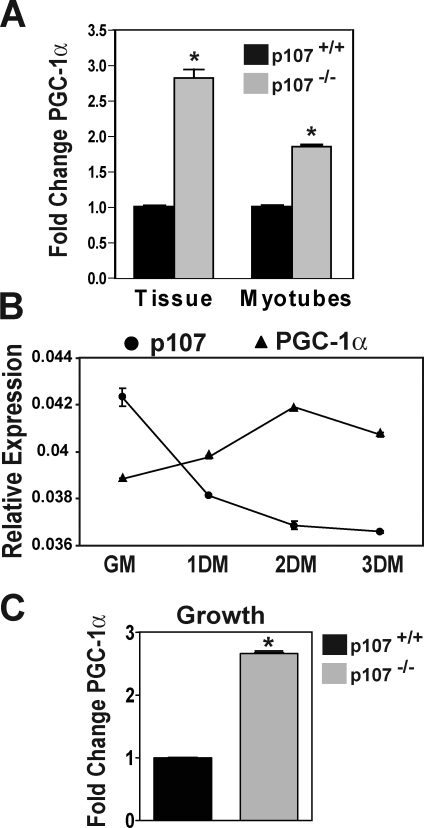
Importantly, PGC-1α mRNA levels in p107−/− TA muscle were significantly (P < 0.004) higher by almost threefold as compared with wild-type littermate controls (Fig. 5 A). We next confirmed if the higher levels of PGC-1α in p107−/− mice were specific to myogenic cells within the tissue. This was accomplished by isolating primary myoblasts from wild-type and mutant mice and inducing myogenic differentiation. Levels of PGC-1α mRNA were significantly (P < 0.008) elevated by about twofold in differentiated p107−/− myotubes (Fig. 5 A). Indeed, p107 and PGC-1α displayed an inverse correlation in the level of expression during growth and differentiation, which supports the assertion that p107 negatively regulates PGC-1α transcription (Fig. 5 B). Isolated primary myoblasts lacking p107 in growth media also exhibited an ∼2.5-fold increase in PGC-1α mRNA (Fig. 5 C). Collectively, these results strongly support the conclusion that the pro-oxidative nature of p107−/− skeletal muscle is a consequence of a cell-autonomous loss of p107 and subsequent up-regulation of PGC-1α transcription.
Figure 5.
Elevated PGC-1α expression in p107−/− skeletal muscle. (A) Graphical representation of the relative fold change expression for PGC-1α in wild-type and p107−/− TA muscle (Tissue) and differentiated primary myoblasts (Myotubes) using qRT-PCR. Asterisks denote significance (P < 0.004 and P < 0.008 for tissue and myotubes, respectively; n = 3 for both p107+/+ and p107−/−). (B) Graphical representation of the relative expression, using qRT-PCR for PGC-1α and p107 in a time course of differentiation, for primary myoblasts. GM, growth media; 1DM, 2DM, and 3DM, 1, 2 and 3 d in differentiation media, respectively. Note the reciprocal expression pattern for p107 and PGC-1α through differentiation (n = 3). (C) Graphical representation of the relative fold change expression for PGC-1α in wild-type and p107−/− myoblasts in growth using qRT-PCR. Asterisks denote significance (P < 0.004; n = 3 for both p107+/+ and p107−/−). Error bars indicate SD.
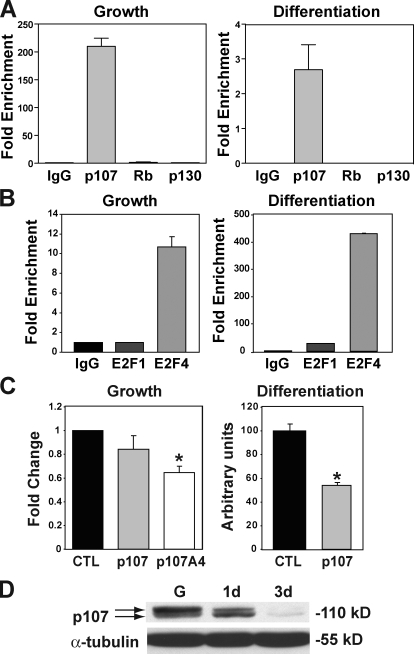
Examination of the sequence of the PGC-1α proximal promoter revealed multiple potential p107/E2F binding sites. Therefore, we tested whether p107 binds to the PGC-1α proximal promoter by quantitative chromatin immunoprecipitation (ChIP). Assays were performed using primers that amplified a 311-bp fragment located −134 to −445 bp upstream of the PGC-1α transcriptional start site (Fig. 6 A). ChIP analysis revealed robust binding of p107 in proliferating myoblasts and differentiated myotubes. In contrast, neither Rb nor p130 exhibited notable binding under growth or differentiation conditions (Fig. 6 A).
Figure 6.
PGC-1α transcription is negatively regulated by p107/E2F binding. (A) Quantitative ChIP assays for control immunoglobulin (IgG), p107, Rb, and p130 to the PGC-1α promoter on a fragment −134 to −445 bp upstream of the transcription start in growth (Growth) and 48 h after differentiation (Differentiation). Results are given as fold enrichment relative to IgG (n = 3). (B) Quantitative ChIP assays for control immunoglobulin (IgG), E2F1, and E2F4 to the PGC-1α promoter on a fragment −134 to −445 bp upstream of the transcription start in growth (Growth) and 48 h after differentiation (Differentiation). Results are given as fold enrichment relative to IgG (n = 3). (C) Underphosphorylated p107 represses PGC1α promoter activity in growth and differentiation. C2C12 cells were transfected with a luciferase reporter driven by the 3.1-kb region upstream of the ATG start site of the PGC-1α gene with empty vector (CTL), or with p107HA and p107A4HA in growth. Cells were also transfected with the luciferase reporter alone (CTL) or with p107HA and differentiated for 2 d. Note that the p107 mutant p107A4HA that is deficient in Cdk2 phosphorylation is able to significantly repress the promoter activity in growth and full-length p107 in differentiation (asterisks denote significance of the mean, P < 0.00000687 and P < 0.0000196, respectively; n = 9). Error bars indicate SD. (D) Western blot for p107 in growth (G) and after 1 (1d) and 3 (3d) days differentiation of C2C12 cells. Note that p107 becomes hypophosphorylated as differentiation progresses.
Rb family factors such as p107 are recruited to promoters to repress transcription through binding to DNA-binding transcription factors such as E2F1 and E2F4 (Mulligan and Jacks, 1998). Therefore, we used ChIP to test whether E2F1 and E2F4 bind the PGC-1α proximal promoter (Fig. 6 B). Notably, quantitative ChIP assays revealed that E2F4 bound the PGC-1α promoter in proliferating myoblasts and differentiated myotubes, whereas E2F1 interacted to a lesser degree only after differentiation. Therefore, these results are consistent with the notion that p107 controls PGC-1α activity at the transcriptional level through binding of E2F-4 and E2F1 in sites located in the proximal promoter.
To investigate the ability of p107 to negatively regulate PGC-1α transcription, we used transient transcription assays using a luciferase reporter under the control of the PGC-1α 3.1-kb proximal promoter. Overexpression of p107 significantly repressed the activity of the PGC-1α proximal promoter at 48 h after differentiation (Fig. 6 C). Expression of p107A4a, a nonphosphorylatable form of p107 (K657-L660 replaced by alanine), significantly repressed PGC-1α promoter activity in growth media. Notably, p107A4 cannot be phosphorylated by Cdk2 (Leng et al., 2002). Therefore, these data suggest that p107 regulates the PGC-1α transcription in a Cdk2 phosphorylation–dependent manner (Fig. 6, C and D). Thus, overexpression of p107 in growth displayed a modest repression effect on PGC-1α transcription likely caused by the high level of Cdk2 activity (Fig. 6 D).
Collectively, these results demonstrate that p107 controls PGC-1α activity at the transcriptional level, dependent on the phosphorylation status of p107 and binding to E2F4 at a site likely located within the proximal promoter.
Ectopic expression of p107 influences fiber type determination
Our experimental results suggested the hypothesis that p107 negatively regulates PGC-1α transcription in skeletal muscle to modulate the oxidative state. To directly test this hypothesis, we ectopically expressed p107 in skeletal muscle to ask whether p107 overexpression was sufficient to influence the fiber type character in vivo.
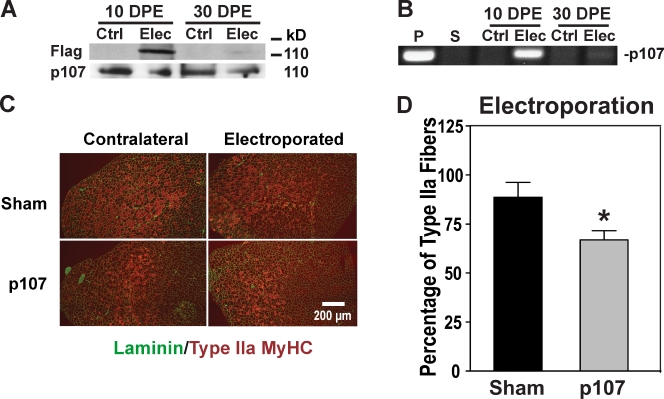
Cytomegalovirus (CMV) expression plasmids were electroporated directly into the TA muscle using conditions that result in high-level expression in ∼80% of the fibers (Le Grand et al., 2009). The expression of Flag-tagged p107 was readily visible at 10 d and 30 d after electroporation by Western blotting and PCR analysis (Fig. 7, A and B). 21 d after transfer, the TA muscles from electroporated and nonelectroporated contralateral legs from each mouse were isolated, and the percentage of muscle fibers expressing type IIa MyHC was enumerated (Fig. 7 C).
Figure 7.
Ectopic expression of p107 influences fiber type determination. (A) Western blot for endogenous p107 and Flag-tagged p107 of electroporated (Elec) Flag-p107 and contralateral untreated (Ctrl) TAs at 10- and 30-d postelectroporation (DPE). (B) PCR detection of electroporated Flag-p107 (Elec) and contralateral untreated (Ctrl) TA muscles after 10- and 30-d DPE. P, Flag-p107; S, empty vector (sham). (C) Type IIa MyHC (red) and Laminin (green) fluorescence immunostaining of representative sections for nonelectroporated and contralateral electroporated TA muscle with empty vector (Sham) or with p107. Note the decreased levels of type IIa oxidative fibers for the p107 electroporated muscle. (D) p107 influences the fiber type character of skeletal muscle in vivo. Graphical representation of the percentage of muscle fibers expressing type IIa MyHC for electroporated TA muscle with empty vector (Sham) or with p107 from their nonelectroporated contralateral TA. Note the significantly reduced oxidative type IIa fibers for p107 expressing TAs. The asterisk denotes significance (P < 0.03; n = 3). Error bars indicate SD.
Our results revealed that when the cmv-p107 plasmid was electroporated into muscle, the number of oxidative type IIa fibers was significantly reduced relative to TA muscles electroporated with empty vector (sham; Fig. 7 D). Therefore, ectopic overexpression of p107 within skeletal muscle fibers directly promotes a less oxidative myofiber character in a cell-autonomous manner.
Discussion
Our experiments support a role for p107 in controlling fiber type specification at the level of the coactivator PGC-1α. The pro-oxidative fiber type composition corresponds to significant increases in the expression of PGC-1α, a key regulator of mitochondrial gene expression and oxidative metabolism (Puigserver et al., 1998; Wu et al., 1999; Vega et al., 2000). Recent studies have described elevated levels of PGC-1α during endurance exercise training that is correlated to fiber type switching from glycolytic to oxidative fibers (Pilegaard et al., 2003; Russell et al., 2003; Akimoto et al., 2005). Indeed, transgenic overexpression of PGC-1α promotes the enrichment of pro-oxidative type I and type IIa fibers in type II muscle groups (Puigserver et al., 1998; Wu et al., 1999; Lin et al., 2002; Pilegaard et al., 2003; Russell et al., 2003).
Our results suggest that p107 functions to control PGC-1α activity by modulating its expression. p107 acts as a repressor to PGC-1α transcriptional control that is verified by the significantly higher levels of the coactivator that are found in p107-null mice. Quantitative ChIP assays revealed robust interaction of p107 to sequences in the PGC-1α proximal promoter in both proliferating myoblasts and in differentiated myocytes. In addition, transient transcription assays using luciferase reporters found that p107 overexpression inhibits transcription from a 3.1-kb proximal promoter of mouse PGC-1α in a Cdk-dependent manner. Indeed, p107 in a hypophosphorylated state when Cdks are inactivated 48 h after differentiation, and a mutant form that was deficient in modification by Cdk phosphorylation, were both significantly more efficient at repression.
Previously, we had documented the interaction and inhibition of the PGC-1α promoter activity by Rb in 3T3-L1 preadipocytes (Scimè et al., 2005). However, binding of Rb is not apparent in myoblasts. In BAT, there is evidence for enhanced RB phosphorylation, presumably by Cdks in cold-induced neodifferentiation when PGC-1α levels are increased (Hansen et al., 2004). In myoblasts, the PGC-1α promoter appears to be modulated directly by p107 interacting with bound transcription factors that are responsive to upstream signals unique to myogenic cells. The differences in control of the two tissue types are currently being investigated.
These results imply that myogenic and adipogenic control mechanisms for PGC-1α expression are similar, in that they involve the Rb family. Indeed, recent data suggest that brown adipocytes and skeletal muscle are closely related (Seale et al., 2008). Lineage tracing experiments found that Myf5, a factor once thought to arise only in the myogenic lineage, is also present in precursors of brown adipocytes. Moreover, PRD1-BF1-RIZ (PRDM16) controls a bidirectional cell fate switch between myoblasts and brown adipocytes.
The classical role for the Rb family is control of the cell cycle by direct repression of the E2F family of transcription factors, but they also have a function in differentiation and development. In vivo studies deleting Rb before and after myogenic differentiation reveal that it plays a crucial role in the switch from proliferation to differentiation rather than the maintenance of the terminally differentiated state (Huh et al., 2004). Until now, no data had existed for the role of the Rb family member p107 in skeletal muscle differentiation and maintenance. Our results provide for the first time a function for p107 in skeletal muscle that acts by controlling the pro-oxidative coactivator PGC-1α.
It is not known in what setting p107 controls slow type fiber formation, but many possibilities can be hypothesized. During muscle regeneration, the quiescent satellite cell, the primary adult myogenic stem cell, is activated and undergoes several proliferative cycles as myoblasts before fusing to form myotubes, which are the building blocks of myofibers (Scimè and Rudnicki, 2006). A subset of the myogenic precursors that are not committed to the myogenic program divide symmetrically to replenish the satellite stem cell pool (Le Grand et al., 2009).
The level where p107 functions in the myogenic developmental program remains to be determined. p107 might function during the myogenic specification of satellite cells to influence the fiber type choice of the resulting myoblast. Hence, inactivating p107 would predetermine that satellite cells differentiate into oxidative myofibers, as is the case in early embryogenesis (Stockdale, 1997). However, in skeletal muscle, p107 might affect fiber type determination at the level of myoblast commitment to fiber fate. Thus, differentiation of myoblasts with higher levels of p107 might promote less oxidative fiber type cells and conversely lower levels of more oxidative fibers. Indeed, myoblasts are not homogenous; e.g., myogenic progenitors within a muscle are specified to reflect the phenotype of the fiber type of origin (Wehrle et al., 1994; Rosenblatt et al., 1996; Kalhovde et al., 2005; Huang et al., 2006). p107 has been shown to bind with E2F4 at specific myogenic promoters that might potentially influence the outcome of myogenesis (Parakati and DiMario, 2005). Finally, p107 might control fiber type switching at the level of the differentiated fiber as a member of a sensing pathway that modulates signal transduction and gene transcription in the presence of metabolites, or by specific neuronal impulses (Cameron-Smith et al., 2003). This is consistent with the occupation and control of the PGC-1α promoter with p107. p107 might be involved in one of the multiple signaling pathways that are reprogrammed during muscle fiber type determination and/or switching. Increasing p107 in this circumstance might cause the dysregulation of a variety of metabolic genes, including decreasing expression levels of PGC-1α, contributing to the enhanced glycolytic nature of muscle fibers.
Understanding the underlying mechanisms of skeletal muscle fiber type determination can lead to effective prevention and treatment strategies for the escalating obesity crisis in society. Indeed, the identification of a central role for p107 in determining muscle fiber character raises the possibility that this pathway may be amenable for therapeutic manipulation for the treatment of obesity.
Materials and methods
HF diet
Experimental procedures were performed on mice with the Balb/c genetic background, including those with a targeted deletion of p107 (LeCouter et al., 1998). Procedures were performed according to the guidelines of the University of Ottawa. Fat pads were carefully dissected and weighed from 10-wk- and 1-yr-old mice unless stated. Adiposity was determined by dividing the total adipose tissue weight by the weight of the animal. Mice fed an HF diet that was made up of 60% fat (Harlan) for 2 mo were weighed every 3 d.
qPCR
For real-time qRT-PCR (qRT-PCR), RNA was first extracted from TA muscle of p107−/− and wild-type mice ranging from 12 to 16 wk in age using the RNeasy Fibrous Tissue kit (QIAGEN), or from primary myoblasts using the RNeasy kit (QIAGEN). 500 ng of RNA was reverse transcribed using the GeneAmp kit (Applied Biosystems). 25 ng of reverse-transcribed RNA was used for PCR analysis. Primer sets and conditions for PGC-1α, UCP-2, UCP-3 Glut4, Glut5, FABP3, and CPT1b have been described previously (Hansen et al., 2004; Lau et al., 2004). Primer sets for p107 were as follows: right primer, 5′-CAATGCTATAATGTGCCCAA-3′; and left primer, 5′-TAGGATTCCGCATACAAGAT-3′. Assays were performed on a qPCR system (MX 4000 [Stratagene] using SYBR green PCR Master mix [Sigma-Aldrich]). All reactions were run at least in triplicate and the results were graphed by normalizing to β-actin.
Histology
H&E and NADH reductase staining of tissue sections with NBT were performed according to standard protocol. Fiber type enumeration was accomplished by counting fluorescent stained fibers from frozen cross sections of TA, diaphragm, and pectoralis muscles. Antibodies used were mouse anti–type I MyHC (BF-34), anti–type IIa MyHC (SC-21), and anti–type IIb MyHC (BF-F3); and rabbit anti-Laminin (Sigma-Aldrich). Secondary antibodies used were Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 conjugated to specific IgG types (Invitrogen).
Microscopy
Images were obtained at room temperature using a microscope (Axioskop; Carl Zeiss, Inc.) with 10× NA 0.30 Plan-Neofluar (Carl Zeiss, Inc.) or 20× NA 0.50 Plan-Neofluar (Carl Zeiss, Inc.) objectives, and a microscope (Axioplan2; Carl Zeiss, Inc.) with a 20× NA 0.75 Plan-Apochromat (Carl Zeiss, Inc.) objective. Digital images were captured using Axiovision (Carl Zeiss, Inc.) and were processed with Photoshop (Adobe). ImageJ software was used for measuring the cross-sectional area of white adipocytes in tissue sections (n = 3).
In vivo electroporation
For the in vivo electro transfer, 40 µg of plasmid DNA in 0.9% NaCl or 0.9% NaCl (saline) was injected directly into one TA muscle for each mouse (2-mo-old BALB/c) that had been exposed by an incision through the skin. Immediately after injection, electric stimulation by a pulse generator (ECM 830; BTX) of 100 V for six pulses, with a fixed duration of 20 ms and an interval of 200 ms using 5-mm needle electrodes (BTX), was applied directly to the TA. 21 d after transfer, the electroporated and untreated contralateral TAs from each mouse were dissected and treated as with histology to enumerate the percentage of type IIa fibers. A total of three mice was used for each treatment.
Indirect calorimetry
Whole body oxygen consumption (VO2) and carbon dioxide production (VCO2) of mice was measured using an open circuit four-chamber indirect calorimetry system with automatic temperature and light controls (Columbus Instruments). All mice had ad libitum access to water, with the HF diet mice having ad libitum access to HF pellets and the chow diet fed mice having ad libitum access to a chow diet. Mice were placed in respiration chambers and data were recorded before and after 1 and 2 mo on an HF diet for a 24 h period with temperature at 24°C and light between 0700–1900 h. The RER was determined by the ratio of the volume of carbon dioxide eliminated per minute to the volume of oxygen taken into the lungs during the same time period over a 24 h period.
Quantitative ChIP
Quantitative ChIP was performed on growing and 2-d differentiated C2C12 myoblasts with an assay kit (Millipore) according to the manufacturer’s instructions. The promoter was immunoprecipitated by anti–rabbit p107 sc-318, RB sc-50, p130 sc-317, E2F1 sc-193, E2F4 sc-1082, and control Ig antibody (all from Santa Cruz Biotechnology, Inc.). ChIP primer sequences amplified a 311-bp fragment corresponding to −134 to −445 bp within the PGC-1α upstream promoter region (right primer, 5′-CCCCCGATTTTCTTTCTCTC-3′; and left primer, 5′-GGCTTGCTTGCTTTACAAGG-3′). The PCR conditions for quantitative ChIP were 40 cycles for 30 s at 95°C denaturation, 30 s at 56°C annealing, and 30 s at 72° extension.
Time course and Western blotting
For the differentiation time-course, the myoblast cell line C2C12 was differentiated in differentiation media: DME containing 2% horse serum or primary myoblasts with DME containing 5% horse serum for the stated times. For Western blotting, cells were lysed with Ripa buffer (0.5% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, and 50 mM Tris-HCl, pH 7.5), then the lysate was loaded onto a 7.5% polyacrylamide gel and blotted according to standard protocol with p107 c-20 (Santa Cruz Biotechnology, Inc.) or M2 anti-Flag (Sigma-Aldrich).
siRNA knockdown
For siRNA knockdown of p107, myoblasts were isolated from hind-limb muscles as previously described (Huh et al., 2004) and plated on collagen-coated dishes in Ham’s F10 medium supplemented with 20% FBS and 5 ng/ml of basic FGF (Invitrogen). At 80% confluency, the cells were transfected with either nonsilencing or p107 siRNA (TRCN0000218550; Sigma-Aldrich) with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. The myoblasts were differentiated 24 h after transfection in DME containing 5% horse serum for 3 d. The cells were lysed and equal amounts of protein were used for Western blotting with the following antibodies: anti-skeletal myosin (M7523; Sigma-Aldrich), MyHCIIa (A4.74; Developmental Studies Hybridoma Bank), MyHCIIb (BF-F3; American Type Culture Collection), and p107 (sc-318; Santa Cruz Biotechnology, Inc.). Quantification of Western blots was performed using the ImageJ software.
Luciferase assay
For promoter analysis, C2C12 cells were transfected using Superfect (QIAGEN) according to the manufacturer’s instructions. Cells were harvested 24 h after transfection or re-fed after transfection with differentiation media for 48 h before harvesting. The 3.1-kb proximal PGC1α–promoter–luciferase construct was provided by M. Czubryt (University of Manitoba, Winnipeg, Manitoba, Canada; Czubryt et al., 2003). Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega), with the pcDNA3 vector used as an empty vector control and for equilibrating transfection inputs. The Cdk2-deficient phosphorylation mutant p107A4, pcDNA-p107A4HA, was provided by W. Harper (Harvard Medical School, Boston, MA; Leng et al., 2002).
Fatty acid oxidation
The measurement of fatty acid oxidation ex vivo was done essentially as described previously (Dyck et al., 1997). In brief, gastrocnemius skeletal muscle was collected from knockout and wild-type animals and cut into small pieces of ∼50 mg. The tissue was treated in a 24-well plate with 1.5 ml of Krebs-Henseleit buffer, pH 7.4, containing 4% fatty acid–free bovine serum albumin (Sigma-Aldrich), 10 mM glucose, and 0.3 mM palmitate (9-10-3H, 5 mCi/ml) for 1.5 h at 37°C. Extraction was performed on 500 µl of buffer with the addition of 2.5 ml of chloroform/methanol (2:1) and 1 ml of 2M KCl:2M HCl to separate the organic from the aqueous phase. The mixture was vortexed at 3,000 g for 5 min, and 1 ml of aqueous phase was re-extracted. After final extraction, a 500-µl aliquot containing 3H2O was taken for scintillation counting, and the amount of palmitic acid oxidized was calculated as described previously (Dyck et al., 1997).
Online supplemental material
Fig. S1. shows that mice lacking p107 exhibit a lean metabolic phenotype. Graphical representations of the serum triglycerides, leptin, insulin, and free fatty acids from control and p107−/− mice are shown. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005076/DC1.
Acknowledgments
We thank Dr. Patrick Seale for critical reading of the manuscript.
M.A. Rudnicki holds the Canada Research Chair in Molecular Genetics and is an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by grants to M.A. Rudnicki from the Canadian Institutes of Health Research (MOP12080), and the Canada Research Chair program. C.F. Bentzinger is supported by a grant from the Swiss National Science Foundation (PBBSP3-127870).
Footnotes
Abbreviations used in this paper:
- BAT
- brown adipose tissue
- ChIP
- chromatin immunoprecipitation
- CPT1b
- carnitine palmitoyltransferase Ib
- FABP3
- fatty acid binding protein 3
- HF
- high fat
- MyHC
- myosin heavy chain
- NBT
- nitro-blue tetrazolium
- PGC-1α
- peroxisome proliferator–activated receptor gamma co-activator-1α
- qRT-PCR
- quantitative RT-PCR
- RER
- respiratory exchange ratio
- TA
- tibialis anterior
- UCP
- uncoupling protein
- WAT
- white adipose tissue
References
- Akimoto T., Pohnert S.C., Li P., Zhang M., Gumbs C., Rosenberg P.B., Williams R.S., Yan Z. 2005. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 280:19587–19593 10.1074/jbc.M408862200 [DOI] [PubMed] [Google Scholar]
- Baldi A., Esposito V., De Luca A., Fu Y., Meoli I., Giordano G.G., Caputi M., Baldi F., Giordano A. 1997. Differential expression of Rb2/p130 and p107 in normal human tissues and in primary lung cancer. Clin. Cancer Res. 3:1691–1697 [PubMed] [Google Scholar]
- Bassel-Duby R., Olson E.N. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37 10.1146/annurev.biochem.75.103004.142622 [DOI] [PubMed] [Google Scholar]
- Cameron-Smith D., Burke L.M., Angus D.J., Tunstall R.J., Cox G.R., Bonen A., Hawley J.A., Hargreaves M. 2003. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. Am. J. Clin. Nutr. 77:313–318 [DOI] [PubMed] [Google Scholar]
- Czubryt M.P., McAnally J., Fishman G.I., Olson E.N. 2003. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA. 100:1711–1716 10.1073/pnas.0337639100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais G.R., Tancredi R.G., Zierler K.L. 1976. Free fatty acid oxidation by forearm muscle at rest, and evidence for an intramuscular lipid pool in the human forearm. J. Clin. Invest. 58:421–431 10.1172/JCI108486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck D.J., Peters S.J., Glatz J., Gorski J., Keizer H., Kiens B., Liu S., Richter E.A., Spriet L.L., van der Vusse G.J., Bonen A. 1997. Functional differences in lipid metabolism in resting skeletal muscle of various fiber types. Am. J. Physiol. 272:E340–E351 [DOI] [PubMed] [Google Scholar]
- Hallenborg P., Feddersen S., Madsen L., Kristiansen K. 2009. The tumor suppressors pRB and p53 as regulators of adipocyte differentiation and function. Expert Opin. Ther. Targets. 13:235–246 10.1517/14712590802680141 [DOI] [PubMed] [Google Scholar]
- Hansen J.B., Jørgensen C., Petersen R.K., Hallenborg P., De Matteis R., Bøye H.A., Petrovic N., Enerbäck S., Nedergaard J., Cinti S., et al. 2004. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl. Acad. Sci. USA. 101:4112–4117 10.1073/pnas.0301964101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey M.S., Carey J.O., Azevedo J.L., Houmard J.A., Pories W.J., Israel R.G., Dohm G.L. 1995. Skeletal muscle fiber composition is related to adiposity and in vitro glucose transport rate in humans. Am. J. Physiol. 268:E453–E457 [DOI] [PubMed] [Google Scholar]
- Huang Y.C., Dennis R.G., Baar K. 2006. Cultured slow vs. fast skeletal muscle cells differ in physiology and responsiveness to stimulation. Am. J. Physiol. Cell Physiol. 291:C11–C17 10.1152/ajpcell.00366.2005 [DOI] [PubMed] [Google Scholar]
- Huh M.S., Parker M.H., Scimè A., Parks R., Rudnicki M.A. 2004. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166:865–876 10.1083/jcb.200403004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulver M.W., Berggren J.R., Cortright R.N., Dudek R.W., Thompson R.P., Pories W.J., MacDonald K.G., Cline G.W., Shulman G.I., Dohm G.L., Houmard J.A. 2003. Skeletal muscle lipid metabolism with obesity. Am. J. Physiol. Endocrinol. Metab. 284:E741–E747 [DOI] [PubMed] [Google Scholar]
- Hulver M.W., Berggren J.R., Carper M.J., Miyazaki M., Ntambi J.M., Hoffman E.P., Thyfault J.P., Stevens R., Dohm G.L., Houmard J.A., Muoio D.M. 2005. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2:251–261 10.1016/j.cmet.2005.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhovde J.M., Jerkovic R., Sefland I., Cordonnier C., Calabria E., Schiaffino S., Lømo T. 2005. “Fast” and “slow” muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. J. Physiol. 562:847–857 10.1113/jphysiol.2004.073684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E. 2005. Skeletal muscle fat oxidation: timing and flexibility are everything. J. Clin. Invest. 115:1699–1702 10.1172/JCI25758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., Goodpaster B., Wing R.R., Simoneau J.A. 1999. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am. J. Physiol. 277:E1130–E1141 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Hickner R.C., Cortright R.L., Dohm G.L., Houmard J.A. 2000. Lipid oxidation is reduced in obese human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 279:E1039–E1044 [DOI] [PubMed] [Google Scholar]
- Lau P., Nixon S.J., Parton R.G., Muscat G.E. 2004. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J. Biol. Chem. 279:36828–36840 10.1074/jbc.M404927200 [DOI] [PubMed] [Google Scholar]
- LeCouter J.E., Kablar B., Hardy W.R., Ying C., Megeney L.A., May L.L., Rudnicki M.A. 1998. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol. Cell. Biol. 18:7455–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F., Jones A.E., Seale V., Scimè A., Rudnicki M.A. 2009. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 4:535–547 10.1016/j.stem.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott C., Vidal-Puig A.J. 2004. Lipotoxicity, an imbalance between lipogenesis de novo and fatty acid oxidation. Int. J. Obes. Relat. Metab. Disord. 28:S22–S28 10.1038/sj.ijo.0802854 [DOI] [PubMed] [Google Scholar]
- Leng X., Noble M., Adams P.D., Qin J., Harper J.W. 2002. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol. Cell. Biol. 22:2242–2254 10.1128/MCB.22.7.2242-2254.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillioja S., Young A.A., Culter C.L., Ivy J.L., Abbott W.G., Zawadzki J.K., Yki-Järvinen H., Christin L., Secomb T.W., Bogardus C. 1987. Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. J. Clin. Invest. 80:415–424 10.1172/JCI113088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 418:797–801 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- Lin J., Handschin C., Spiegelman B.M. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361–370 10.1016/j.cmet.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P.A. 2003. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 17:2299–2301 [DOI] [PubMed] [Google Scholar]
- Marra M., Scalfi L., Contaldo F., Pasanisi F. 2004. Fasting respiratory quotient as a predictor of long-term weight changes in non-obese women. Ann. Nutr. Metab. 48:189–192 10.1159/000079556 [DOI] [PubMed] [Google Scholar]
- Mulligan G., Jacks T. 1998. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 14:223–229 10.1016/S0168-9525(98)01470-X [DOI] [PubMed] [Google Scholar]
- Nyholm B., Qu Z., Kaal A., Pedersen S.B., Gravholt C.H., Andersen J.L., Saltin B., Schmitz O. 1997. Evidence of an increased number of type IIb muscle fibers in insulin-resistant first-degree relatives of patients with NIDDM. Diabetes. 46:1822–1828 10.2337/diabetes.46.11.1822 [DOI] [PubMed] [Google Scholar]
- Parakati R., DiMario J.X. 2005. Dynamic transcriptional regulatory complexes, including E2F4, p107, p130, and Sp1, control fibroblast growth factor receptor 1 gene expression during myogenesis. J. Biol. Chem. 280:21284–21294 10.1074/jbc.M410744200 [DOI] [PubMed] [Google Scholar]
- Pette D., Staron R.S. 2001. Transitions of muscle fiber phenotypic profiles. Histochem. Cell Biol. 115:359–372 [DOI] [PubMed] [Google Scholar]
- Pilegaard H., Saltin B., Neufer P.D. 2003. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J. Physiol. 546:851–858 10.1113/jphysiol.2002.034850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 92:829–839 10.1016/S0092-8674(00)81410-5 [DOI] [PubMed] [Google Scholar]
- Reue K., Phan J. 2006. Metabolic consequences of lipodystrophy in mouse models. Curr. Opin. Clin. Nutr. Metab. Care. 9:436–441 10.1097/01.mco.0000232904.82038.db [DOI] [PubMed] [Google Scholar]
- Rosenblatt J.D., Parry D.J., Partridge T.A. 1996. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 60:39–45 10.1046/j.1432-0436.1996.6010039.x [DOI] [PubMed] [Google Scholar]
- Russell A.P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Dériaz O. 2003. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 52:2874–2881 10.2337/diabetes.52.12.2874 [DOI] [PubMed] [Google Scholar]
- Ryder J.W., Bassel-Duby R., Olson E.N., Zierath J.R. 2003. Skeletal muscle reprogramming by activation of calcineurin improves insulin action on metabolic pathways. J. Biol. Chem. 278:44298–44304 10.1074/jbc.M304510200 [DOI] [PubMed] [Google Scholar]
- Scimè A., Rudnicki M.A. 2006. Anabolic potential and regulation of the skeletal muscle satellite cell populations. Curr. Opin. Clin. Nutr. Metab. Care. 9:214–219 10.1097/01.mco.0000222102.21385.7d [DOI] [PubMed] [Google Scholar]
- Scimè A., Grenier G., Huh M.S., Gillespie M.A., Bevilacqua L., Harper M.E., Rudnicki M.A. 2005. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2:283–295 10.1016/j.cmet.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H.M., Erdjument-Bromage H., et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 454:961–967 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale F.E. 1997. Mechanisms of formation of muscle fiber types. Cell Struct. Funct. 22:37–43 10.1247/csf.22.37 [DOI] [PubMed] [Google Scholar]
- Tanner C.J., Barakat H.A., Dohm G.L., Pories W.J., MacDonald K.G., Cunningham P.R., Swanson M.S., Houmard J.A. 2002. Muscle fiber type is associated with obesity and weight loss. Am. J. Physiol. Endocrinol. Metab. 282:E1191–E1196 [DOI] [PubMed] [Google Scholar]
- Thyfault J.P., Kraus R.M., Hickner R.C., Howell A.W., Wolfe R.R., Dohm G.L. 2004. Impaired plasma fatty acid oxidation in extremely obese women. Am. J. Physiol. Endocrinol. Metab. 287:E1076–E1081 10.1152/ajpendo.00177.2004 [DOI] [PubMed] [Google Scholar]
- Ukropcova B., McNeil M., Sereda O., de Jonge L., Xie H., Bray G.A., Smith S.R. 2005. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J. Clin. Invest. 115:1934–1941 10.1172/JCI24332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R.B., Huss J.M., Kelly D.P. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20:1868–1876 10.1128/MCB.20.5.1868-1876.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle U., Düsterhöft S., Pette D. 1994. Effects of chronic electrical stimulation on myosin heavy chain expression in satellite cell cultures derived from rat muscles of different fiber-type composition. Differentiation. 58:37–46 10.1046/j.1432-0436.1994.5810037.x [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Spiegelman B.M. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 98:115–124 10.1016/S0092-8674(00)80611-X [DOI] [PubMed] [Google Scholar]
- Zurlo F., Lillioja S., Esposito-Del Puente A., Nyomba B.L., Raz I., Saad M.F., Swinburn B.A., Knowler W.C., Bogardus C., Ravussin E. 1990. Low ratio of fat to carbohydrate oxidation as predictor of weight gain: study of 24-h RQ. Am. J. Physiol. 259:E650–E657 [DOI] [PubMed] [Google Scholar]