ERK8 prevents genome instability by blocking the association of the HDM2 E3 ubiquitin ligase with the genome-protecting protein PCNA.
Abstract
Proliferating cell nuclear antigen (PCNA) acts as a scaffold, coordinator, and stimulator of numerous processes required for faithful transmission of genetic information. Maintaining PCNA levels above a critical threshold is essential, but little is known about PCNA protein turnover. We now show that ERK8 (extracellular signal-regulated kinase 8) is required for PCNA protein stability. ERK8 contains a conserved PCNA-interacting protein (PIP) box. Chromatin-bound ERK8 (ERK8CHROMATIN) interacts via this motif with PCNACHROMATIN, which acts as a platform for numerous proteins involved in DNA metabolism. Silencing ERK8 decreases PCNA levels and increases DNA damage. Ectopic expression of PCNA blocks DNA damage induced by ERK8 loss. ERK8 prevents HDM2-mediated PCNA destruction by inhibiting the association of PCNA with HDM2. This regulation is physiologically relevant as ERK8 activity is inhibited in transformed mammary cells. Our results reveal an unanticipated mechanism to control PCNA levels in normal cycling mammary epithelial cells and implicate ERK8 in the regulation of genomic stability.
Introduction
The physiological functions of ERK8 (extracellular signal-regulated kinase 8) are unknown. It is most closely related to ERK1/2 and ERK5 by virtue of a Thr-Glu-Tyr (T-E-Y) activation motif that must be phosphorylated in order for the kinase to be active (Abe et al., 2002). Unlike ERK1/2, ERK8 and ERK5 contain C-terminal extensions. ERK8 appears to be a rapidly evolving kinase (Coulombe and Meloche, 2007). The C-terminal tail of ERK8 is ∼51% conserved between primates and other mammals, in contrast to ERK5 in which the tail is ∼96% conserved. Given the paucity of information on ERK8, we investigated its function.
Proliferating cell nuclear antigen (PCNA) is a key player in a variety of DNA metabolic pathways in which it coordinates and regulates the functions of numerous proteins that perform enzymatic reactions on DNA (Moldovan et al., 2007). PCNA exists as a homotrimer that encircles DNA and provides a platform for its interacting partners. In addition to facilitating complex assembly, PCNA is able to stimulate or inhibit the activity of some of its binding partners (Gomes and Burgers, 2000; Azam et al., 2001). Intensive efforts have revealed the docking domains that facilitate interaction with PCNA (Moldovan et al., 2007; Gilljam et al., 2009; Havens and Walter, 2009). It is hypothesized that PCNA controls the myriad processes involved in genomic stability by direct physical association with its numerous binding partners (Moldovan et al., 2007). A decrease in PCNA protein levels prevents the formation of complexes necessary for DNA metabolism and results in catastrophic consequences (Jaskulski et al., 1988; Henderson et al., 1994). Thus, it is essential for cell survival to maintain PCNA protein levels at a critical threshold. PCNA is covalently modified by ubiquitin (Andersen et al., 2008; Das-Bradoo et al., 2010) and SUMO (small ubiquitin-like modifier; Hoege et al., 2002), but these modifications are thought to control the association with binding partners and not in the regulation of degradation. Thus, little is known about the regulation of PCNA protein turnover.
We have discovered that ERK8 controls PCNA levels in normal cycling cells by preventing its destruction via human homologue of murine double minute (HDM2). Loss of ERK8 resulted in genomic instability by decreasing PCNA levels beyond a tolerated limit. ERK8 was active in a mammary epithelial cell line and in primary mammary cells but was inactivated in breast cancer cell lines. These data argue that ERK8 control of PCNA levels is physiologically relevant, as ERK8 activity is inhibited in transformed cells.
Results
ERK8 regulates the cell cycle by maintaining PCNA levels
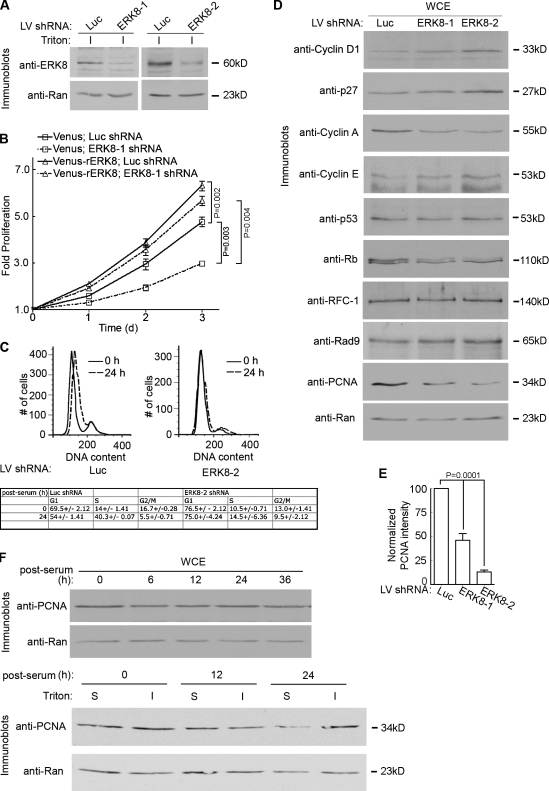
We investigated potential physiological functions for ERK8 by silencing its expression in the human breast epithelial line, MCF-10A. Successful silencing of endogenous ERK8, which was localized predominantly to a detergent-insoluble fraction, was observed with two different short hairpin RNAs (shRNAs; Fig. 1 A). Proliferation was measured starting at 2 d after transduction. Based on the doubling time of the control cells (31.9 ± 0.08 h), the transduced cells doubled approximately once before measuring the proliferation rate. Knockdown of ERK8 decreased proliferation approximately twofold, which was prevented by expression of a mutant ERK8 resistant to silencing (rERK8; Fig. 1 B and Fig. S1 A). In agreement with the silencing experiments, ectopic expression of wild-type ERK8 enhanced proliferation (Fig. 1 B). The most effective knockdown of ERK8 occurred 4–5 d after transduction, and therefore, the majority of experiments were performed using cells transduced in this time frame. To investigate the mechanism by which ERK8 controls the cell cycle, we performed flow cytometry using cells synchronized by serum and growth factor depletion, which resulted in ∼70% of the control cells in G1. The addition of serum and growth factors to the control cells resulted in a threefold increase in the number of cells in S phase after 24 h (Fig. 1 C). In contrast to these results, no change in the cell cycle distribution after release from starvation was observed when ERK8 was silenced (Fig. 1 C). Collectively, these results demonstrate that ERK8 is a limiting factor in regulating the proliferation rate of MCF-10A cells and controls entry into S phase.
Figure 1.
ERK8 regulates proliferation of MCF-10A cells. (A) Analysis of endogenous ERK8 knockdown in MCF-10A cells transduced with shRNA to luciferase (Luc) or one of two different ERK8 targeting sequences for 5 d. The detergent-insoluble fraction (I) of the transduced lysates was normalized for Ran expression and immunoblotted for ERK8. (B) Scatter plot showing the rate of proliferation of transduced MCF-10A cells. Analysis was started 2 d after transduction. Mean is shown (n = 5, quadruplicates), and error bars indicate SEM. (C) A representative flow cytometric analysis of cell cycle progression in MCF-10A cells transduced as in A. Transduced cells were identified by GFP fluorescence, and DNA was stained with DRAQ5. The times indicated are sampling times after the cells were arrested by serum and growth factor depletion, followed by release into the cell cycle by the addition of complete media. The table summarizes the results (mean ± range [n = 2]) generated by ModFit LT (Verity Software House). (D) Analysis of whole cell extracts (WCE) of MCF-10A cells transduced as in A. The lysates were normalized for Ran expression and immunoblotted for regulators of the cell cycle and DNA damage. (E) Quantitation of the relative amount of PCNA in comparison to Ran and normalized to the control, which was transduced with luciferase shRNA. Mean is shown (n = 6), and error bars indicate SEM. (F) Cell cycle analysis of PCNA in whole cell extracts and detergent-insoluble (I) and -soluble (S) fractions. MCF-10A cells were arrested and released into the cell cycle as in C. LV, lentivirus.
While analyzing cell cycle markers, we noticed that loss of ERK8 resulted in increases in cyclin D1, cyclin E, and p27 and a decrease in cyclin A levels compared with the control (Fig. 1 D). Furthermore, retinoblastoma (Rb) appeared as a doublet in the immunoblot analysis, and loss of ERK8 reduced the level of the higher molecular weight form (Fig. 1 D). These observations are consistent with a reduction in the phosphorylated form of Rb. Collectively, these changes provide further evidence that loss of ERK8 results in a cell cycle block in the G1/S phase and are in agreement with our flow cytometry results. But, unexpectedly, loss of ERK8 resulted in a decrease in the total level of PCNA (Fig. 1, D and E; and see Fig. 4 B). Silencing ERK8 did not result in higher molecular weight forms of PCNA that could account for the decrease in the unmodified form (Figs. 1 D and 2 E). The decrease in PCNA was not correlated with a particular stage in the cell cycle, as total PCNA levels did not change during the cell cycle (Fig. 1 F). In agreement with the literature (Naryzhny and Lee, 2001), the subcellular distribution of PCNA changed during the cell cycle such that during S phase, PCNA preferentially partitioned into a detergent-insoluble fraction (Fig. 1 F and Fig. S1 B). Loss of ERK8 appeared to specifically reduce PCNA levels as we did not observe a decrease in the level of Rad9, a component of the heterotrimeric complex 9-1-1, which, like PCNA, encircles DNA and is involved in DNA damage repair (Fig. 1 D; St Onge et al., 1999). Moreover, the level of RFC-1 (replication factor C-1), a subunit of the clamp loader that loads PCNA onto DNA (Waga and Stillman, 1998), was not changed (Fig. 1 D). We conclude that silencing ERK8 results in a specific decrease in PCNA protein levels.
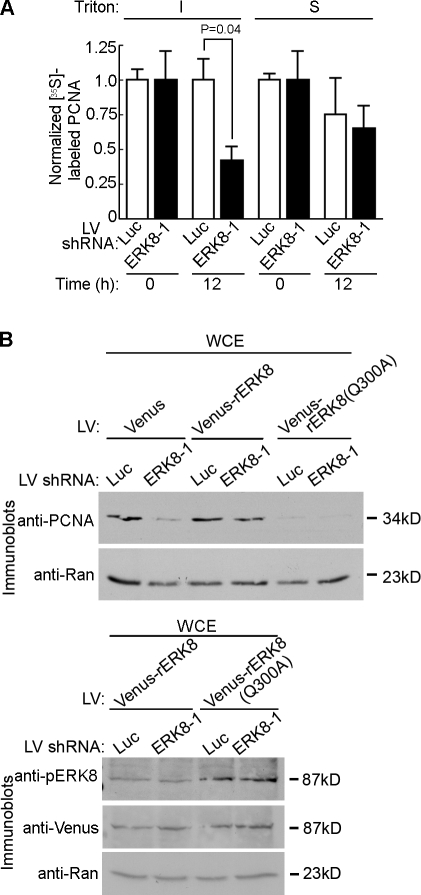
Figure 4.
The association of ERK8 with PCNA inhibits PCNA protein turnover. (A) Time course of PCNA degradation by [35S]Met pulse-chase labeling. MCF-10A cells transduced for 5 d were depleted of intracellular Met, labeled with [35S]Met for 4 h, washed, and incubated in excess cold Met for 0 or 12 h. Lysates were separated into insoluble (I) and soluble (S) fractions, and each fraction was immunoprecipitated with an anti-PCNA antibody. The amount of [35S]Met labeling was divided by the relative level of immunoprecipitated PCNA and then normalized to the control, which was transduced with luciferase (Luc) shRNA and analyzed at 0 h after the addition of excess, cold Met. Mean is shown (n = 3, duplicates), and error bars indicate SEM. (B) Rescue of PCNA levels by ectopic expression of resistant ERK8 (rERK8). MCF-10A cells were transduced with resistant ERK8 constructs or Venus control, followed by a second transduction with control or ERK8-specific shRNA. Lysates were taken 5 d after transduction, normalized for Ran expression, and immunoblotted for PCNA. The bottom panel shows the expression levels and activation of the Venus-ERK8(Q300A) in comparison with wild type. IP, immunoprecipitation; LV, lentivirus; WCE, whole cell extract.
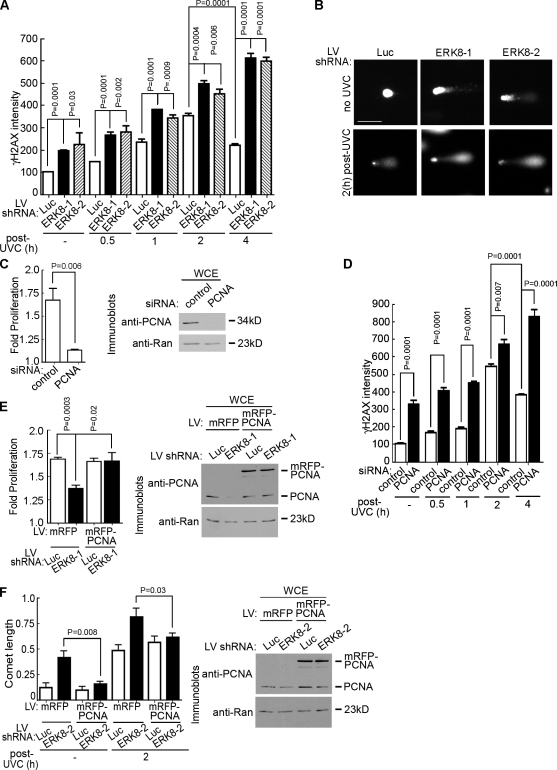
Figure 2.
ERK8 regulates DNA repair by a PCNA-dependent mechanism. (A) Analysis of DNA damage by γ-H2AX immunofluorescence in ERK8 knockdown cells. MCF-10A cells transduced for 5 d were treated with or without (−) 20 J/m2 UVC. The times indicated refer to the length of time after irradiation. At the indicated time, the cells were treated with detergent, fixed, and immunostained with an anti–γ-H2AX antibody and an anti–mouse fluorescent secondary antibody. Nuclei were stained with DRAQ5. The intensity of γ-H2AX staining was determined and normalized to the levels obtained in the control cells, which were transduced with luciferase (Luc) shRNA and not irradiated. Mean is shown (n = 2, duplicates, ≥35 cells/condition), and error bars indicate SEM. (B) Analysis of DNA damage by comet assay of cells transduced as in A, without irradiation or 2 h after UVC treatment. DNA was visualized by staining with Sybr green (Trevigen, Inc.). Bar, 50 µm. (C) Rate of proliferation of MCF-10A cells transfected with control or PCNA-specific siRNA. The rate of proliferation was determined over 48 h, starting at 2 d after transfection. Mean is shown (n = 4, sextuplicate), and error bars indicate SEM. The right panel shows the extent of PCNA knockdown 4 d after transfection in lysates normalized for Ran. (D) Analysis of DNA damage by γ-H2AX immunofluorescence in PCNA knockdown cells. Cells transfected as in C were treated as in A, and the intensity of γ-H2AX staining was determined and normalized to the levels obtained in the control cells, which were transfected with control siRNA and not irradiated. Mean is shown (n = 2, duplicates, ≥40 cells/condition), and error bars indicate SEM. (E) Rescue of proliferation by ectopic expression of PCNA in ERK8 knockdown cells. MCF-10A cells were transduced with mRFP or mRFP-PCNA and then transduced a second time with control or ERK8-specific shRNA. The rate of proliferation was determined over 48 h, starting at 4 d after transduction. Mean is shown (n = 4, quadruplicate), and error bars indicate SEM. The right panel shows the level of mRFP-PCNA (∼62 kD) in comparison with endogenous PCNA (∼34 kD). (F) Comet assay of MCF-10A cells transduced as in E. Transduced cells were treated as in B, and tail length was measured. Mean is shown (n = 2, duplicates), and error bars indicate SEM. The right panel shows the level of mRFP-PCNA (∼62 kD) in comparison with endogenous PCNA (∼34 kD). LV, lentivirus; WCE, whole cell extract.
To determine whether the reduction of PCNA levels caused by ERK8 depletion is physiologically significant, we asked whether silencing ERK8 increases DNA damage, as PCNA is essential for genomic integrity (Umar et al., 1996; Gary et al., 1997; Hoege et al., 2002; Unk et al., 2002; Zhang et al., 2003; Xia et al., 2005). Staining with an antibody to phospho-Ser139 in H2AX (γ-H2AX), a marker of DNA double-stranded breaks (Rogakou et al., 1999; Sedelnikova et al., 2003), resulted in a twofold increase in γ-H2AX staining in ERK8-depleted cells compared with the control, even in the absence of extrinsic genotoxic stress (Fig. 2 A and Fig. S2 A). PCNA acts to repair DNA in response to irradiation with UVC radiation (Essers et al., 2005). Therefore, we determined whether loss of ERK8 would further increase DNA damage in response to UVC irradiation. In response to UVC, both ATR (ataxia telangiectasia and Rad3 related) and Chk2 were activated, although loss of ERK8 reduced the level of Chk2 activation (Fig. S2 B). However, the intensity of γ-H2AX staining increased in parallel in the silenced and control cells up to 2 h after UVC treatment (Fig. 2 A and Fig. S2 A). Strikingly, however, although at 4 h the amount of γ-H2AX staining decreased in the control cells, it remained elevated when ERK8 was absent. We confirmed that the increased γ-H2AX staining was a result of DNA breaks using the comet assay. Silencing ERK8 in the absence of extrinsic genotoxic stress resulted in an approximately threefold increase in comet tail length compared with the control (Fig. 2, B and F). Moreover, in response to UVC treatment, the knockdown of ERK8 resulted in longer tail lengths compared with the control (Fig. 2, B and F). We conclude that ERK8 is not required for the initial response to DNA damage but is required for genomic stability most likely through its ability to maintain PCNA protein levels.
If the effects of ERK8 are mainly caused by a decrease in PCNA levels, depleting PCNA should phenocopy the knockdown of ERK8. Consistent with our observations with ERK8, we observed that silencing PCNA inhibited proliferation of MCF-10A cells (Fig. 2 C). Moreover, in the absence of extrinsic genotoxic stress, loss of PCNA resulted in an increased level of γ-H2AX staining compared with the control (Fig. 2 D and Fig. S2 C). Furthermore, at 4 h, although DNA repair started to occur in the control cells, the level of γ-H2AX staining remained elevated in the absence of PCNA. These results are consistent with our hypothesis that ERK8 controls genomic integrity via regulation of PCNA protein levels. To further test our hypothesis that ERK8 acts primarily through PCNA, we asked whether the ectopic expression of PCNA would reverse the decrease in proliferation caused by reduced ERK8 levels. MCF-10A cells were transduced with mRFP-tagged PCNA (mRFP-PCNA) or the mRFP control and then transduced a second time with ERK8-specific or control shRNA. In agreement with our earlier results, silencing ERK8 reduced proliferation; however, importantly, the overexpression of PCNA after silencing ERK8 prevented the decrease in proliferation (Fig. 2 E). Additionally, the forced expression of PCNA alleviated the DNA damage caused by loss of ERK8 (Fig. 2 F and Fig. S2 D). Collectively, these data support the surprising conclusion that ERK8 is essential to maintain PCNA expression at a level that prevents DNA replication stress in normal cycling cells (Halazonetis et al., 2008).
ERK8 interacts and controls PCNA turnover via a PCNA-interacting protein (PIP) box
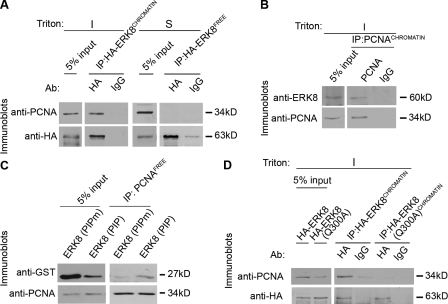
To determine how ERK8 controls PCNA levels, we asked whether ERK8 and PCNA physically associate. A fraction of wild-type Venus-tagged ERK8 (Venus-ERK8) and endogenous PCNA associated with chromatin after a detergent and high salt wash extraction, which indicates that there is a pool of ERK8 and PCNA that has a high affinity for chromatin (Fig. S3 A). Based on these observations, we used a modified chromatin immunoprecipitation assay and found that, in the presence of DNase I, chromatin-bound HA-tagged ERK8 (HA-ERK8CHROMATIN) associated with PCNACHROMATIN (Fig. 3 A). Furthermore, endogenous PCNACHROMATIN associated with endogenous ERK8CHROMATIN (Fig. 3 B). A fraction of PCNA and ERK8 was not associated with chromatin, and HA-ERK8 obtained from this soluble fraction (HA-ERK8FREE) and PCNAFREE did not interact (Fig. 3 A). Most of the interacting partners for PCNA interact through a conserved motif called the PIP box (Warbrick, 1998; Moldovan et al., 2007). We identified a putative PIP box in ERK8, specifically the sequence from 297 to 308 (QALQHPYVQRFH; ERK8 (PIP)), where bold indicates a match with the conserved motif (Fig. S3 B). The spacing between the Q and the V is usually two residues; however, DNA polymerase β has a three-residue spacing, like ERK8 (Kedar et al., 2002). A GST fusion protein containing the putative ERK8 PIP box (GST–ERK8 (PIP)) interacted with PCNAFREE (Fig. 3 C), but a mutant PIP box, in which the residue corresponding to Gln300 was mutated to Ala (GST–ERK8 (PIPm)) did not interact. Further confirmation that ERK8 contains a PIP box was shown by mutation of Gln300 to Ala within the context of the full-length kinase (ERK8(Q300A)). This mutant did not associate with PCNACHROMATIN even though it was able to bind chromatin (Fig. 3 D and Fig. S3 C). Collectively, we conclude that ERK8CHROMATIN associates with PCNACHROMATIN via the PIP box.
Figure 3.
ERK8 interacts with PCNA via a PIP box. (A and B) Chromatin was immunoprecipitated with antibodies against ectopically expressed HA-tagged ERK8 (A) or endogenous PCNA (B), treated with DNase I, and analyzed by immunoblotting. A fraction of the input is shown for comparison to the level of associated PCNA or ERK8. (C) GST–ERK8 (PIP) fusion proteins were immunoprecipitated with antibodies to PCNA. The soluble fraction of MCF-10A lysates was incubated with GST-PIP box fusions containing the wild-type (QALQHPYVQRFH) or mutant ERK8 PIP (QALAHPYVQRFH) box. PCNA was immunoprecipitated and electrophoresized, and associating proteins were analyzed by immunoblotting. A fraction of the input is shown for comparison to the level of associated GST–ERK8 (PIP). (D) Chromatin was immunoprecipitated with antibodies (Ab) against ectopically expressed HA-tagged wild type or ERK8(Q300A) mutant, treated with DNase I, and analyzed by immunoblotting. A fraction of the input is shown for comparison to the level of associated PCNA. I, insoluble fraction; IP, immunoprecipitation; S, soluble fraction.
We next asked whether ERK8 controls PCNA protein stability by pulse-chase labeling with [35S]Met. The degradation rate of PCNAFREE, which does not interact with ERK8, was similar in the presence and absence of ERK8 (Fig. 4 A and Fig. S3 D). However, silencing ERK8 increased the destruction of PCNACHROMATIN by ∼60% compared with the control. Consistent with these observations, total PCNA in ERK8-depleted cells was reduced approximately twofold relative to the control after a 12-h treatment with the protein synthesis inhibitor cycloheximide (Fig. S3 E). Thus, loss of ERK8 enhances PCNACHROMATIN turnover. Mutation of Tyr211 has been reported to destabilize PCNA (Wang et al., 2006). However, we did not detect any Tyr phosphorylation of PCNA (Fig. S3 F). These results are in agreement with orthophosphate labeling analysis of PCNA, which also did not detect phosphorylation of PCNA (Naryzhny and Lee, 2004). We propose that the increase in PCNACHROMATIN turnover caused by the loss of ERK8 indirectly decreases PCNAFREE levels because of continued recruitment of PCNAFREE to the chromatin in an effort to maintain critical cellular functions (Fig. 5 A). This hypothesis is consistent with the literature in which increasing the extent of DNA damage leads to depletion of PCNAFREE (Mortusewicz and Leonhardt, 2007).
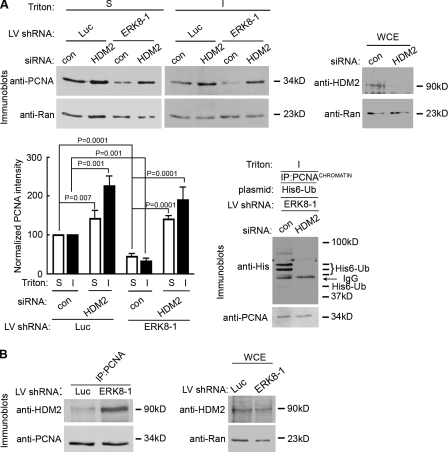
Figure 5.
ERK8 regulates the association of PCNA with HDM2. (A) Silencing HDM2 enhances PCNA levels in the absence or presence of ERK8. MCF-10A cells were transduced with control (con) or ERK8-specific shRNA, followed by transfection with control or HDM2 siRNA. Lysates were taken 5 d after transduction and divided into insoluble (I) and soluble (S) fractions, normalized for Ran levels, and immunoblotted for PCNA. The top right panel shows the extent of HDM2 knockdown in lysates normalized for Ran levels. Quantitation of the relative amount of PCNA in comparison to Ran and normalized to the appropriate control, which was transduced with luciferase (Luc) shRNA and transfected with control siRNA. Mean is shown (n = 3), and error bars indicate SEM. Silencing HDM2 decreased levels of ubiquitinated PCNA. MCF-10A cells were transduced with ERK8-specific shRNA followed by transfection with a plasmid encoding His6-Ub and control or HDM2 siRNA. PCNA was immunoprecipitated 5 d after transduction, and the normalized PCNA immunoprecipitates were analyzed for ubiquitination. The IgG band is from the immunoprecipitating antibody. (B) Loss of ERK8 increases the association of HDM2 with PCNA. MCF-10A cells were transduced with control or ERK8-specific shRNA for 5 d. PCNA was immunoprecipitated, and the normalized PCNA immunoprecipitates were analyzed for HDM2. The right panel shows the level of HDM2 expression in lysates normalized for Ran. IP, immunoprecipitation; LV, lentivirus; WCE, whole cell extract.
We next asked whether the interaction of ERK8 with PCNA is necessary for stabilization of PCNA. MCF-10A cells were transduced with Venus-rERK8, Venus–rERK8 PIP mutant (Venus-rERK8(Q300A)), or the Venus control and then transduced a second time with ERK8-specific or control shRNA. Expression of resistant ERK8 prevented the decrease in PCNA levels caused by silencing of endogenous ERK8 (Fig. 4 B). However, importantly, the ERK8 PIP mutant was unable to rescue PCNA levels even though the level of rERK8(Q300A) and its ability to associate with the chromatin was similar to that of the wild type (Fig. 4 B and Fig. S3 C). Furthermore, the mutant was as active as the wild type, as shown by the anti-pERK8 antibody (Fig. 4 B). This antibody recognizes the dual phosphorylation of the T-E-Y motif, which must be phosphorylated for members of the ERK1/2 family to be active (Her et al., 1993). Therefore, the pERK8 levels reflect the amount of active kinase. Surprisingly, expression of the ERK8 PIP mutant caused an at least twofold decrease in PCNA levels even in the presence of endogenous ERK8. These results indicate that ERK8(Q300A) acts as a dominant-negative, most likely by competing with endogenous ERK8 for binding to chromatin, thereby blocking association of the endogenous ERK8 with PCNA, which permits PCNA destruction. Collectively, our data support a model in which ERK8CHROMATIN stabilizes PCNA by inhibiting access to PCNACHROMATIN of a destruction factor. This is the first report of a PCNA-binding partner that regulates PCNA stability during normal cell cycling (Izumi et al., 2001; Wang et al., 2006; Yu et al., 2009).
ERK8 inhibits the interaction of PCNA with HDM2
PCNA is able to act as a platform to mediate the degradation of some of its binding partners via a specialized PIP box (Havens and Walter, 2009). However, information on how PCNA turnover is regulated in normal cycling cells is lacking. In response to DNA damage to UV, PCNA destruction has been shown to occur via the 26S proteasome pathway (Yu et al., 2009). Therefore, we reasoned that an E3 ligase, which is able to compete for ERK8 binding to PCNA, may also control PCNA turnover in normal cycling cells. HDM2 is the only currently known E3 ligase that interacts with PCNA through a PIP box (Banks et al., 2006), and therefore, it could possibly compete with ERK8 for binding to PCNA. However, it is not known whether HDM2 targets PCNA for destruction. We observed that silencing HDM2 resulted in an increase in PCNA levels (Fig. 5 A). These results suggest that HDM2 regulates PCNA turnover in normal cycling cells. However, as HDM2 regulates p53 levels, knockdown of HDM2 will result in an increase in p53 levels and disruption of the cell cycle (Martin et al., 1995). To investigate whether elevated p53 levels could increase PCNA levels, we used the inhibitor nutlin-3 (Vassilev et al., 2004). As expected by disrupting the association of HDM2 and p53, the levels of both proteins increased substantially (Fig. S4). We also observed that nutlin-3 altered the electrophoretic mobility of Rb to generate a single immunoreactive band (Fig. S4), which is consistent with the unphosphorylated form of Rb. Unphosphorylated Rb binds and inhibits E2F (Stevens and La Thangue, 2003), which can result in decreased expression of some E2F-regulated genes such as PCNA (Black et al., 2005). However, treatment with nutlin-3 did not alter the levels of PCNA. These results indicate that the increase in PCNA levels in response to the loss of HDM2 is independent of p53 and the phosphorylation status of Rb. To investigate whether HDM2 facilitates PCNA turnover in the absence of ERK8, we silenced both HDM2 and ERK8. Strikingly, the decrease in PCNA levels caused by the loss of ERK8 was reversed by also silencing HDM2 expression (Fig. 5 A). From this data, we would predict that the loss of HDM2 would prevent ubiquitination of PCNA. To test this hypothesis, we silenced ERK8 and then transfected with control or HDM2 siRNA. Additionally, we transfected with a plasmid encoding a six-His tag fused to ubiquitin (His6-Ub). Loss of HDM2 decreased the levels of ubiquitinated PCNA (Fig. 5 A). Knockdown of ERK8 did not alter the levels of HDM2 (Fig. 5 B). However, silencing ERK8 increased the association of PCNA with HDM2 (Fig. 5 B). We conclude that ERK8 inhibits PCNA degradation by inhibiting its association with HDM2, which facilitates PCNA degradation via the 26S proteasome pathway.
ERK8 is activated by chromatin binding
It has been reported that ERK8 activity is increased in response to various DNA-damaging agents (Klevernic et al., 2009). Using identical conditions as reported by Klevernic et al. (2006, 2009), we observed changes in ERK8 activity only in response to 1 mM H2O2 (Fig. S5 A). We did not observe changes in ERK8 protein levels with any of the DNA-damaging reagents tested. Activation of ERK1/2 was dose dependent on H2O2, but ERK8 activation was observed only at 1 mM H2O2 (Fig. S5 B). This concentration of H2O2 was extremely toxic and resulted in cell death within 3 h, and therefore, it is difficult to determine the physiological relevance of this observation. We conclude that in MCF-10A cells, DNA-damaging agents do not enhance activation of ERK8, and basal levels of ERK8 activation are sufficient to stabilize PCNA levels.
Active ERK8, as shown by pERK8, is mainly present in the insoluble fraction (Fig. S5 C). These results are consistent with the immunofluorescence data that show that a kinase-dead mutant of ERK8 (ERK8(K42A)) does not associate with the chromatin (Fig. S3 A). This mutant has the essential Lys residue that is necessary for interaction with ATP mutated to Ala (Abe et al., 2002). It is possible that the interaction of PCNA activates ERK8, like Chk1 (Scorah et al., 2008). However, pERK8 levels are similar for wild type and the ERK8(Q300A) mutant, which does not bind PCNA (Figs. 3 D and 4 B). It is also unlikely that ERK8 is recruited to chromatin by PCNA because, in contrast to PCNA, soluble ERK8 does not redistribute to the insoluble fraction in response to UVC (Fig. S5 C). Furthermore, the ERK8(Q300A) mutant associates with chromatin but not with PCNA. Additionally, ERK8 and PCNA only partially colocalize as 42% ± 10.9 of the nuclei positive for Venus-ERK8 costain for PCNA, whereas in nuclei positive for PCNA, 22% ± 5.4 of the cells costain with ERK8. Thus, ERK8 must be able to interact with chromatin in a PCNA-independent manner.
To investigate the mechanism by which ERK8 associates with the chromatin, we analyzed deletion mutants. ERK8 (1–475) was active, associated with chromatin, and stimulated proliferation similar to that of the wild type (Fig. 6, A and B). However, further deletions of the C terminus generated inactive constructs that did not target to chromatin (Fig. 6 A). We identified a repeating PXXXP motif, which is highly conserved among mammals, located between residues 371 and 398 (Fig. S5 D). Unexpectedly, mutations within this motif (ERK8 (P390A/P398A)) substantially increased the level of activation, as indicated by pERK8, compared with the wild type (Fig. 6 C). However, the mutant also showed defective chromatin association (Fig. 6 D and Fig. S5 E). We conclude that the PXXXP motif acts as an autoinhibitory regulator of kinase activity. We propose that the mutation mimics the conformational change that occurs upon association of ERK8 with chromatin, which relieves autoinhibition. We conclude that binding to chromatin activates ERK8, independently of PCNA binding.
Figure 6.
ERK8 activity is regulated by autoinhibition. (A) Representative images of wild-type and mutant ERK8CHROMATIN. MCF-10A cells were transduced with Venus-ERK8 constructs. Transduced cells were treated with detergent and a high salt wash and fixed. Venus-ERK8 constructs were detected by direct fluorescence. The right panel shows immunoblots of lysates normalized for Venus expression. The black line indicates that intervening lanes were removed. WCE, whole cell extract. Bar, 50 µm. (B) Graph of the proliferation rate of transduced MCF-10A cells between day 2 and 4 after transduction. Mean is shown (n = 7, quadruplicates), and error bars indicate SEM. (C) Relative activity of wild type and ERK8 (P390A, P398A) as detected by pERK8. Transduced MCF-10A lysates were divided into insoluble (I) and soluble (S) fractions and normalized to the levels of active ERK1/2, and total expression of Venus-tagged constructs and pERK8 was determined. Active ERK1/2 levels were detected by dual phosphorylation of its T-E-Y motif. (D) The percentage of the total nuclei that stained for chromatin-bound Venus-ERK8 constructs after a detergent and high salt wash extraction. The total number of nuclei was determined by staining with DRAQ5. Venus-ERK8 constructs were detected by direct fluorescence. Mean is shown (n = 2, ≥50 cells/condition), and error bars indicate SEM. LV, lentivirus.
ERK8 stabilizes PCNA in primary mammary epithelial cells
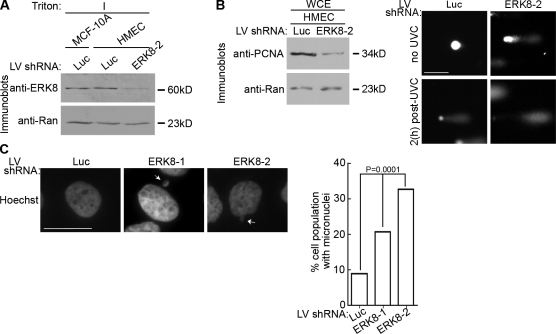
ERK8 was detected in the detergent-insoluble fraction of primary human mammary epithelial (HME) cells at levels similar to MCF-10A cells (Fig. 7 A). Importantly, silencing ERK8 in HME cells decreased PCNA levels (Fig. 7 B), and this reduction in PCNA was accompanied by a substantial increase in the frequency of DNA breaks for both untreated and UVC-irradiated HME cells (Fig. 7 B). The fold increase in comet length in HME cells ranged from ∼2–10-fold in the absence of UVC, and this range probably reflects the genetic differences between patient samples. Thus, the regulation of PCNA by ERK8 is not an artifact of the MCF-10A immortalized cell line but occurs also in primary mammary cells.
Figure 7.
ERK8 is critical for genomic integrity in mammary epithelial cells. (A) Analysis of endogenous ERK8 knockdown in HME cells (HMEC) transduced with control or ERK8 shRNA for 5 d. The detergent-insoluble fraction (I) of the transduced lysates was normalized for Ran expression. (B) Analysis of DNA damage by comet assay of transduced HME cells treated without irradiation or 2 h after UVC treatment. The left panel shows lysates from transduced HME cells that were normalized for Ran expression and immunoblotted. (C) Analysis of DNA morphology by Hoechst staining of transduced MCF-10A cells kept in long-term culture (∼2 wk). Arrows indicate micronuclei. The percentage of the total cell population that contained micronuclei was determined. Mean is shown (≥500 cells), and error bars indicate SEM. Luc, luciferase; LV, lentivirus. Bars: (B) 50 µm; (C) 10 µm.
ERK8 is inactive in transformed breast lines
Our results predict that loss of ERK8 will lead to genomic instability. To test this prediction, we silenced ERK8 in MCF-10A cells and maintained the cells in culture for 2 wk. Analysis of the nuclei of surviving cells showed an at least threefold increase in fragmented nuclei with the more potent ERK8 shRNA (Fig. 7 C). As transformed cells are often genetically unstable (Hoeijmakers, 2001), we investigated whether ERK8 was functional in human breast cancer lines. MCF-7 cells contained endogenous ERK8, whereas ERK8 was not detected in T47-D cells (Fig. 8 A). As the levels of endogenous active ERK8 were below the detection of the pERK8 antibody, we ectopically expressed ERK8 in the breast lines. ERK8 was only active in MCF-10A cells (Fig. 8 B). To confirm this analysis, we silenced ERK8 in MCF-7 cells and found that PCNA levels were not altered because of the loss of ERK8 (Fig. 8 C). These results further suggest that ERK8 is inactive in MCF-7 cells as only the active form of ERK8 regulates PCNA turnover. These observations are consistent with the literature in which PCNA degradation via the 26S proteasome pathway was not observed in transformed cells in the absence of extrinsic genotoxic stress (Izumi et al., 2001; Wang et al., 2006; Yu et al., 2009). Collectively, these data argue that ERK8 control of PCNA levels is physiologically relevant, as ERK8 activity is inhibited or lost in transformed cells.
Figure 8.
Breast cancer cells do not have active ERK8. (A) Analysis of endogenous ERK8 levels in breast cancer cell lines. Whole cell extracts (WCE) from different breast cell lines were normalized for Ran. For comparison, MCF-10A cells were transduced with luciferase (Luc) or ERK8 shRNA, and the insoluble (I) fraction was analyzed 5 d after transduction. (B) Lysates from the various breast lines were transduced with Venus-ERK8, normalized to expression of Venus-ERK8, and immunoblotted for active ERK8 (pERK8). (A and B) The black lines indicate that intervening lanes were removed. (C) Loss of ERK8 does not decrease PCNA levels in MCF-7 cells. MCF-7 cells were transduced with luciferase or ERK8 shRNA for 5 d. Lysates were normalized to Ran and immunoblotted. (D) Model of ERK8 regulation of PCNA levels. Active ERK8 preferentially binds to the chromatin through its binding partners. ERK8CHROMATIN and PCNACHROMATIN interact via the ERK8 PIP box. Loss of ERK8 binding to PCNA via silencing or mutation of the PIP domain results in the increased recruitment of HDM2, which enhances PCNA turnover. Autoinhibition is alleviated by mutating Pro390 and Pro398. However, these mutations interfere with chromatin binding. LV, lentivirus.
Discussion
We conclude that ERK8 is as a key controller of genomic stability in mammary epithelial cells via its ability to regulate the formation of complexes containing PCNA. In untransformed mammary cells, ERK8 prevents the destruction of PCNA that is mediated via HDM2 (Fig. 8 D). Remarkably, a twofold reduction in PCNA levels is sufficient to cause extensive DNA damage and mitotic failure even in the absence of extrinsic genotoxic stress. We would speculate that in breast cancer cells, there is a mechanism to prevent HDM2 from associating with PCNA in the absence of active ERK8. Our results also suggest that loss of ERK8 activity is important in the transformation process.
We have identified two important functional domains in ERK8, the PIP box and the PXXXP motif. The PIP box is located within the kinase domain, and the PXXXP motif is in the C-terminal extension. To identify the region within the kinase domain that contains the PIP box, we modeled the ERK8 kinase domain to that of ERK2 and found that the PIP box is adjacent to the conserved docking (CD) domain. The CD domain contributes but is not solely responsible for the interactions of the MAPK superfamily with their docking partners (Tanoue et al., 2000, 2001; Zhang et al., 2003). A corresponding PIP box was not found in other members of the ERK1/2 or p38MAPK families but was present in JNK1/3 (Fig. S5 F). It is not known whether JNK1/3 can interact with PCNA. The CD domain in ERK8 is unusual in that it contains a Cys residue, whereas all other members of the MAPK superfamily have an acidic residue in this position. It is possible that the Cys residue regulates the ability of ERK8 to interact with PCNA. The PXXXP motif functions to regulate the association of ERK8 with chromatin and thereby indirectly regulates association with PCNA as only ERK8CHROMATIN interacts with PCNA. PCNA is known to interact with a plethora of proteins that contain PIP boxes and the mechanism by which these interactions are regulated is unknown.
It is intriguing that a rapidly evolving kinase would control genomic stability via PCNA, which is a highly conserved protein. Curiously, mammary epithelial cells are susceptible to transformation by loss of BRCA1, which is also important in DNA metabolism (Saal et al., 2008). We speculate that because mammary epithelial cells are subjected to proliferative signals throughout the reproductive life of the animal that additional layers of regulation are required to ensure their genomic integrity.
Materials and methods
Plasmids and lentiviral production
Constructs used to generate lentivirus, including pSPAX2, pLVTHM, and pMD2G, were provided by D. Trono (Swiss Federal Institute of Technology, Lausanne, Switzerland; Zufferey et al., 1997). pLV-Venus and pLV-mRFP lentivirus constructs were provided by I.G. Macara (University of Virginia, Charlottesville, VA; McCaffrey and Macara, 2009). pCMV5HIS6-Ub was provided by D. Wotton (University of Virginia). pWPI was purchased from Addgene. Lentiviral production was performed as described previously (McCaffrey and Macara, 2009) but titered using MCF-10A cells.
The short hairpin sequence including the ERK8-targeting shRNAs (in bold) are (ERK8-1) 5′-GATCCCCACATTTACCTGGTGTTTGATTCAAGAGATCAAACACCAGGTAAATGTTTTTTGGAAA-3′ and (ERK8-2) 5′-GATCCCCGACAGATGCCCAGAGAACATTCAAGAGATGTTCTCTGGGCATCTGTCTTTTTGGAAA-3′. The short hairpin sequence including the luciferase-targeting shRNAs (in bold) is 5′-GATCCCCCGTACGCGGAATACTTCGATTCAAGAGATCGAAGTATTCCGCGTACGTTTTTGGAA-3′ (Malliri et al., 2004). GST-PIP and GST-PIPm were generated by subcloning into pGEX2T (GE Healthcare) the annealed oligonucleotides with the upper strand sequences 5′-GATCCCAGGCACTGCAGCACCCCTACGTGCAGAGGTTCCACTGCCCCTGAG-3′ and 5′-GATCCCAGGCACTGGCGCACCCCTACGTGCAGAGGTTCCACTGCCCCTGAG-3′. All shRNA, deletion, and mutant constructs were verified by sequencing (Biomolecular Research Core, University of Virginia).
Cell culture and treatment
MCF-10A, MCF-7, and T47D were cultured as indicated by the American Type Culture Collection. Primary HME cells were purified from tissue (McCaffrey and Macara, 2009) and cultured (Eisinger-Mathason et al., 2008). For irradiation, the cells were washed with PBS and exposed to 20 J/m2 UVC. Comet assay was performed according to the manufacturer (Trevigen, Inc.) and electrophoresed under alkaline conditions (275 mA for 30 min at 4°C). Cell fractionation was performed as described previously (Avkin et al., 2006). Transient transfection was performed with Lipofectamine 2000 (Invitrogen) reagent using nontargeting siRNA #1, human MDM2 SMARTpool siRNA, or human PCNA SMARTpool siRNA (Thermo Fisher Scientific). The following chemicals were used: nutlin-3 (EMD); hydroxyurea, etoposide, cisplatin, and MMS (methyl methane sulphonate; Sigma-Aldrich); and H2O2 (Thermo Fisher Scientific) and MNNG (N-methyl-N’-N-nitrosoguanidine; TCI America).
Immunodetection and immunofluorescence
Primary antibodies used were monoclonal anti-p27, rabbit anti–RFC-1, monoclonal anti-GST, rabbit anti-HA, monoclonal anti-HDM2, monoclonal anti-p53, and monoclonal anti-PCNA (Santa Cruz Biotechnology, Inc.); monoclonal anti–cyclin D1, monoclonal anti-Rb, rabbit anti–phospho-Ser428 ATR, rabbit anti–phospho-Thr68 Chk2, and rabbit anti-Chk2 (Cell Signaling Technology); monoclonal anti–phospho-Tyr, rabbit anti–phospho-Ser, rabbit anti–cyclin E, and monoclonal anti–γ-H2AX (Millipore); rabbit anti–phospho-Thr (Invitrogen); monoclonal anti-Rad9 and rabbit anti–phospho-MAPK (pThr202/pTyr204; Thermo Fisher Scientific); rabbit anti-ERK8 (Abgent); monoclonal anti-PCNA (Dako); monoclonal anti-Ran (BD); monoclonal anti–cyclin A (Novocastra); goat anti-GFP and rabbit anti-GFP (Venus; Abcam); and monoclonal anti-HA (12CA5; Lymphocyte Culture Center, University of Virginia). Secondary antibodies used were goat anti–mouse, goat anti–rabbit, and donkey anti–goat (Jackson ImmunoResearch Laboratories, Inc.).
Detection of PCNA and Venus-tagged chromatin-binding proteins was adapted from Sporbert et al. (2002). In brief, cells were plated on coverslips, washed twice with ice-cold PBS, and permeabilized for 30 s with ice-cold cytoskeleton buffer (10 mM Pipes, pH 6.8, 250 mM sucrose, 50 mM NaCl, 2 mM MgCl2, 2 mM EGTA, and 0.1% Triton X-100). The permeabilized cells were washed with ice-cold wash buffer (4.3 mM Na2HPO4, 1.5 mM KH2PO4, 520 mM NaCl, and 2.66 mM KCl) and fixed in ice-cold methanol at −20°C for 5 min. PCNA was detected by indirect fluorescence with a mouse monoclonal anti-PCNA antibody (clone PC-10; Dako) and an anti–mouse IgG antibody conjugated with Alexa Fluor 546 (Invitrogen). Direct fluorescence was observed with Venus fusion proteins.
Detection of γ-H2AX was adapted from Jørgensen et al. (2007). Cells were plated on coverslips, washed twice in ice-cold PBS, followed by extraction for 10 min on ice with Triton buffer (20 mM Hepes, pH 7.4, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, and 0.5% Triton X-100), and fixed with 4% paraformaldehyde for 10 min. γ-H2AX was detected by indirect fluorescence with phospho-Ser139 histone H2A.X (clone JBW301; Millipore) primary antibody and an anti–mouse IgG antibody conjugated with Alexa Fluor 546. DNA was stained with Hoechst (Sigma-Aldrich) or DRAQ5 (Axxora).
Densitometry analysis for immunodetection was performed using ImageJ (National Institutes of Health) or ImageQuant (GE Healthcare) software. Indirect and direct fluorescent images were obtained at room temperature with a laser-scanning microscope (510/Meta/FCS; Carl Zeiss, Inc.) using a 40× NA 1.3 oil immersion lens. Images were acquired using LSM-FCS software (Carl Zeiss, Inc.) and quantitated using Openlab 3.1.4 (PerkinElmer) and ImageJ software. Images were processed in Photoshop version CS3 (Adobe).
Proliferation and cell cycle assays
Cells were analyzed for proliferation using a Luminescent Cell Viability Assay (Promega). Fixed cells (2% paraformaldehyde, pH 7.4, and 0.05% Triton X-100) were incubated with 40 µM DRAQ5, washed, and analyzed on a FACSCalibur (BD) using FlowJo 6.4.2 software (Tree Star, Inc.; Flow Cytometry Core Facility, University of Virginia).
Immunoprecipitation
Cells were washed twice with ice-cold PBS. The soluble fraction was obtained by incubating the cells (5 min at 4°C) with cytoskeleton buffer with 1 µM microcystin and phosphatase inhibitor cocktail (Sigma-Aldrich). The insoluble fraction was treated with sonication buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 1 µM microcystin, and phosphatase inhibitor cocktail). The insoluble fraction was sonicated (15 pulses of 5 s with 15-s cool down on setting 5) with a cup horn sonicator (Misonix). The sonicated insoluble fraction was incubated with 3 mM MgCl2 and 400 U RNase-free DNase I/10 µg DNA (New England Biolabs, Inc.) for 1 h at 4°C. Soluble and insoluble fractions were incubated with 2 µg HA, PCNA, or IgG antibodies for 3 h at 4°C in thermomixer (900 rpm). In some experiments 1 µg GST, GST-PIP, or GST-PIPm was incubated with the soluble fraction. Approximately 140 µl of Dynabeads sheep anti–mouse secondary antibody (Invitrogen) was washed with immunoprecipitation buffer and incubated with lysate for 1 h at 4°C in a thermomixer (900 rpm). The immunoprecipitates were washed five times in the appropriate immunoprecipitation buffer, three times high salt buffer (10 mM Hepes, pH 7.4, 300 mM NaCl, 10 mM KCl, and 1 mM EDTA), and three times with the immunoprecipitation buffer. The beads were boiled in 2× SDS lysis buffer (0.12 M Tris-Cl, pH 6.8, 4% SDS, 20% glycerol, 0.2 M DTT, and 0.008% bromophenol blue), heated to 100°C for 5 min
Pulse-chase labeling
Cells were washed and preincubated for 4 h with cold labeling media (Met-free DMEM/F12 [Sigma-Aldrich], 5% dialyzed horse serum, 0.5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 µg/ml insulin, and 5.75 µM cold Met). l-[35S]Met (200 µCi/ml; 800 Ci/mmol; PerkinElmer) was added and incubated for 4 h. After washing with labeling media plus 2 mM cold Met, the cells were replaced in their normal growth media and harvested for immunoprecipitation.
Statistical analysis
Data are presented as mean ± SEM; Student’s two-tailed t test was used for comparisons, with P < 0.05 considered significant.
Online supplemental material
Fig. S1 shows that ERK8 regulates the cell cycle. Fig. S2 shows that loss of ERK8 increases DNA damage. Fig. S3 shows that ERK8 regulates PCNA turnover. Fig. S4 shows that inhibition of p53 degradation does not alter PCNA protein levels. Fig. S5 examines the activation, levels, and subcellular distribution of ERK8 in response to DNA-damaging agents. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002124/DC1.
Acknowledgments
This work was supported by grant GM084386 (to D.A. Lannigan), the Robert R. Wagner Fellowship in Cancer Research (to A.L. Groehler), grant T32CA009109-30 (to A.L. Groehler), and the Patients and Friends of the University of Virginia Cancer Center research fund (to D.A. Lannigan).
Footnotes
Abbreviations used in this paper:
- CD
- conserved docking
- HME
- human mammary epithelial
- PCNA
- proliferating cell nuclear antigen
- PIP
- PCNA-interacting protein
- Rb
- retinoblastoma
- shRNA
- short hairpin RNA
References
- Abe M.K., Saelzler M.P., Espinosa R., III, Kahle K.T., Hershenson M.B., Le Beau M.M., Rosner M.R. 2002. ERK8, a new member of the mitogen-activated protein kinase family. J. Biol. Chem. 277:16733–16743 10.1074/jbc.M112483200 [DOI] [PubMed] [Google Scholar]
- Andersen P.L., Xu F., Xiao W. 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18:162–173 10.1038/cr.2007.114 [DOI] [PubMed] [Google Scholar]
- Avkin S., Sevilya Z., Toube L., Geacintov N., Chaney S.G., Oren M., Livneh Z. 2006. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 22:407–413 10.1016/j.molcel.2006.03.022 [DOI] [PubMed] [Google Scholar]
- Azam N., Vairapandi M., Zhang W., Hoffman B., Liebermann D.A. 2001. Interaction of CR6 (GADD45gamma) with proliferating cell nuclear antigen impedes negative growth control. J. Biol. Chem. 276:2766–2774 10.1074/jbc.M005626200 [DOI] [PubMed] [Google Scholar]
- Banks D., Wu M., Higa L.A., Gavrilova N., Quan J., Ye T., Kobayashi R., Sun H., Zhang H. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle. 5:1719–1729 [DOI] [PubMed] [Google Scholar]
- Black E.P., Hallstrom T., Dressman H.K., West M., Nevins J.R. 2005. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. USA. 102:15948–15953 10.1073/pnas.0504300102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P., Meloche S. 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta. 1773:1376–1387 10.1016/j.bbamcr.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Das-Bradoo S., Nguyen H.D., Wood J.L., Ricke R.M., Haworth J.C., Bielinsky A.K. 2010. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat. Cell Biol. 12:74–79 10.1038/ncb2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason T.S., Andrade J., Groehler A.L., Clark D.E., Muratore-Schroeder T.L., Pasic L., Smith J.A., Shabanowitz J., Hunt D.F., Macara I.G., Lannigan D.A. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell. 31:722–736 10.1016/j.molcel.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Theil A.F., Baldeyron C., van Cappellen W.A., Houtsmuller A.B., Kanaar R., Vermeulen W. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 25:9350–9359 10.1128/MCB.25.21.9350-9359.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R., Ludwig D.L., Cornelius H.L., MacInnes M.A., Park M.S. 1997. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 272:24522–24529 10.1074/jbc.272.39.24522 [DOI] [PubMed] [Google Scholar]
- Gilljam K.M., Feyzi E., Aas P.A., Sousa M.M., Müller R., Vågbø C.B., Catterall T.C., Liabakk N.B., Slupphaug G., Drabløs F., et al. 2009. Identification of a novel, widespread, and functionally important PCNA-binding motif. J. Cell Biol. 186:645–654 10.1083/jcb.200903138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes X.V., Burgers P.M. 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19:3811–3821 10.1093/emboj/19.14.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Havens C.G., Walter J.C. 2009. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell. 35:93–104 10.1016/j.molcel.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D.S., Banga S.S., Grigliatti T.A., Boyd J.B. 1994. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 13:1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her J.H., Lakhani S., Zu K., Vila J., Dent P., Sturgill T.W., Weber M.J. 1993. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem. J. 296:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- Izumi M., Yatagai F., Hanaoka F. 2001. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 276:48526–48531 10.1074/jbc.M101463200 [DOI] [PubMed] [Google Scholar]
- Jaskulski D., deRiel J.K., Mercer W.E., Calabretta B., Baserga R. 1988. Inhibition of cellular proliferation by antisense oligodeoxynucleotides to PCNA cyclin. Science. 240:1544–1546 10.1126/science.2897717 [DOI] [PubMed] [Google Scholar]
- Jørgensen S., Elvers I., Trelle M.B., Menzel T., Eskildsen M., Jensen O.N., Helleday T., Helin K., Sørensen C.S. 2007. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179:1337–1345 10.1083/jcb.200706150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar P.S., Kim S.J., Robertson A., Hou E., Prasad R., Horton J.K., Wilson S.H. 2002. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J. Biol. Chem. 277:31115–31123 10.1074/jbc.M201497200 [DOI] [PubMed] [Google Scholar]
- Klevernic I.V., Stafford M.J., Morrice N., Peggie M., Morton S., Cohen P. 2006. Characterization of the reversible phosphorylation and activation of ERK8. Biochem. J. 394:365–373 10.1042/BJ20051288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevernic I.V., Martin N.M., Cohen P. 2009. Regulation of the activity and expression of ERK8 by DNA damage. FEBS Lett. 583:680–684 10.1016/j.febslet.2009.01.011 [DOI] [PubMed] [Google Scholar]
- Malliri A., van Es S., Huveneers S., Collard J.G. 2004. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J. Biol. Chem. 279:30092–30098 10.1074/jbc.M401192200 [DOI] [PubMed] [Google Scholar]
- Martin K., Trouche D., Hagemeier C., Sørensen T.S., La Thangue N.B., Kouzarides T. 1995. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 375:691–694 10.1038/375691a0 [DOI] [PubMed] [Google Scholar]
- McCaffrey L.M., Macara I.G. 2009. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 23:1450–1460 10.1101/gad.1795909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G.L., Pfander B., Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell. 129:665–679 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Mortusewicz O., Leonhardt H. 2007. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 8:81 10.1186/1471-2199-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryzhny S.N., Lee H. 2001. Protein profiles of the Chinese hamster ovary cells in the resting and proliferating stages. Electrophoresis. 22:1764–1775 [DOI] [PubMed] [Google Scholar]
- Naryzhny S.N., Lee H. 2004. The post-translational modifications of proliferating cell nuclear antigen: acetylation, not phosphorylation, plays an important role in the regulation of its function. J. Biol. Chem. 279:20194–20199 10.1074/jbc.M312850200 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 10.1083/jcb.146.5.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L.H., Gruvberger-Saal S.K., Persson C., Lövgren K., Jumppanen M., Staaf J., Jönsson G., Pires M.M., Maurer M., Holm K., et al. 2008. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 40:102–107 10.1038/ng.2007.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorah J., Dong M.Q., Yates J.R., III, Scott M., Gillespie D., McGowan C.H. 2008. A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function. J. Biol. Chem. 283:17250–17259 10.1074/jbc.M800369200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova O.A., Pilch D.R., Redon C., Bonner W.M. 2003. Histone H2AX in DNA damage and repair. Cancer Biol. Ther. 2:233–235 [DOI] [PubMed] [Google Scholar]
- Sporbert A., Gahl A., Ankerhold R., Leonhardt H., Cardoso M.C. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365 10.1016/S1097-2765(02)00729-3 [DOI] [PubMed] [Google Scholar]
- St Onge R.P., Udell C.M., Casselman R., Davey S. 1999. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell. 10:1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C., La Thangue N.B. 2003. E2F and cell cycle control: a double-edged sword. Arch. Biochem. Biophys. 412:157–169 10.1016/S0003-9861(03)00054-7 [DOI] [PubMed] [Google Scholar]
- Tanoue T., Adachi M., Moriguchi T., Nishida E. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110–116 10.1038/35000065 [DOI] [PubMed] [Google Scholar]
- Tanoue T., Maeda R., Adachi M., Nishida E. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20:466–479 10.1093/emboj/20.3.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A., Buermeyer A.B., Simon J.A., Thomas D.C., Clark A.B., Liskay R.M., Kunkel T.A. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 87:65–73 10.1016/S0092-8674(00)81323-9 [DOI] [PubMed] [Google Scholar]
- Unk I., Haracska L., Gomes X.V., Burgers P.M., Prakash L., Prakash S. 2002. Stimulation of 3′—>5′ exonuclease and 3′-phosphodiesterase activities of yeast apn2 by proliferating cell nuclear antigen. Mol. Cell. Biol. 22:6480–6486 10.1128/MCB.22.18.6480-6486.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 303:844–848 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751 10.1146/annurev.biochem.67.1.721 [DOI] [PubMed] [Google Scholar]
- Wang S.C., Nakajima Y., Yu Y.L., Xia W., Chen C.T., Yang C.C., McIntush E.W., Li L.Y., Hawke D.H., Kobayashi R., Hung M.C. 2006. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8:1359–1368 10.1038/ncb1501 [DOI] [PubMed] [Google Scholar]
- Warbrick E. 1998. PCNA binding through a conserved motif. Bioessays. 20:195–199 [DOI] [PubMed] [Google Scholar]
- Xia L., Zheng L., Lee H.W., Bates S.E., Federico L., Shen B., O’Connor T.R. 2005. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J. Mol. Biol. 346:1259–1274 10.1016/j.jmb.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Yu Y., Cai J.P., Tu B., Wu L., Zhao Y., Liu X., Li L., McNutt M.A., Feng J., He Q., et al. 2009. Proliferating cell nuclear antigen is protected from degradation by forming a complex with MutT Homolog2. J. Biol. Chem. 284:19310–19320 10.1074/jbc.M109.015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhou B., Zheng C.F., Zhang Z.Y. 2003. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 278:29901–29912 10.1074/jbc.M303909200 [DOI] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875 10.1038/nbt0997-871 [DOI] [PubMed] [Google Scholar]