In response to tissue stiffening, fibroblasts increase production of extracellular matrix while decreasing production of matrix-degrading enzymes and the fibrosis inhibitor prostaglandin E2.
Abstract
Tissue stiffening is a hallmark of fibrotic disorders but has traditionally been regarded as an outcome of fibrosis, not a contributing factor to pathogenesis. In this study, we show that fibrosis induced by bleomycin injury in the murine lung locally increases median tissue stiffness sixfold relative to normal lung parenchyma. Across this pathophysiological stiffness range, cultured lung fibroblasts transition from a surprisingly quiescent state to progressive increases in proliferation and matrix synthesis, accompanied by coordinated decreases in matrix proteolytic gene expression. Increasing matrix stiffness strongly suppresses fibroblast expression of COX-2 (cyclooxygenase-2) and synthesis of prostaglandin E2 (PGE2), an autocrine inhibitor of fibrogenesis. Exogenous PGE2 or an agonist of the prostanoid EP2 receptor completely counteracts the proliferative and matrix synthetic effects caused by increased stiffness. Together, these results demonstrate a dominant role for normal tissue compliance, acting in part through autocrine PGE2, in maintaining fibroblast quiescence and reveal a feedback relationship between matrix stiffening, COX-2 suppression, and fibroblast activation that promotes and amplifies progressive fibrosis.
Introduction
Fibrosis is a pathobiological process common to many human diseases characterized by the progressive replacement of normal parenchymal tissue with collagen-rich extracellular matrix (Desmoulière et al., 2003). Fibrogenesis occurs through the aberrant accumulation and activation of myofibroblasts, which deposit extracellular matrix components and remodel the tissue (Kuhn and McDonald, 1991; Zhang et al., 1994), culminating in increased tissue mechanical stiffness (Gross and Hunninghake, 2001; Tomasek et al., 2002). In its most severe clinical manifestations, fibrosis can reach a progressive and self-sustaining phase, the cause of which remains largely mysterious.
Much attention has focused on the role that soluble inflammatory and fibrogenic mediators play in the initiation and progression of fibrosis (Wynn, 2008). However, recent observations have highlighted the fact that variations in matrix stiffness potently alter fibroblast morphology, proliferation, TGF-β signaling, and myofibroblast activation (Arora et al., 1999; Wang et al., 2000; Paszek et al., 2005; Goffin et al., 2006; Wipff et al., 2007). These findings raise the intriguing possibility that stiffening of the mechanical environment, long regarded only as an outcome of fibrosis, might play a pivotal role in driving the cellular behaviors that promote, amplify, and perpetuate fibrosis. In the rat liver, macroscale measurements of organ stiffness during onset of carbon tetrachloride–induced fibrosis indicate that stiffening occurs rapidly and may even precede measurablefibrosis, suggesting a possible role for stiffening in the initial stages of fibrosis (Georges et al., 2007). In the lung, fibrotic lesions are heterogeneously distributed, and although it is known that fibrosis alters gross tissue mechanics (Bachofen and Scherrer, 1967), measurements of stiffness at the local microscale relevant to resident cells are not available. Thus, the role of matrix stiffening in promoting lung fibrogenesis remains untested.
To assess the role that pathophysiological matrix stiffening plays in the promotion of fibrogenesis, we designed experiments to measure the stiffness of normal and fibrotic lung tissue and investigated key fibrogenic behaviors of human lung fibroblasts grown on substrates spanning this stiffness range. Our results demonstrate a pivotal role for matrix stiffening in promotion of fibroblast proliferation and matrix synthesis and uncover stiffness-driven suppression of COX-2 (cyclooxygenase-2) expression and PGE2 (prostaglandin E2) synthesis as a key link between matrix stiffening and fibroblast activation. We further show that PGE2 and Rho kinase (ROCK) exert mutually antagonistic effects on fibroblasts, leading to stable and diametrically opposed quiescent and activated states influenced by the stiffness of the underlying matrix. Because matrix stiffening promotes self-reinforcing effects on fibroblast matrix synthesis, our findings show how feedback signaling in the context of mutually antagonistic signaling cascades can switch fibroblasts from a quiescent state to self-sustaining activation supportive of progressive fibrosis.
Results
Stiffness of normal and fibrotic lung tissue
To establish baseline data on the homogeneity and variance of lung parenchymal tissue stiffness at a scale relevant to resident cells, we prepared thin slices of fresh murine lung tissue and applied atomic force microscopy (AFM) microindentation to map local elastic properties (Fig. 1). To compare normal and fibrotic tissue stiffness, samples were harvested 14 d after intratracheal administration of either saline or bleomycin. Administration of the latter is well recognized to induce pulmonary fibrosis in a heterogeneous pattern that mimics aspects of human pulmonary fibrosis (Moore and Hogaboam, 2008). Consistent with prior work in mice, bleomycin-treated lungs exhibited focal increases in expression of collagen I (Fig. 1 A) and the myofibroblast marker α–smooth muscle actin (α-SMA; not depicted; Zhang et al., 1994; Pena et al., 2007; Moore and Hogaboam, 2008). Stiffness mapping was performed in areas devoid of major airways or blood vessels (Fig. 1 A); in bleomycin-treated lungs, mapping was restricted (except where noted) to areas of grossly evident fibrosis.
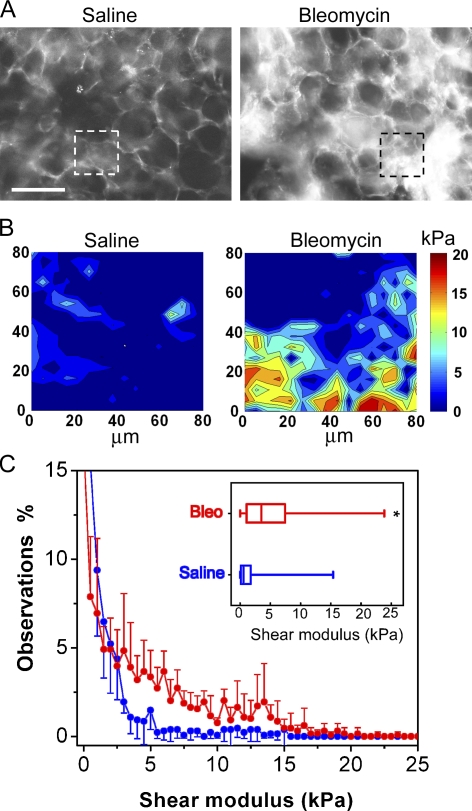
Figure 1.
Fibrosis focally increases stiffness of lung parenchymal tissue. (A) Immunostaining of collagen I in fresh, unfixed saline (left)- and bleomycin (right)-treated mouse lung parenchyma. Bar, 100 µm. (B) Representative elastographs of saline (left)- and bleomycin (right)-treated mouse lung parenchyma. Maps were made from tissue in the respective regions of interest identified in A. The color bar indicates shear modulus. Darkest blue corresponds to shear modulus values between 0.05 and 2 kPa. (C) Occurrence frequency analysis of stiffness in saline- and bleomycin (Bleo)-treated lung tissue. Data indicate mean ± SD of stiffness measurements pooled from five animals each for normal and fibrotic groups in two independent bleomycin injection experiments. (Inset) Stiffness increases in fibrotic lung parenchyma presented as box and whisker plots, in which the vertical line within each box represents the median, the limits of each box represent the interquartile range, and the whiskers represent the maximum and minimum values. *, P < 0.0001 comparing stiffness of fibrotic versus normal lung parenchyma using the Mann-Whitney-Wilcoxon test.
Elastographs from AFM microindentation mapping demonstrated striking differences in the range and distribution of tissue stiffness in normal and fibrotic lung parenchyma (Fig. 1 B and Fig. S1). The parenchymal tissue from saline-treated lungs was highly compliant (median shear modulus ∼0.5 kPa), with only rare portions of the mapping area exhibiting stiffness >3 kPa. These observations, the first made on intact lung parenchyma at the microscale, are consistent with a long-standing model of lung mechanics in which alveolar walls are assumed to offer negligible contributions to lung elasticity compared with the extracellular matrix–rich connective tissue sheath that connects the alveolar ducts, airways, and pleura (Wilson and Bachofen, 1982). In bleomycin-treated lungs, regions rich in collagen I expression exhibited an overall increase in shear modulus to a median 3 kPa, with highly localized increases in stiffness to >15 kPa (Fig. 1 C and Fig. S1 B). These focal changes in tissue stiffness far exceed the two- to threefold increases in stiffness observed previously in tissue strips from bleomycin-treated lungs (Ebihara et al., 2000). Local increases in stiffness were restricted to collagen-enriched regions; when we examined the tissue stiffness of bleomycin-treated lungs in areas devoid of fibrosis, the shear modulus was similar to that observed in saline-treated lungs (Fig. S1 C). Together, the elasticity mapping results emphasize the focal nature of stiffness changes in early fibrotic lesions and establish an unexpectedly large range in tissue stiffness between highly compliant normal lung parenchyma and locally stiffened fibrotic lesions.
Regulation of fibroblast accumulation by matrix stiffness
To elucidate the effects of variations in stiffness on lung fibroblast biology, we designed and fabricated gradient stiffness hydrogel substrates by using photopolymerization of polyacrylamide through gradient masks, an approach adapted from Wong et al. (2003). The resulting hydrogels exhibited a 1D gradient in shear moduli spanning 0.1–50 kPa over a distance of ∼6 mm (Fig. S2 A). The stiffness range was selected to encompass and surpass that observed in the murine bleomycin model to account for the increasing stiffness of extensively cross-linked mature fibrotic lesions (Dunn et al., 1985). To permit cell attachment, the inert polyacrylamide surface was derivatized with collagen I using a heterobifunctional cross-linker. The uniformity of the collagen I surface density was confirmed by immunostaining with collagen I primary antibody, followed by secondary antibody conjugated to fluorescently labeled beads that interact only with surface-available antigens (Fig. S2; Pelham and Wang, 1997).
Human lung fibroblasts (CCL-151) attached uniformly across the stiffness gradient, with only modest variability in the degree of cell spreading apparent at 4 h (Fig. 2 A). However, incubation of fibroblasts on these stiffness gradients for 120 h in the presence of 10% FBS gave rise to striking patterns of stiffness-dependent fibroblast morphology and accumulation (Fig. 2 A). Increasing stiffness was accompanied by prominent transitions in fibroblast morphology from attenuated, rounded cells at low stiffness to an appearance marked by multiple dendritic processes at intermediate stiffness (Fig. 2 A). At stiffness levels of 20–50 kPa, fibroblasts aligned in parallel swirls of spindle-shaped cells typical of patterns observed in 3D fibroblastic foci in vivo (Kuhn and McDonald, 1991; Gross and Hunninghake, 2001) and on standard rigid cell culture substrates. The crossover from net cell loss at low stiffness to cell accumulation at high stiffness identified an equilibrium point where cell migration, proliferation, and death were in balance under the given culture conditions (Fig. 2 B). Intriguingly, we observed this intersection to occur reproducibly at shear moduli in the range of 0.5–3 kPa, which is only slightly higher than the median shear modulus measured in normal lung tissue. The pattern of stiffness-dependent accumulation was attenuated when serum was diluted (Fig. 2 C), indicating that both soluble and mechanical factors were necessary elements in the response. However, addition of the fibrogenic cytokine TGF-β1 did not alter the pattern of cell accumulation anywhere along the stiffness gradient (Fig. 2 C). Together, these results suggest that the compliance of the normal lung itself protects against fibroblast proliferation, even in the presence of a growth-promoting or fibrogenic-soluble environment, but that only relatively small changes in stiffness are needed to shift lung fibroblasts into a mode of rapid accumulation.
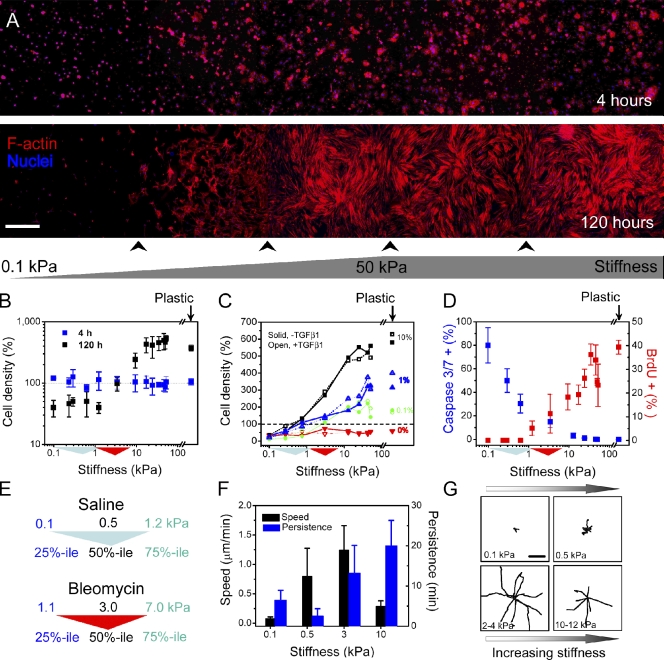
Figure 2.
Fibroblasts preferentially accumulate across stiffness gradient. (A) Human lung fibroblasts stained with phalloidin (red) to visualize F-actin and Hoechst 33342 (blue) to visualize nuclei. Cells attach uniformly across the stiffness gradient 4 h after seeding. After 120 h, fibroblasts accumulate preferentially to stiffest region, and cell morphology transitions from round to spread. Panorama images were generated by imaging the entire gel width along the stiffness gradient and then tiling five to seven adjacent pictures. Arrowheads below the image indicate stitching positions. (B) Cell density is constant across the gradient after 4 h, but by 120 h, cells dramatically accumulate at stiffness >3 kPa and are lost below this stiffness level (note logarithmic scale). Density values are normalized to the global mean at 4 h. (C) Serum but not TGF-β1 is required for stiffness-dependent cell accumulation. Serial dilutions of serum gradually attenuate the stiffness-dependent accumulation behavior (black, 10% FBS; blue, 1% FBS; green, 0.1% FBS; red, 0% FBS). 2 ng/ml exogenous TGF-β1 shows little effect on cell density (open symbols and dotted lines) at any serum concentration. (B and C) Dashed lines indicate that cell density values are normalized to the global average obtained at 4 h. (D) Quantification of substrate stiffness effects on fibroblast apoptosis (blue indicates percentage of cells exhibiting caspase 3/7 activity after 24-h serum deprivation) and proliferation (red indicates percentage of nuclei positive for BrdU incorporation in 10% FBS). Standard deviation is from three independent experiments. (E) In panels B–D, colored triangles along the x axis are used to indicate the interquartile and median stiffness values of lung parenchyma from saline (cyan)- or bleomycin (red)-treated mice. (F) Fibroblast migration speed and persistence vary along substrate stiffness as measured with time-lapse video microscopy. Error bars indicate SD from 12 cells for each condition from two independent experiments. (G) Fibroblast migration tracks on stiffness gradient gels obtained from time-lapse video microscopy. Digital images were taken every 2 min for a total of 5 h per experiment. Each wind rose plot shows centroid tracks from 7–10 representative cells from each indicated stiffness region, with the initial position of each track superimposed at a common origin. Bars: (A) 500 µm; (G) 50 µm.
To elucidate the cellular processes contributing to the stiffness-dependent accumulation of fibroblasts, we compared cell proliferation, apoptosis, and migration across the stiffness gradient. BrdU-positive cells, indicative of ongoing DNA synthesis, were absent from areas of stiffness equivalent to normal median lung stiffness levels even in the presence of 10% FBS but were detected with increasing frequency as substrate stiffness increased (Fig. 2 D and Fig. S3 A). In contrast, serum depletion for 24 h revealed the opposite pattern of staining for active caspases 3/7, end effectors of apoptosis (Fig. 2 D and Fig. S3 B), with decreased staining observed as substrate stiffness increased. Consistent with stiffness-dependent protection from apoptosis, a similar but more muted distribution of active caspase 3/7 was observed across the stiffness gradient in the presence of serum (unpublished data). These results concur with and considerably extend prior findings in 2D (Wang et al., 2000) and 3D (Fringer and Grinnell, 2001) systems by demonstrating that a critical shift in bias from fibroblast apoptosis to proliferation occurs within the pathophysiological stiffness range measured in normal and fibrotic lung parenchyma.
Because cells have previously been shown to preferentially migrate toward stiffer regions when cultured on substrates with sharp stiffness gradients (Lo et al., 2000), we measured lung fibroblast migration at various locations along the stiffness gradient. Although fibroblast migration speed and persistence varied with substrate stiffness (Fig. 2 F), consistent with prior observations using vascular smooth muscle cells (Peyton and Putnam, 2005), no preferential migration relative to the direction of the stiffness gradient was observed (Fig. 2 G). A similar lack of preferential migration has been observed in vascular smooth muscle cells on stiffness gradients of similar slope (Isenberg et al., 2009). Although we cannot rule out a role for stiffness-directed migration in the steeper stiffness gradients found in the intact lung, in the current experiments, the predominant factor in stiffness-dependent accumulation appeared to be the shift in balance between apoptosis and proliferation. Subsequent observations of fibroblast accumulation in wells of discrete stiffness, where migration along a stiffness gradient was not a factor, revealed similar patterns to those observed in the stiffness gradient, which is consistent with a dominant role for proliferation over migration (see Matrix stiffness promotes fibrogenesis through COX-2 suppression).
Regulation of fibroblast matrix synthesis by stiffness
Fibroblasts are the major source for excess deposition of extracellular matrix components, including collagen I, in fibrotic disorders (Hinz et al., 2007). We observed a gradual increase in procollagen I immunofluorescence staining in cells along a continuous stiffness gradient (Fig. 3, A and B; and Fig. S3 C), starting from no detectable procollagen I signal at physiological median stiffness (∼0.5 kPa). Analysis of mRNA from fibroblasts grown on discrete stiffness gels demonstrated parallel increases in COL1A1 and COL3A1 transcripts (encoding collagen I[α1] and collagen III[α1] chains) with increasing stiffness and a prominent decrease in expression of MMP1 (encoding collagenase-1; Fig. 3 C). Although stiffness-mediated expression of collagen gene expression has been noted previously (Li et al., 2007), our results extend this observation by demonstrating enhanced procollagen protein expression and parallel decreases in gene expression of a key collagenolytic enzyme with increasing stiffness. Further supporting a matrix synthetic effect of increasing stiffness, we found that net secretion of collagen increased on a per cell basis as matrix stiffness increased on discrete stiffness gels (Fig. 3 D).
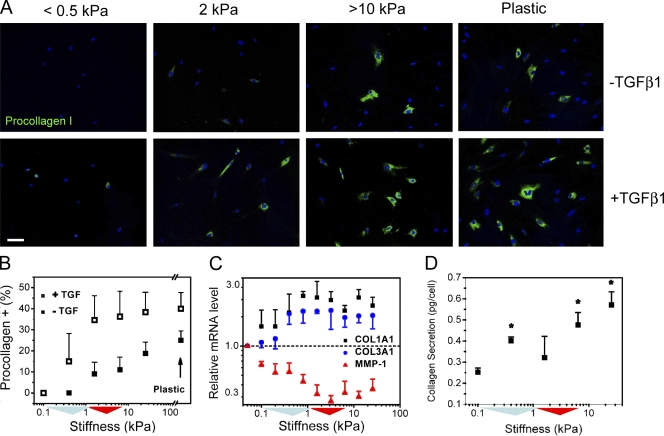
Figure 3.
Collagen synthesis increases across stiffness gradient. (A) Gradual increase of procollagen I protein expression in cells along the stiffness gradient. Nuclei are counterstained with Hoechst 33342 (blue). 2 ng/ml exogenous TGF-β1 shifts procollagen I protein expression to lower stiffness and enhances expression on stiffer substrate regions (10% FBS). Bar, 100 µm. (B) Quantification of substrate stiffness effects on procollagen I protein expression in the presence (open symbols) and absence (closed symbols) of 2 ng/ml exogenous TGF-β1 for cells grown in 10% FBS. (C) Expression of COL1A1 and COL3A1 transcripts increase with substrate stiffness, whereas MMP1 transcripts decrease with increasing stiffness for cells grown in 0.1% FBS. The dashed line indicates that the gene expression values are normalized to the values from the softest substrate. Error bars indicate SD from four replicate samples from one representative experiment. (D) Net collagen secretion per cell from fibroblasts grown on discrete stiffness gels over 72 h, cells grown in 1% FBS. *, P < 0.05 compared with 0.1 kPa. (B and D) Error bars indicate SD from two independent experiments. (B–D) Colored triangles along the x axis are used to indicate the interquartile and median stiffness values of lung parenchyma from saline (cyan)- or bleomycin (red)-treated mice.
Supplementation of culture medium with exogenous TGF-β1 for 3 d shifted procollagen I protein expression to as low as 0.5 kPa (Fig. 3 A) and increased expression across the entire stiffness gradient (Fig. 3 B). However, the positive relationship between stiffness and procollagen I expression was maintained across the pathophysiological stiffness range even in the presence of supplemental TGF-β1. This finding demonstrates cooperative enhancement of matrix synthesis through the action of soluble and mechanical signals and echoes previous examples of cooperative interactions between soluble and mechanical signals in myofibroblast activation (Arora et al., 1999; Goffin et al., 2006). Together, these results demonstrate that physiological levels of lung tissue stiffness act as a powerful brake on procollagen I synthesis but that either exogenous TGF-β or matrix stiffening alone can overcome this physiological brake, with enhanced augmentation of matrix synthesis when both stimuli are present.
Stiffness-dependent fibroblast gene expression
To more rigorously test the assertion that increasing stiffness promotes expression of a fibrogenic cellular program, we performed a microarray analysis of RNA harvested from normal human lung fibroblasts (NHLFs) grown on five discrete stiffness conditions spanning 100–25,600 Pa in fourfold increments. Cells grown from three independent human donors were used, and we focused our attention on genes with cross-donor rank correlation among stiffness conditions of >0.4, limiting analysis to the 2,462 probes (representing 1,829 unique genes) expressed in a consistent fashion among donors. We further restricted our attention to genes with a minimum within-donor coefficient of variation (of their exponentiated, base 2, robust multichip average [RMA] signal) across stiffness conditions of ≥0.2, resulting in a selection of 192 probes (150 of which have a gene assignment), representing 124 unique genes which we classified as responsive to alterations in stiffness across all three donors. Hierarchical clustering based on linear correlation across the three donors identified two major clusters of genes, those whose expression generally increased with stiffness (Fig. S4 A) and those that decreased (Fig. S4 C) with increasing stiffness.
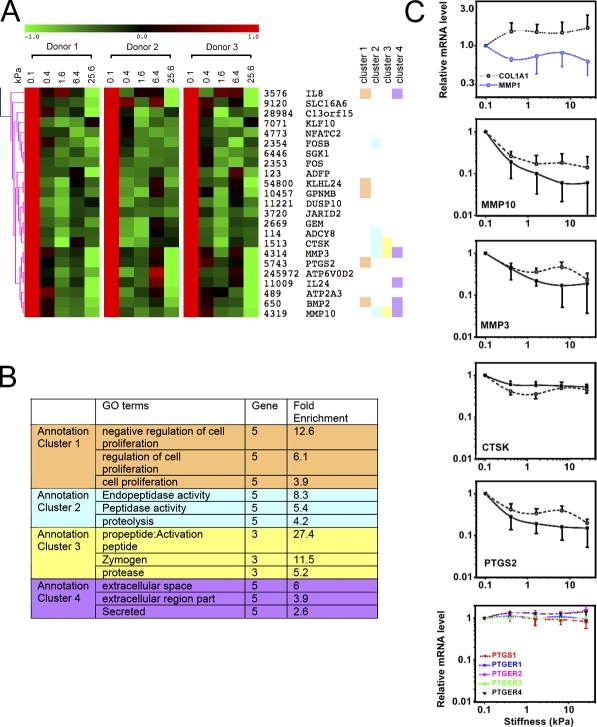
Ontological analysis of the cluster of genes increasing with stiffness identified highly significant enrichment for mitosis and microtubule motor activity (Fig. S4 B), which is consistent with our prior measurement of enhanced cell division across increasing matrix stiffness (Fig. 2 D). A similar analysis of the subcluster of genes most prominently attenuated with increasing stiffness (Fig. 4 A) identified negative regulation of proliferation as a highly enriched ontology, as well as several functional annotations related to propeptide activation and proteolytic activity in the extracellular space (Fig. 4 B). These ontologies were conspicuous for their inclusion of gene members MMP3, MMP10, and cathepsin K (CTSK), all of which are proteolytic for matrix proteins, including gelatin and fibrillar collagens. CTSK in particular has been shown to play a key role in protecting against lung fibrosis (Bühling et al., 2004). The microarray analysis also confirmed the changes in COL1A1 and MMP1 noted previously (Fig. 3 C), although the changes were modest relative to the strongest signals emerging from the microarray analysis (Fig. 4 C). Analysis by quantitative PCR (qPCR) confirmed the changes in transcript levels for MMP10, MMP3, and CTSK, along with PTGS2 across stiffness conditions (Fig. 4 C). Collectively, these data suggest that matrix stiffening drives a broad gene expression program in support of fibroblast proliferation and selective but coordinated suppression of genes encoding collagen-degrading enzymes. Coupled with our demonstration that stiffness enhances procollagen synthesis and suppresses MMP1 expression (Figs. 3 and 4), our findings are consistent with a potentially devastating adverse feedback loop in which progressive matrix stiffening begets enhanced matrix deposition and decreased matrix degradation by fibroblasts, promoting self-sustaining fibrosis.
Figure 4.
Stiffness drives coordinated expression of a fibrogenic gene program. (A) Gene expression cluster of down-regulated transcripts in response to increasing substrate stiffness from NHLFs (0.1% FBS). Each row displays relative expression levels from three donors for each gene, which has a minimum coefficient variance of 0.2 per donor. Columns within each donor represent gene expression for stiffness effect at 0.1, 0.4, 1.6, 6.4, and 25.6 kPa. Genes are listed with their EntrezID, followed by their symbols according to the HUGO nomenclature. Hierarchical clustering is based on linear correlation across the three donors. (B) Top four annotation clusters from functional annotation clustering analysis of genes in A. Corresponding genes within each annotation cluster are marked with the same color next to the gene name in A. (C) Selected stiffness-regulated gene expression from array analysis. In panels with both solid and dashed lines, dashed lines represent array values averaged from three donors, and solid lines represent replicate analysis by real time qPCR using cells from donor 1. Data shown are means relative to the values at the stiffness 0.1 kPa. The final panel shows microarray data from three donors for PTGS1 and prostanoid receptors PTGER1–4. Error bars on dashed lines indicate SD from three donors; error bars on solid lines indicate SD from three independent experiments using cells from donor 1.
Because fibroblasts interacting with matrices of increasing stiffness have been shown to alter activation of local matrix-incorporated TGF-β (Wipff et al., 2007), we sought to compare stiffness response genes to the transcriptional changes evoked by treatment with exogenous TGF-β. We analyzed a previously published dataset reporting TGF-β1–responsive genes in NHLFs (Kapoun et al., 2006), identifying 1,176 unique genes responsive to this stimulus. Although there are subtle differences in culture conditions and passage number that should be noted in comparing our results with the TGF-β–responsive gene list, of 124 stiffness-responsive genes, only 33 genes were identified as also responsive to TGF-β1 (Fig. S4). Strikingly, among these 33 genes, expression levels of 18 were driven in opposite directions by the two stimuli (e.g., increasing with stiffness and decreasing with TGF-β). This comparative analysis strongly suggests that the response to substrate stiffness identified in our microarray analysis is largely independent from a TGF-β–driven transcriptional program.
Matrix stiffness promotes fibrogenesis through COX-2 suppression
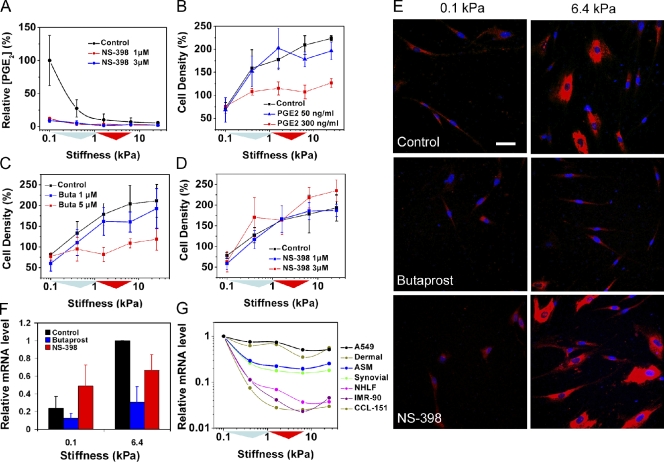
One intriguing candidate gene that emerged from the microarray analysis was PTGS2 (Fig. 4 C), which encodes the COX-2 enzyme. The COX enzymes convert arachidonic acid into prostaglandin H2, which is further processed into various prostaglandins, including PGE2. Notably, PGE2 is the most abundant prostaglandin in the airways, is present at high concentrations in healthy lungs, and is the predominant prostaglandin product of alveolar epithelial cells and lung fibroblasts (Ozaki et al., 1987; Huang and Peters-Golden, 2008). We confirmed that increasing matrix stiffness dramatically attenuates PTGS2 expression by qPCR (>80% reduction; Fig. 4 C), and extended this to show greatly diminished autocrine levels of PGE2 present in the supernatants of cultured fibroblasts on matrices of increasing stiffness (Fig. 5 A). The PGE2 levels were largely dependent on COX-2 activity, as selective inhibition of COX-2 with NS-398 (IC50 for COX-2 is 1.8 µM and for COX-1 is 75 µM) reduced soluble PGE2 levels by 90–100% at low stiffness. We did not observe stiffness-dependent changes in expression of either COX-1 (PTGS1) or prostanoid receptor (PTGER1–4) levels (Fig. 4 C), further confirming that suppression of COX-2 expression was the dominant factor in the attenuation of PGE2 levels with increasing stiffness.
Figure 5.
PGE2 regulates fibroblast responses to increasing stiffness. (A) Relative PGE2 levels in NHLF cell culture supernatants with and without selective COX-2 inhibitor (NS-398). (B–D) NHLF accumulation in discrete stiffness wells (1% FBS) and modification by exogenous PGE2, EP2 agonist (butaprost; Buta), and COX-2 inhibitor (NS-398), respectively. (E) Immunostaining for procollagen I in NHLFs (1% FBS). EP2 agonist (5 µM butaprost) abrogates the stiffness effect on procollagen I expression, whereas COX-2 selective inhibitor (3 µM NS-398) is unable to augment procollagen I expression at low stiffness. Bar, 50 µm. (F) Relative mRNA levels for COL1A1, assessed by qPCR, on 0.1 and 6.4 kPa substrates (0.1% FBS) with and without 5 µM butaprost or 3 µM NS-398. (A–D and F) Error bars indicate SD from three independent experiments for each panel. (G) COX-2 (PTGS2) mRNA levels decrease across stiffness, as measured by qPCR, in A549 human lung epithelial cells, primary human dermal, synovial and lung (NHLF) fibroblasts, and airway smooth muscle (ASM) cells, as well as CCL-151 and IMR90 lung fibroblast cell lines (0.1% FBS). (A–D and G) Colored triangles along the x axis are used to indicate the interquartile and median stiffness values of lung parenchyma from saline (cyan)- or bleomycin (red)-treated mice.
PGE2 has been shown to suppress fibroblast proliferation and collagen synthesis by signaling through the EP2 prostanoid receptor (Choung et al., 1998; Huang et al., 2007). We confirmed that suppression of PGE2 levels was a necessary step in stiffness-induced fibroblast accumulation by treating with exogenous PGE2 (Fig. 5 B) or the EP2 receptor agonist butaprost (EC50 5 µM; Fig. 5 C). Both treatments largely equilibrated fibroblast spatial density across a range of discrete matrix stiffness conditions from 100 to 25,600 Pa in a dose-dependent fashion. Interestingly, suppression of endogenous COX-2 activity with NS-398 had little effect on fibroblast accumulation and was unable by itself to significantly enable proliferation on physiological stiffness conditions (Fig. 5 D). Thus, suppression of COX-2 and PGE2 alone, in the absence of increased matrix stiffness, is insufficient to engage fibroproliferation, indicating a dominant role for stiffness in holding this process in check.
We found similar effects of COX-2 and PGE2 on procollagen synthesis across discrete matrix stiffness conditions. Exogenous PGE2 (not depicted) and treatment with butaprost were both able to suppress stiffness-induced increases in procollagen expression, whereas NS-398 alone was unable to engage substantial procollagen synthesis on physiological stiffness substrates (Fig. 5 E and Fig. S5). These results were largely mirrored at the message level by changes in COL1A1 (Fig. 5 F).
Although the suppressive effects of COX-2 and PGE2 on fibrogenesis and fibroproliferation are best known in the context of lung fibrosis (Vancheri et al., 2004; Huang and Peters-Golden, 2008), limited evidence also implicates this axis in fibrogenesis in other tissues and contexts (Hui et al., 2006; Sandulache et al., 2007; Liu et al., 2009). Therefore, we tested whether the stiffness-dependent suppression of PTGS2 was a feature common to multiple cell types. We found that increasing matrix stiffness suppressed PTSG2 expression to varying degrees in primary human dermal fibroblasts, synovial fibroblasts, airway smooth muscle cells, and the A549 human lung epithelial cell line, along with similar changes in NHLF, CCL-151, and IMR90 human lung fibroblasts (Fig. 5 G). These findings suggest that stiffness-dependent suppression of COX-2 expression and prostaglandin signaling may have potentially broad relevance.
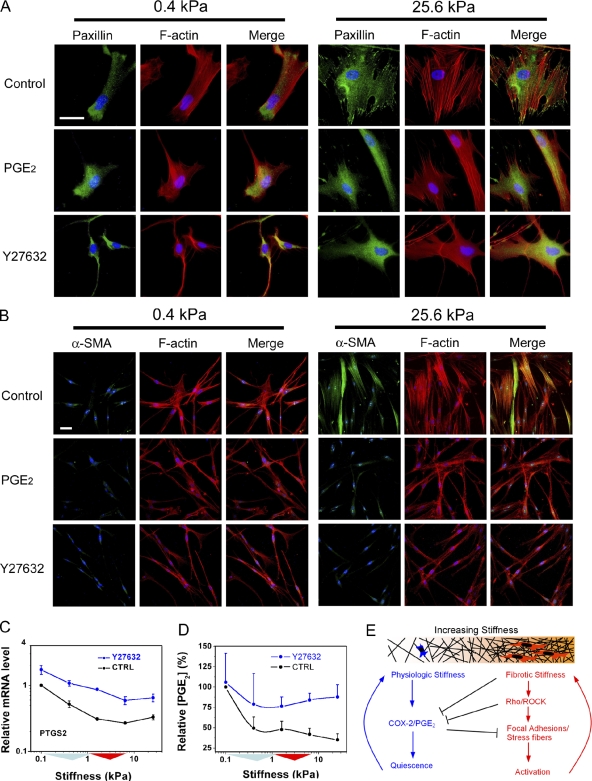
Numerous studies link the Rho/ROCK pathway to matrix stiffness responses (Paszek et al., 2005; Peyton and Putnam, 2005; Assoian and Klein, 2008), although none have examined the interrelationship of stiffness, Rho/ROCK, and prostaglandin signaling. We observed that increasing matrix stiffness triggers dramatic changes in the organization of focal adhesions and actin stress fibers (Fig. 6 A), along with α-SMA staining (Fig. 6 B), which is consistent with prior work (Yeung et al., 2005; Goffin et al., 2006; Li et al., 2007). Strikingly, the addition of exogenous PGE2 had little effect on these structures at low stiffness but dramatically attenuated their presence at high stiffness (Fig. 6, A and B), resulting in indistinguishable cell morphologies, actin cytoskeletal organization, and adhesion structures between the two stiffness extremes in the presence of exogenous PGE2. The morphological and cytoskeletal responses to exogenous PGE2 were sustained for up to 24 h (unpublished data) and were comparable in scope with those induced by blocking ROCK activity with Y27632, suggesting an antagonistic relationship between PGE2 and ROCK.
Figure 6.
ROCK and PGE2 exert mutually antagonistic effects on fibroblasts. (A) Increasing stiffness enhances organization of filamentous F-actin cytoskeleton (red) and triggers formation of paxillin-positive focal adhesions (green). The effect of stiffness on formation of these cellular structures is reversed with 30 ng/ml exogenous PGE2, in a fashion identical to treatment with 33 µM of the ROCK inhibitor Y27632 (1% FBS). (B) Increasing stiffness drives expression and assembly of α-SMA (green) into F-actin stress fibers (red). The effect of stiffness on α-SMA is reversed with 30 ng/ml exogenous PGE2, in a fashion identical to treatment with 33 µM of the ROCK inhibitor Y27632 (1% FBS). (C) 33 µM Y27632 increases expression of COX-2 (PTGS2) mRNA across matrix stiffness conditions (0.1% FBS), relative to untreated cells (CTRL). Error bars indicate SD from three replicate samples from one representative experiment. (D) 33 µM Y27632 equilibrates endogenous PGE2 levels across matrix stiffness conditions (0.1% FBS), relative to untreated cells (CTRL). Error bars indicate SD from three independent experiments. (C and D) Colored triangles along the x axis are used to indicate the interquartile and median stiffness values of lung parenchyma from saline (cyan)- or bleomycin (red)-treated mice. (E) Results are consistent with mutually antagonistic and self-reinforcing effects of Rho/ROCK and COX-2/PGE2 signaling in fibroblasts. Under normal conditions, autocrine PGE2 and PGE2 contributions from surrounding cells, coupled with the compliant nature of the normal matrix, maintain fibroblasts in a quiescent state. Increasing matrix stiffness suppresses endogenous COX-2 and shifts fibroblasts into an activated state. Subsequent matrix synthesis generates positive feedback on matrix synthesis, leading to persistent fibroblast activation and progressive matrix stiffening. Bars, 50 µm.
We also found that blocking ROCK with Y27632 increased COX-2 expression across the entire range of matrix stiffness tested (Fig. 6 C) and largely equilibrated the autocrine levels of PGE2 present across matrix stiffness conditions (Fig. 6 D). Thus, endogenous ROCK activity suppresses COX-2 expression and PGE2 synthesis in lung fibroblasts, whereas exogenous PGE2 mimics ROCK inhibition. Collectively, these findings demonstrate that Rho/ROCK and COX-2/PGE2 signaling exert mutually antagonistic effects on fibroblast morphology, cytoskeletal structure, and focal adhesions. Our results suggest that the relative balance between these antagonistic pathways defines two fibroblast phenotypes: active (high Rho and low COX-2) and quiescent (low Rho and high COX-2), with normal lung stiffness maintaining the quiescent phenotype and matrix stiffening promoting fibroblast activation (Fig. 6 E). Because these fibroblast states lead to self-reinforcing effects on matrix synthesis and degradation (Figs. 2–4), matrix stiffening and the accompanying shift in balance between quiescence and activation represent a critical step toward self-sustaining fibrosis.
Discussion
With the use of AFM microindentation, we found that emerging fibrotic lesions in the murine model of bleomycin-induced fibrosis exhibit median stiffness sixfold higher than surrounding normal lung parenchyma. The increases in stiffness far exceed previous macroscale measurements of fibrotic stiffening of the lung (Ebihara et al., 2000), and their highly localized nature is consistent with the patchy nature of both the bleomycin model and human disease (Selman et al., 2001; Moore and Hogaboam, 2008). Although prior work has established that fibroblast proliferation and myofibroblast differentiation vary with matrix stiffness (Arora et al., 1999; Wang et al., 2000; Goffin et al., 2006; Li et al., 2007), our work shows for the first time that fibroblast proliferation and procollagen I synthesis are completely suppressed at the median stiffness of normal lung tissue, even in the presence of serum, suggesting a dominant role for normal lung compliance in maintaining the fibroblast quiescence typically observed in vivo but, notably, not in cell culture on rigid surfaces. Across the pathophysiological range identified in emerging fibrotic lesions, we observed that matrix stiffness progressively activates fibroblasts to proliferate and for the first time documented a coordinated program of enhanced collagen synthesis and suppression of transcripts encoding matrix-degrading proteases. Together, these findings establish that stiffness within the pathophysiological range exerts profound effects on the key aspects of fibroblast biology central to lung fibrosis. Based on these findings, we propose that an adverse feedback cycle of matrix stiffening, fibroblast proliferation, and net matrix synthesis underlies the relentless progressive nature of severe lung fibrosis.
Our study identifies suppression of COX-2 expression and PGE2 synthesis as necessary elements in the fibroblast proliferative and matrix synthetic responses to increasing matrix stiffness. Although various eicosanoids have been demonstrated to play both positive and negative roles in animal models of fibrosis (Lovgren et al., 2006; Oga et al., 2009), COX-2 and PGE2 levels are known to be reduced in the lungs of patients with fibrosis relative to healthy individuals (Borok et al., 1991; Xaubet et al., 2004), and fibroblasts isolated from fibrotic lungs are known to be deficient in PGE2 synthesis (Wilborn et al., 1995; Vancheri et al., 2000; Keerthisingam et al., 2001). Our data provide the first physiological explanation for these deficiencies. Exogenous PGE2 is well appreciated to suppress fibroblast proliferation and activation (Lama et al., 2002; Huang et al., 2007; Sandulache et al., 2007; Thomas et al., 2007; Weinberg et al., 2009) and limit experimental lung fibrosis (Failla et al., 2009), but our data expand on this by demonstrating that endogenous PGE2 is secreted at high levels by fibroblasts growing on physiological stiffness matrices, and this capacity is gradually lost as matrix stiffness increases. However, notably, on soft substrates, pharmacological treatment to suppress COX-2 activity alone was insufficient to engage fibroblast proliferation and collagen synthesis, demonstrating that redundant mechanisms protect against spontaneous fibroblast activation when cells are resident in an environment as compliant as the normal lung. These findings are supported by animal models in which genetic deletion of COX-2 alone was insufficient to initiate fibrosis in the lung but did increase susceptibility to and severity of experimentally induced lung fibrosis (Keerthisingam et al., 2001; Bonner et al., 2002; Hodges et al., 2004). Collectively, our findings document a double layer of protection against fibrosis in the lung through the innate suppressive effects of the lung’s mechanical compliance, coupled with the abundant PGE2 production it engenders.
TGF-β is widely acknowledged to play a key role in driving fibrogenesis through its effects on activation of fibroblasts to myofibroblasts and stimulation of matrix synthesis (Tomasek et al., 2002). Recent work indicates that fibroblasts grown for several days on matrices of varying stiffness activate matrix-bound TGF-β in a stiffness-dependent fashion (Wipff et al., 2007). We were surprised to find through microarray analysis that the early (48 h) fibroblast genomic response to stiffness is both supportive of proliferation and suppressive of collagen degradation but largely diverges from responses driven by TGF-β. Nowhere was this divergence more clear than in the case of COX-2. Although we found that increasing matrix stiffness suppressed COX-2 expression and downstream synthesis of PGE2, TGF-β has been shown to stimulate COX-2 expression and PGE2 production (Diaz et al., 1989; McAnulty et al., 1995; Keerthisingam et al., 2001; Kapoun et al., 2006). Because PGE2 acts to suppress TGF-β–driven responses in fibroblasts (Kolodsick et al., 2003; Sandulache et al., 2007; Thomas et al., 2007), the COX-2/PGE2 stimulation by TGF-β is thought to dampen cellular responses to this fibrogenic cytokine and prevent overexuberant fibrotic reactions. Our data raise the possibility that fibroblasts resident on stiff matrices experience enhanced responses to TGF-β because of stiffness-dependent down-regulation of a usual brake on TGF-β signaling. This may account, in part, for the emergence of TGF-β responses that we and others (Arora et al., 1999; Goffin et al., 2006) observe on matrices of increasing stiffness.
How variations in matrix stiffness are transduced by cells remains an area of intense investigation. Several important molecular players have emerged, including integrins (Paszek et al., 2005; Friedland et al., 2009), focal adhesion proteins such as FAK (Pelham and Wang, 1997; Paszek et al., 2005; Assoian and Klein, 2008), and cytoskeletal tension driven by the action of myosin II (Engler et al., 2006). Our results indicate that an antagonistic relationship between ROCK and COX-2 activities shapes fibroblast responses to matrix stiffness, wherein ROCK suppresses COX-2 expression and activity, whereas the COX-2 downstream product PGE2 acts in a fashion similar to a ROCK inhibitor in disassembling the focal adhesions and stress fibers that form as matrix stiffness increases. This mutually antagonistic relationship sets up two feedback loops, both self-reinforcing, that drive fibroblasts to one of two extremes, activated (high Rho and low COX-2) or quiescent (low Rho and high COX-2). We speculate that in the lung, abundant PGE2, produced by fibroblasts and surrounding epithelial cells (Ozaki et al., 1987; Wilborn et al., 1995; Lama et al., 2002), normally maintains the quiescent phenotype but that lung injury and matrix stiffening act in part through effects on PGE2 levels to tilt the balance toward persistent, self-sustaining fibroblast activation (Fig. 6 D). Although we have not pinpointed the precise molecular interactions between Rho/ROCK and COX-2/PGE2 in these experiments, PGE2 is known to signal through cAMP in lung fibroblasts (Choung et al., 1998; Huang et al., 2007), and cAMP and downstream protein kinase A have been shown to both inactivate Rho and lead to cofilin-mediated actin depolymerization in multiple cell types (Dong et al., 1998; Lang et al., 1996; Murthy et al., 2003; Goeckeler and Wysolmerski, 2005), which is consistent with our observations.
How increasing matrix stiffness regulates expression of COX-2 is unknown. Recent findings indicate that diminished expression of COX-2 in fibroblasts isolated from fibrotic lungs is caused by defective histone acetylation in the COX-2 promoter (Coward et al., 2009). Epigenetic regulation of COX-2 is consistent with the observations that certain stimuli can provoke long-lasting changes in COX-2 expression that persist through multiple cell divisions (Korn, 1983) and could explain how deficiencies in PGE2 secretion remain stable in serially passaged cells in culture (Wilborn et al., 1995; Vancheri et al., 2000; Keerthisingam et al., 2001). Although not yet well established, early evidence suggests that changes in matrix stiffness and cell shape do indeed regulate global levels of histone acetylation (Le Beyec et al., 2007). Further studies will be needed to elucidate the molecular links connecting matrix stiffness to gene expression and to test the role that epigenetic regulation plays in stiffness-dependent cellular behaviors.
In conclusion, we demonstrate for the first time that the median stiffness of normal lung tissue strongly inhibits the fibroblast functions at the core of fibrosis and elucidate a novel mechanism linking matrix stiffening to fibroblast activation in part through suppression of an endogenous COX-2/PGE2 inhibitory pathway. Our data imply that fibrotic stiffening of a critical mass and extent, once established, is sufficient to generate a self-sustaining fibrotic process by virtue of its activation of fibroblasts, suppression of autoinhibitory COX-2/PGE2, and subsequent feedback amplification of stiffness through net matrix synthesis. Importantly, our results imply that fibroblasts within a stiffened fibrotic lesion are primed to engage in further fibrogenic activities. The instability inherent in the presence of a mechanobiological profibrotic feedback loop could help to explain the variable course of the disease and rapid declines in lung function seen in exacerbations of lung fibrosis (Martinez et al., 2005; Collard et al., 2007). Based on the fact that similar stiffness levels and cellular behaviors are observed during fibrogenesis in other soft tissues (Georges et al., 2007; Li et al., 2007; Wells, 2008), we propose that the mechanobiological feedback loop detailed in this study may represent a common feature of progressive fibrosis. Improved recognition of how constraints are broken to engage this feedback loop may prove critical for developing new prevention or early intervention strategies, whereas further elucidation of the mechanisms linking matrix stiffness to ongoing fibroblast activation may lead to new therapeutic opportunities for the treatment of progressive fibrosis.
Materials and methods
Animals and bleomycin administration
C57BL/6 mice (The National Cancer Institute Animal Production Program) were instilled intratracheally with 0.05 U bleomycin (Gensia Sicor Pharmaceuticals Inc.) in 50 µl sterile saline or with 50 µl sterile saline for control mice, with lung tissue harvested 14 d later (Tager et al., 2008). All experiments were performed in accordance with National Institutes of Health guidelines and protocols approved by the Massachusetts General Hospital Subcommittee on Research Animal Care, and all mice were maintained in a specific pathogen-free environment certified by the American Association for Accreditation of Laboratory Animal Care.
Lung parenchyma strip preparation and elastomapping
Mouse lung parenchyma strips were prepared by inflating lungs with 2% low gel point agarose at 37°C (50 ml/kg body weight) followed by cooling to 4°C. Tissue strips of 5 × 5 mm in length and width and 400 µm in thickness were cut with a razor blade from subpleural regions, excluding larger airways, and then washed in PBS at 37°C to remove the agarose (Bergner and Sanderson, 2003). The samples were placed on poly-l-lysine–coated coverslips. Samples were mechanically characterized using an atomic force microscope (MFP-3D; Asylum Research) by performing microindentation using a sphere-tipped probe (Novascan) with a diameter of 5 µm and a nominal spring constant of ∼60 pN/nm. The cantilever spring constant was further confirmed by the thermal fluctuation method (Thundat et al., 1994). The AFM system was calibrated by following the manufacturer’s instruction before each indentation measurement. Force-indentation profiles were acquired at an indentation rate of 20 µm/s separated by 5 µm spatially in a 16 × 16 sample grid covering an 80 × 80–µm area. Shear modulus at each point on the grid was calculated from fitting force-indentation data using a Hertz sphere model (Richert et al., 2004), and resulting shear modulus data were plotted in contour maps. For lung tissue, a Poisson’s ratio of 0.4 (Butler et al., 1986) was used to convert elastic modulus (E) to shear modulus (G), using the relationship E = 2 × (1 + ν) × G.
Immunofluorescence imaging
Lung tissue strips were immunostained after blocking with 5% goat serum without permeabilization. Human lung fibroblasts (CCL-151; American Type Culture Collection) cultured on stiffness gradients for 5 d or NHLFs (Lonza) cultured in 96-well plates containing individual discrete stiffness gels for 2 d were immunostained after fixation, permeabilization, and blocking with 5% goat serum. Primary antibodies used were obtained from Sigma-Aldrich (collagen I mAb COL-1, Paxillin mAb PXC-10, α-SMA mAb, 1A4) and Fitzgerald (ProCollagen I mAb PCIDG10). Secondary antibodies were obtained from Invitrogen (Alexa Fluor 488 or 546 goat anti–mouse IgG2a and Alexa Fluor 488 or 546 goat anti–mouse IgG1) and Polysciences (goat anti–mouse IgG–coated fluorescent yellow-green 1.0-µm microspheres). F-actin and nuclei were stained with Alexa Fluor 546–phalloidin and Hoechst 33342 (Invitrogen), respectively. Fluorescence images were obtained with a fluorescent microscope (TE300; Nikon). The final image contrast was adjusted with MetaMorph 6.1 (MDS Analytical Technologies) by setting the background to the fluorescence intensity level of the samples in the corresponding secondary antibody-only control images. Panorama images were assembled with Photoshop 6.0 (Adobe).
Stiffness gradient substrate preparation
An aqueous solution containing 12% acrylamide, 0.3% bisacrylamide, and 3.0 mg/ml Irgacure 2959 (photoinitiator; Ciba) was pipetted between two 22 × 22–mm coverslips and polymerized by exposure to UV light (365 nm; 7.0 mW/cm2) for ∼6.5 min at 30°C. Linear stiffness gradients were obtained by modulating the intensity of UV light exposure through a filter mask with a clear to 100% gradient in opacity (Wong et al., 2003). Stiffness was measured by AFM and converted from elastic modulus to shear modulus assuming a Poisson’s ratio of 0.3 (Li et al., 1993). The conversion from shear to elastic (Young’s) modulus is thus E = 2.6 × G.
Cell culture and cell density measurements
Lung fibroblasts (CCL-151) were cultured in Kaighn’s Modification of Ham’s F12 medium (F12K) supplemented with 10% FBS (Lonza), 100 U/ml penicillin, and 100 µg/ml streptomycin. Gradient stiffness gels were seeded at 50 cells/mm2 in 6-well plates. After 4 h, gels were gently washed with fresh medium and then moved into new wells. Cells were cultured for 5 d before fixation for immunostaining. For TGF-β1 treatment, 2 ng/ml TGF-β1 was added at day 2 and day 4. To generate panorama images, the entire gel width was imaged along the stiffness gradient by tiling 5–7 adjacent pictures. Cell density was determined by counting nuclei numbers on fluorescent images with MetaMorph 6.1. Density values were normalized to the global mean value from 4-h attachment. NHLFs were seeded at a density of 50 cell/mm2 in 96-well plates containing gels with five individual discrete stiffnesses: 0.1, 0.4, 1.6, 6.4, and 25.6 kPa. Cells were cultured with F12K containing 1% FBS at 37°C and 5% CO2. After 48 h, cell density was determined using CyQuant NF Cell Proliferation Assay (Invitrogen). Density values were normalized to the global mean value from 4-h attachment. For drug treatments, PGE2, butaprost, or NS-398 was added after 4-h initial attachment. In general, NHLFs or CCL-151 cells were cultured in 0.1% FBS for microarray and qPCR analysis to minimize differences in transcript levels caused by variations in proliferation and cell cycle progression. NHLFs were cultured in 1% FBS for proliferation, procollagen expression, PGE2, and collagen secretion analysis; CCL-151 cells grew more slowly and were cultured in 10% FBS for proliferation and procollagen expression analyses.
Proliferation and apoptosis assays
Fibroblasts were cultured on stiffness gradients for 4 d and then exposed to 50 µM BrdU for 24 h, fixed, permeabilized with 0.5% Triton X-100, treated with 4N hydrochloric acid for 15 min, and then neutralized with 0.1 M sodium borate, pH 8.5, for 20 min. Cells were then stained with anti-BrdU antibody (Alexa Fluor 546 mAb IgG1; Invitrogen) to identify replicating cells and with Hoechst 33342 to determine total cell number. For measurement of apoptosis, fibroblasts cultured on stiffness gradients for 4 d were serum-starved for 24 h and then stained with Image-iT Live green Caspase Detection kit (Invitrogen) to detect caspase 3/7 activity.
Migration assay
Cell migration data were generated using a time-lapse videomicroscopy system consisting of an inverted microscope (Leica) with 10× phase-contrast objective, charge-coupled device camera, and automated data acquisition software (MetaMorph 6.1). A temperature and CO2 control system was used for the maintenance of cell viability. Cells grown on stiffness gradients for 24 h were imaged every 2 min for 5 h. Cell xy-centroids and displacements were determined using MetaMorph 6.1 cell tracking module. Only cells that did not divide, touch other cells, or leave the image field during the experiment were used for data analysis. Single cell speed was derived by dividing total cell path length by the total time of migration. Single cell persistence was derived from nonlinear least-squares regression using individual cell speed and mean-squared cell displacement as described previously (Harms et al., 2005).
Discrete stiffness gel preparation
Glass-bottom 96-well plates (Matrical) were treated with a 0.4% aqueous solution of g-methacryloxypropyltrimethoxysilane (Acros Organics). Solutions containing 0.075% ammonium persulfate, 0.15% tetramethylethylenediamine, and variable ratios of acrylamide/bisacrylamide (Bio-Rad Laboratories) were delivered into the well plate. A 96-pin block with affixed, hydrophobic glass squares corresponding to the diameter of the wells was inserted, sandwiching the polymerization solutions between two glass surfaces. Gel thickness was controlled by placing 100-µm-thick spacers in the corner wells. After polymerization, the gel surface was derivatized with heterobifunctional cross-linker Sulfo-SANPAH (Thermo Fisher Scientific; Pelham and Wang, 1997). Monomeric collagen (PureCol) diluted in PBS at 100 µg/ml was delivered to each well and incubated for 4 h at room temperature. The well plate was rinsed in PBS and UV-sterilized before cell seeding.
Microarray analysis
NHLFs from three independent donors were seeded at a density of 50 cell/mm2 in 96-well plates containing gels with five individual discrete stiffnesses: 0.1, 0.4, 1.6, 6.4, and 25.6 kPa. Cells were allowed to attach to the gels in serum-free F12K media for 4 h and incubated at 37°C and 5% CO2. Media was then replaced with F12K containing 0.1% FBS to minimize differences in transcript levels caused by variations in proliferation and cell cycle progression. After 48 h, cells were lysed with 50 µl RNA lysis buffer per well (QIAGEN). Replicate stiffness wells were pooled, and RNA was isolated (RNeasy mini kit; QIAGEN) and quantified with RiboGreen reagent (Invitrogen). RNA extracts were used to generate labeled cRNA and hybridized to GeneChip Human Gene 1.0 ST arrays according to the standard protocol (Affymetrix). The resulting raw CEL file data were processed using standard RMA normalization into a numerical data matrix for subsequent mathematical analyses. These data are publicly available at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE22011. Gene expression analysis was limited to 2,462 probes with cross-donor rank correlation among stiffness conditions of >0.4. The analysis was further restricted to 192 probes (150 of which have a gene assignment) representing 124 unique genes with minimum within-donor coefficient of variation (of their exponentiated, base 2, RMA signal) across stiffness conditions of ≥0.2. Hierarchical clustering was performed on the selected 150 probes based on linear correlation across the three donors. Before hierarchical clustering, the stiffness expression signal profile of each gene was standardized to mean zero, variance one within each donor. Ontological analysis was conducted using DAVID functional annotation clustering tool (http://david.abcc.ncifcrf.gov/summary.jsp).
RNA analysis
Lung fibroblasts (CCL-151, NHLFs, and IMR90), normal human dermal fibroblasts, human airway smooth muscle cells, A549 lung epithelial cells, and human synovial fibroblasts were seeded at a density of 50 cells/mm2 in 96-well plates containing individual discrete stiffness gels. Synovial fibroblasts were prepared and cultured from discarded synovial tissues from rheumatoid arthritis patients (American College of Rheumatology criteria), obtained with approval of the Brigham and Women’s Hospital Institutional Review Board from synovectomy or joint replacement procedures (courtesy of Z. Gu and M. Brenner, Harvard Medical School and Brigham and Women’s Hospital, Boston, MA). All cells were allowed to attach to the gels in serum-free F12K media for 4 h and incubated at 37°C and 5% CO2. Media was then replaced with F12K containing 0.1% FBS. After 48 h, cells were lysed with 50 µl of RNA lysis buffer per well (QIAGEN). Replicate stiffness wells were pooled, and RNA was isolated (RNeasy mini kit) and quantified with RiboGreen reagent. 50 ng of each sample was primed with oligo (dT) and reverse transcribed into cDNA using Taqman reverse transcription reagents (Applied Biosystems) at 25°C for 10 min, 37°C for 60 min, and 95°C for 5 min. Equivalent amounts of each cDNA sample were added to Power SYBR green PCR master mix (Applied Biosystems) containing 1 µM each of forward and reverse primer (Table S1) for mRNA amplification. Primers were designed with qPrimerDepot (http://primerdepot.nci.nih.gov) and are intron overlapping. qPCR was performed by incubating at 95°C for 10 min to activate AmpliTaq Gold DNA polymerase (Applied Biosystems) and then cycling 40 times at 95°C for 15 s and 60°C for 1 min. Ct values within each experiment were normalized against GAPDH (glyceraldehyde 3-phosphate dehydrogenase), and mRNA levels were expressed relative to the softest substrate tested.
PGE2 assays
NHLFs were seeded at a density of 50 cells/mm2 in 96-well plates containing individual discrete stiffness gels. Cells were cultured with F12K containing 1% FBS at 37°C and 5% CO2. After 48 h, cell culture media were collected, and the PGE2 enzyme-linked immunosorbent assay was performed according to manufacturer’s instructions (Cayman Chemicals). PGE2 concentrations were normalized to the corresponding cell numbers and then to the values at the lowest stiffness.
Soluble collagen assay
NHLFs were seeded at a density of 50 cell/mm2 in 96-well plates containing gels with five individual discrete stiffnesses: 0.1, 0.4, 1.6, 6.4, and 25.6 kPa. Cells were cultured with F12K containing 1% FBS at 37°C and 5% CO2. After 72 h, cell culture media were collected, and the collagen concentration was measured using Sircol Soluble Collagen Assay kit according to manufacturer’s instructions (Biocolor). Collagen concentrations were normalized to the corresponding cell numbers as measured with CyQuant Assay.
Statistical analysis
Results are expressed as means ± SD. Data were analyzed by nonparametric Mann-Whitney-Wilcoxon test or t test using the Prism statistical program (GraphPad Software, Inc.). P-values <0.05 were considered significant. Error bars depict SD.
Online supplemental material
Fig. S1 shows stiffness maps collected from five saline- and eight bleomycin-treated mouse lungs. Fig. S2 provides mechanical validation of the gradient stiffness system and demonstrates uniform collagen density across the stiffness gradient. Fig. S3 contains panorama fluorescence micrographs showing BrdU, caspase 3/7 activity, and procollagen staining across stiffness gradients. Fig. S4 provides complete hierarchical clustering of matrix stiffness-regulated genes, top over-represented ontological categories among these genes, and relative TGF-β responsiveness of stiffness-regulated genes. Fig. S5 demonstrates the consistent effect of increasing stiffness on procollagen I expression, attenuated by EP2 receptor agonist butaprost. Table S1 provides sequence information for qPCR primers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004082/DC1.
Acknowledgments
We thank T. Shiomi for help with mouse lung isolations; M. Symer for technical assistance with polyacrylamide gel systems; J. Park for qPCR assistance; Z. Gu and M. Brenner for providing synovial fibroblasts; and K. Billiar, J. Wong, J. Fredberg, and J. Drazen for helpful discussions.
This work was supported by National Institutes of Health grants HL-092961, HL-088028, GM-073628 and Scleroderma Foundation Established Investigator Grant to D.J. Tschumperlin; American Thoracic Society Fellows Career Development Award to B.S. Shea; and Pulmonary Fibrosis Foundation, Coalition for Pulmonary Fibrosis/American Thoracic Society, and Nirenberg Center for Advanced Lung Disease grants to A.M. Tager. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation under NSF award ECS-0335765. CNS is part of the Faculty of Arts and Sciences at Harvard University.
Footnotes
Abbreviations used in this paper:
- AFM
- atomic force microscopy
- α-SMA
- α–smooth muscle actin
- CTSK
- cathepsin K
- NHLF
- normal human lung fibroblast
- qPCR
- quantitative PCR
- RMA
- robust multichip average
- ROCK
- Rho kinase
References
- Arora P.D., Narani N., McCulloch C.A. 1999. The compliance of collagen gels regulates transforming growth factor-beta induction of alpha-smooth muscle actin in fibroblasts. Am. J. Pathol. 154:871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assoian R.K., Klein E.A. 2008. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 18:347–352 10.1016/j.tcb.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachofen H., Scherrer M. 1967. Lung tissue resistance in diffuse interstitial pulmonary fibrosis. J. Clin. Invest. 46:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner A., Sanderson M.J. 2003. Airway contractility and smooth muscle Ca(2+) signaling in lung slices from different mouse strains. J. Appl. Physiol. 95:1325–1332 [DOI] [PubMed] [Google Scholar]
- Bonner J.C., Rice A.B., Ingram J.L., Moomaw C.R., Nyska A., Bradbury A., Sessoms A.R., Chulada P.C., Morgan D.L., Zeldin D.C., Langenbach R. 2002. Susceptibility of cyclooxygenase-2-deficient mice to pulmonary fibrogenesis. Am. J. Pathol. 161:459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borok Z., Gillissen A., Buhl R., Hoyt R.F., Hubbard R.C., Ozaki T., Rennard S.I., Crystal R.G. 1991. Augmentation of functional prostaglandin E levels on the respiratory epithelial surface by aerosol administration of prostaglandin E. Am. Rev. Respir. Dis. 144:1080–1084 [DOI] [PubMed] [Google Scholar]
- Bühling F., Röcken C., Brasch F., Hartig R., Yasuda Y., Saftig P., Brömme D., Welte T. 2004. Pivotal role of cathepsin K in lung fibrosis. Am. J. Pathol. 164:2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.P., Nakamura M., Sasaki H., Sasaki T., Takishima T. 1986. Poissons’ ratio of lung parenchyma and parenchymal interaction with bronchi. Jpn. J. Physiol. 36:91–106 10.2170/jjphysiol.36.91 [DOI] [PubMed] [Google Scholar]
- Choung J., Taylor L., Thomas K., Zhou X., Kagan H., Yang X., Polgar P. 1998. Role of EP2 receptors and cAMP in prostaglandin E2 regulated expression of type I collagen alpha1, lysyl oxidase, and cyclooxygenase-1 genes in human embryo lung fibroblasts. J. Cell. Biochem. 71:254–263 [DOI] [PubMed] [Google Scholar]
- Collard H.R., Moore B.B., Flaherty K.R., Brown K.K., Kaner R.J., King T.E., Jr., Lasky J.A., Loyd J.E., Noth I., Olman M.A., et al. 2007. Acute exacerbations of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 176:636–643 10.1164/rccm.200703-463PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward W.R., Watts K., Feghali-Bostwick C.A., Knox A., Pang L. 2009. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol. Cell. Biol. 29:4325–4339 10.1128/MCB.01776-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoulière A., Darby I.A., Gabbiani G. 2003. Normal and pathologic soft tissue remodeling: role of the myofibroblast, with special emphasis on liver and kidney fibrosis. Lab. Invest. 83:1689–1707 10.1097/01.LAB.0000101911.53973.90 [DOI] [PubMed] [Google Scholar]
- Diaz A., Varga J., Jimenez S.A. 1989. Transforming growth factor-beta stimulation of lung fibroblast prostaglandin E2 production. J. Biol. Chem. 264:11554–11557 [PubMed] [Google Scholar]
- Dong J.M., Leung T., Manser E., Lim L. 1998. cAMP-induced morphological changes are counteracted by the activated RhoA small GTPase and the Rho kinase ROKalpha. J. Biol. Chem. 273:22554–22562 10.1074/jbc.273.35.22554 [DOI] [PubMed] [Google Scholar]
- Dunn M.G., Silver F.H., Swann D.A. 1985. Mechanical analysis of hypertrophic scar tissue: structural basis for apparent increased rigidity. J. Invest. Dermatol. 84:9–13 10.1111/1523-1747.ep12274528 [DOI] [PubMed] [Google Scholar]
- Ebihara T., Venkatesan N., Tanaka R., Ludwig M.S. 2000. Changes in extracellular matrix and tissue viscoelasticity in bleomycin-induced lung fibrosis. Temporal aspects. Am. J. Respir. Crit. Care Med. 162:1569–1576 [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. 2006. Matrix elasticity directs stem cell lineage specification. Cell. 126:677–689 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Failla M., Genovese T., Mazzon E., Fruciano M., Fagone E., Gili E., Barera A., La Rosa C., Conte E., Crimi N., et al. 2009. 16,16-Dimethyl prostaglandin E2 efficacy on prevention and protection from bleomycin-induced lung injury and fibrosis. Am. J. Respir. Cell Mol. Biol. 41:50–58 10.1165/rcmb.2007-0438OC [DOI] [PubMed] [Google Scholar]
- Friedland J.C., Lee M.H., Boettiger D. 2009. Mechanically activated integrin switch controls alpha5beta1 function. Science. 323:642–644 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- Fringer J., Grinnell F. 2001. Fibroblast quiescence in floating or released collagen matrices: contribution of the ERK signaling pathway and actin cytoskeletal organization. J. Biol. Chem. 276:31047–31052 10.1074/jbc.M101898200 [DOI] [PubMed] [Google Scholar]
- Georges P.C., Hui J.J., Gombos Z., McCormick M.E., Wang A.Y., Uemura M., Mick R., Janmey P.A., Furth E.E., Wells R.G. 2007. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G1147–G1154 10.1152/ajpgi.00032.2007 [DOI] [PubMed] [Google Scholar]
- Goeckeler Z.M., Wysolmerski R.B. 2005. Myosin phosphatase and cofilin mediate cAMP/cAMP-dependent protein kinase-induced decline in endothelial cell isometric tension and myosin II regulatory light chain phosphorylation. J. Biol. Chem. 280:33083–33095 10.1074/jbc.M503173200 [DOI] [PubMed] [Google Scholar]
- Goffin J.M., Pittet P., Csucs G., Lussi J.W., Meister J.J., Hinz B. 2006. Focal adhesion size controls tension-dependent recruitment of α-smooth muscle actin to stress fibers. J. Cell Biol. 172:259–268 10.1083/jcb.200506179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T.J., Hunninghake G.W. 2001. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 345:517–525 10.1056/NEJMra003200 [DOI] [PubMed] [Google Scholar]
- Harms B.D., Bassi G.M., Horwitz A.R., Lauffenburger D.A. 2005. Directional persistence of EGF-induced cell migration is associated with stabilization of lamellipodial protrusions. Biophys. J. 88:1479–1488 10.1529/biophysj.104.047365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. 2007. The myofibroblast: one function, multiple origins. Am. J. Pathol. 170:1807–1816 10.2353/ajpath.2007.070112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges R.J., Jenkins R.G., Wheeler-Jones C.P., Copeman D.M., Bottoms S.E., Bellingan G.J., Nanthakumar C.B., Laurent G.J., Hart S.L., Foster M.L., McAnulty R.J. 2004. Severity of lung injury in cyclooxygenase-2-deficient mice is dependent on reduced prostaglandin E(2) production. Am. J. Pathol. 165:1663–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.K., Peters-Golden M. 2008. Eicosanoid lipid mediators in fibrotic lung diseases: ready for prime time? Chest. 133:1442–1450 10.1378/chest.08-0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Wettlaufer S.H., Hogaboam C., Aronoff D.M., Peters-Golden M. 2007. Prostaglandin E(2) inhibits collagen expression and proliferation in patient-derived normal lung fibroblasts via E prostanoid 2 receptor and cAMP signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L405–L413 10.1152/ajplung.00232.2006 [DOI] [PubMed] [Google Scholar]
- Hui A.Y., Leung W.K., Chan H.L., Chan F.K., Go M.Y., Chan K.K., Tang B.D., Chu E.S., Sung J.J. 2006. Effect of celecoxib on experimental liver fibrosis in rat. Liver Int. 26:125–136 10.1111/j.1478-3231.2005.01202.x [DOI] [PubMed] [Google Scholar]
- Isenberg B.C., Dimilla P.A., Walker M., Kim S., Wong J.Y. 2009. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys. J. 97:1313–1322 10.1016/j.bpj.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoun A.M., Gaspar N.J., Wang Y., Damm D., Liu Y.W., O’young G., Quon D., Lam A., Munson K., Tran T.T., et al. 2006. Transforming growth factor-beta receptor type 1 (TGFbetaRI) kinase activity but not p38 activation is required for TGFbetaRI-induced myofibroblast differentiation and profibrotic gene expression. Mol. Pharmacol. 70:518–531 10.1124/mol.105.021600 [DOI] [PubMed] [Google Scholar]
- Keerthisingam C.B., Jenkins R.G., Harrison N.K., Hernandez-Rodriguez N.A., Booth H., Laurent G.J., Hart S.L., Foster M.L., McAnulty R.J. 2001. Cyclooxygenase-2 deficiency results in a loss of the anti-proliferative response to transforming growth factor-beta in human fibrotic lung fibroblasts and promotes bleomycin-induced pulmonary fibrosis in mice. Am. J. Pathol. 158:1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodsick J.E., Peters-Golden M., Larios J., Toews G.B., Thannickal V.J., Moore B.B. 2003. Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am. J. Respir. Cell Mol. Biol. 29:537–544 10.1165/rcmb.2002-0243OC [DOI] [PubMed] [Google Scholar]
- Korn J.H. 1983. Fibroblast prostaglandin E2 synthesis. Persistence of an abnormal phenotype after short-term exposure to mononuclear cell products. J. Clin. Invest. 71:1240–1246 10.1172/JCI110873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C., McDonald J.A. 1991. The roles of the myofibroblast in idiopathic pulmonary fibrosis. Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am. J. Pathol. 138:1257–1265 [PMC free article] [PubMed] [Google Scholar]
- Lama V., Moore B.B., Christensen P., Toews G.B., Peters-Golden M. 2002. Prostaglandin E2 synthesis and suppression of fibroblast proliferation by alveolar epithelial cells is cyclooxygenase-2-dependent. Am. J. Respir. Cell Mol. Biol. 27:752–758 [DOI] [PubMed] [Google Scholar]
- Lang P., Gesbert F., Delespine-Carmagnat M., Stancou R., Pouchelet M., Bertoglio J. 1996. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 15:510–519 [PMC free article] [PubMed] [Google Scholar]
- Le Beyec J., Xu R., Lee S.Y., Nelson C.M., Rizki A., Alcaraz J., Bissell M.J. 2007. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp. Cell Res. 313:3066–3075 10.1016/j.yexcr.2007.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Hu Z., Li Y. 1993. Poisson’s ratio in polymer gels near the phase-transition point. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 48:603–606 [DOI] [PubMed] [Google Scholar]
- Li Z., Dranoff J.A., Chan E.P., Uemura M., Sévigny J., Wells R.G. 2007. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 46:1246–1256 10.1002/hep.21792 [DOI] [PubMed] [Google Scholar]
- Liu H., Wei W., Li X. 2009. Celecoxib exacerbates hepatic fibrosis and induces hepatocellular necrosis in rats treated with porcine serum. Prostaglandins Other Lipid Mediat. 88:63–67 10.1016/j.prostaglandins.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Lo C.-M., Wang H.-B., Dembo M., Wang Y.L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovgren A.K., Jania L.A., Hartney J.M., Parsons K.K., Audoly L.P., Fitzgerald G.A., Tilley S.L., Koller B.H. 2006. COX-2-derived prostacyclin protects against bleomycin-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 291:L144–L156 10.1152/ajplung.00492.2005 [DOI] [PubMed] [Google Scholar]
- Martinez F.J., Safrin S., Weycker D., Starko K.M., Bradford W.Z., King T.E., Jr., Flaherty K.R., Schwartz D.A., Noble P.W., Raghu G., Brown K.K. 2005. The clinical course of patients with idiopathic pulmonary fibrosis. Ann. Intern. Med. 142:963–967 [DOI] [PubMed] [Google Scholar]
- McAnulty R.J., Chambers R.C., Laurent G.J. 1995. Regulation of fibroblast procollagen production. Transforming growth factor-beta 1 induces prostaglandin E2 but not procollagen synthesis via a pertussis toxin-sensitive G-protein. Biochem. J. 307:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B.B., Hogaboam C.M. 2008. Murine models of pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 294:L152–L160 10.1152/ajplung.00313.2007 [DOI] [PubMed] [Google Scholar]
- Murthy K.S., Zhou H., Grider J.R., Makhlouf G.M. 2003. Inhibition of sustained smooth muscle contraction by PKA and PKG preferentially mediated by phosphorylation of RhoA. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G1006–G1016 [DOI] [PubMed] [Google Scholar]
- Oga T., Matsuoka T., Yao C., Nonomura K., Kitaoka S., Sakata D., Kita Y., Tanizawa K., Taguchi Y., Chin K., et al. 2009. Prostaglandin F(2alpha) receptor signaling facilitates bleomycin-induced pulmonary fibrosis independently of transforming growth factor-beta. Nat. Med. 15:1426–1430 10.1038/nm.2066 [DOI] [PubMed] [Google Scholar]
- Ozaki T., Rennard S.I., Crystal R.G. 1987. Cyclooxygenase metabolites are compartmentalized in the human lower respiratory tract. J. Appl. Physiol. 62:219–222 [DOI] [PubMed] [Google Scholar]
- Paszek M.J., Zahir N., Johnson K.R., Lakins J.N., Rozenberg G.I., Gefen A., Reinhart-King C.A., Margulies S.S., Dembo M., Boettiger D., et al. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pelham R.J., Jr., Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 94:13661–13665 10.1073/pnas.94.25.13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena A.M., Fabre A., Débarre D., Marchal-Somme J., Crestani B., Martin J.L., Beaurepaire E., Schanne-Klein M.C. 2007. Three-dimensional investigation and scoring of extracellular matrix remodeling during lung fibrosis using multiphoton microscopy. Microsc. Res. Tech. 70:162–170 10.1002/jemt.20400 [DOI] [PubMed] [Google Scholar]
- Peyton S.R., Putnam A.J. 2005. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 204:198–209 10.1002/jcp.20274 [DOI] [PubMed] [Google Scholar]
- Richert L., Engler A.J., Discher D.E., Picart C. 2004. Elasticity of native and cross-linked polyelectrolyte multilayer films. Biomacromolecules. 5:1908–1916 10.1021/bm0498023 [DOI] [PubMed] [Google Scholar]
- Sandulache V.C., Parekh A., Li-Korotky H., Dohar J.E., Hebda P.A. 2007. Prostaglandin E2 inhibition of keloid fibroblast migration, contraction, and transforming growth factor (TGF)-beta1-induced collagen synthesis. Wound Repair Regen. 15:122–133 10.1111/j.1524-475X.2006.00193.x [DOI] [PubMed] [Google Scholar]
- Selman M., King T.E., Pardo A. 2001. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann. Intern. Med. 134:136–151 [DOI] [PubMed] [Google Scholar]
- Tager A.M., LaCamera P., Shea B.S., Campanella G.S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B.A., Kim N.D., et al. 2008. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14:45–54 10.1038/nm1685 [DOI] [PubMed] [Google Scholar]
- Thomas P.E., Peters-Golden M., White E.S., Thannickal V.J., Moore B.B. 2007. PGE(2) inhibition of TGF-beta1-induced myofibroblast differentiation is Smad-independent but involves cell shape and adhesion-dependent signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 293:L417–L428 10.1152/ajplung.00489.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thundat T., Warmack R.J., Chen G.Y., Allison D.P. 1994. Thermal and ambient-induced deflections of scanning force microscope cantilevers. Appl. Phys. Lett. 64:2894–2896 10.1063/1.111407 [DOI] [Google Scholar]
- Tomasek J.J., Gabbiani G., Hinz B., Chaponnier C., Brown R.A. 2002. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 3:349–363 10.1038/nrm809 [DOI] [PubMed] [Google Scholar]
- Vancheri C., Sortino M.A., Tomaselli V., Mastruzzo C., Condorelli F., Bellistrí G., Pistorio M.P., Canonico P.L., Crimi N. 2000. Different expression of TNF-alpha receptors and prostaglandin E(2)Production in normal and fibrotic lung fibroblasts: potential implications for the evolution of the inflammatory process. Am. J. Respir. Cell Mol. Biol. 22:628–634 [DOI] [PubMed] [Google Scholar]
- Vancheri C., Mastruzzo C., Sortino M.A., Crimi N. 2004. The lung as a privileged site for the beneficial actions of PGE2. Trends Immunol. 25:40–46 10.1016/j.it.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Wang H.B., Dembo M., Wang Y.L. 2000. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279:C1345–C1350 [DOI] [PubMed] [Google Scholar]
- Weinberg E., Zeldich E., Weinreb M.M., Moses O., Nemcovsky C., Weinreb M. 2009. Prostaglandin E2 inhibits the proliferation of human gingival fibroblasts via the EP2 receptor and Epac. J. Cell. Biochem. 108:207–215 10.1002/jcb.22242 [DOI] [PubMed] [Google Scholar]
- Wells R.G. 2008. The role of matrix stiffness in regulating cell behavior. Hepatology. 47:1394–1400 10.1002/hep.22193 [DOI] [PubMed] [Google Scholar]
- Wilborn J., Crofford L.J., Burdick M.D., Kunkel S.L., Strieter R.M., Peters-Golden M. 1995. Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J. Clin. Invest. 95:1861–1868 10.1172/JCI117866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.A., Bachofen H. 1982. A model for mechanical structure of the alveolar duct. J. Appl. Physiol. 52:1064–1070 [DOI] [PubMed] [Google Scholar]
- Wipff P.J., Rifkin D.B., Meister J.J., Hinz B. 2007. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J. Cell Biol. 179:1311–1323 10.1083/jcb.200704042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.Y., Velasco A., Rajagopalan P., Pham Q. 2003. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 19:1908–1913 10.1021/la026403p [DOI] [Google Scholar]
- Wynn T.A. 2008. Cellular and molecular mechanisms of fibrosis. J. Pathol. 214:199–210 10.1002/path.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xaubet A., Roca-Ferrer J., Pujols L., Ramírez J., Mullol J., Marin-Arguedas A., Torrego A., Gimferrer J.M., Picado C. 2004. Cyclooxygenase-2 is up-regulated in lung parenchyma of chronic obstructive pulmonary disease and down-regulated in idiopathic pulmonary fibrosis. Sarcoidosis Vasc. Diffuse Lung Dis. 21:35–42 [PubMed] [Google Scholar]
- Yeung T., Georges P.C., Flanagan L.A., Marg B., Ortiz M., Funaki M., Zahir N., Ming W., Weaver V., Janmey P.A. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 60:24–34 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]
- Zhang K., Rekhter M.D., Gordon D., Phan S.H. 1994. Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis. A combined immunohistochemical and in situ hybridization study. Am. J. Pathol. 145:114–125 [PMC free article] [PubMed] [Google Scholar]