Blocking autophagy protects the apoptosis inhibitor dBruce from destruction and promotes nurse cell survival in developing egg chambers.
Abstract
Autophagy is an evolutionarily conserved pathway responsible for degradation of cytoplasmic material via the lysosome. Although autophagy has been reported to contribute to cell death, the underlying mechanisms remain largely unknown. In this study, we show that autophagy controls DNA fragmentation during late oogenesis in Drosophila melanogaster. Inhibition of autophagy by genetically removing the function of the autophagy genes atg1, atg13, and vps34 resulted in late stage egg chambers that contained persisting nurse cell nuclei without fragmented DNA and attenuation of caspase-3 cleavage. The Drosophila inhibitor of apoptosis (IAP) dBruce was found to colocalize with the autophagic marker GFP-Atg8a and accumulated in autophagy mutants. Nurse cells lacking Atg1 or Vps34 in addition to dBruce contained persisting nurse cell nuclei with fragmented DNA. This indicates that autophagic degradation of dBruce controls DNA fragmentation in nurse cells. Our results reveal autophagic degradation of an IAP as a novel mechanism of triggering cell death and thereby provide a mechanistic link between autophagy and cell death.
Introduction
Autophagy is a tightly regulated process that plays a major role in cell growth, development, and tissue homeostasis, serving to maintain a balance between the synthesis, degradation, and subsequent recycling of cellular products (Meléndez and Neufeld, 2008; He and Klionsky, 2009). Interestingly, autophagy can promote both cell survival and cell death under certain circumstances. Dying cells possess autophagic features, but it is unclear whether autophagic activity causes cell death (Gozuacik and Kimchi, 2007). Studies in Drosophila have demonstrated that autophagy plays a crucial role in programmed cell death during development (Meléndez and Neufeld, 2008; McPhee and Baehrecke, 2009).
Oogenesis in insects is a fundamental developmental process. The structural and functional unit of the ovary is the egg chamber, which consists of the oocyte, the nurse cells, and the follicle cells. Programmed cell death during Drosophila oogenesis occurs in the germarium, as well as during mid-oogenesis and late oogenesis. Cell death of nurse and follicle cells is essential for the normal maturation of the egg chambers during the late stages of oogenesis (Pritchett et al., 2009).
It was previously reported that cell death during early oogenesis in Drosophila melanogaster is mediated through autophagy (Hou et al., 2008; Nezis et al., 2009). In the present study, we extend these findings, and we show that autophagy occurs during developmental cell death of the nurse cells in late oogenesis in D. melanogaster. We show that genetic inhibition of autophagy results in late stage egg chambers containing persistent nurse cell nuclei lacking fragmented DNA and in significant attenuation of cleaved caspase-3 expression. Furthermore, we show that the Drosophila inhibitor of apoptosis (IAP) dBruce is degraded by autophagy and regulates nurse cell survival by controlling DNA fragmentation. Together, these data demonstrate that DNA fragmentation and nurse cell death during late oogenesis is mediated by autophagy and through autophagic degradation of the IAP dBruce.
Results and discussion
Autophagy occurs in dying nurse cells during late oogenesis in D. melanogaster
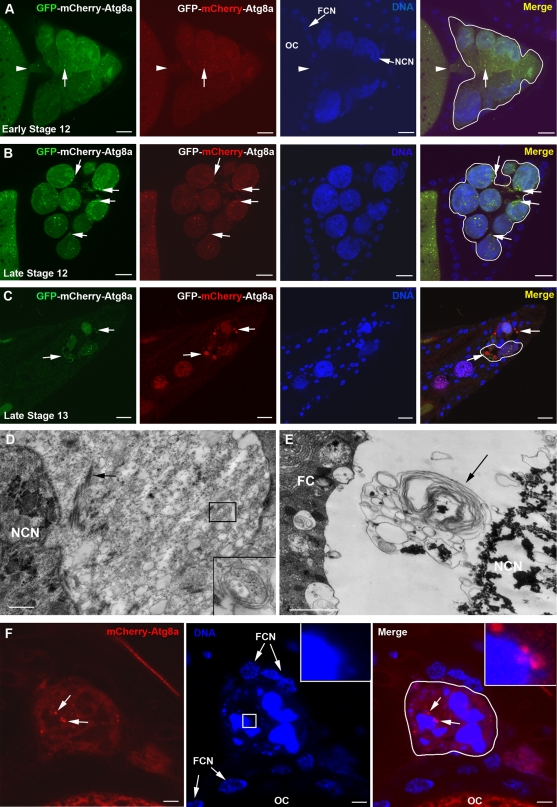
Dying nurse cells exhibit several markers of apoptosis during late oogenesis in D. melanogaster such as caspase activation, chromatin condensation, and DNA fragmentation (Cavaliere et al., 1998; Foley and Cooley, 1998; McCall and Steller, 1998; Nezis et al., 2000; Peterson et al., 2003). To address the role of autophagy in nurse cell death, we generated transgenic flies carrying a UASp-GFP-mCherry-DrAtg8a transgene. The double-tagged Atg8a protein emits yellow (green merged with red) fluorescence in nonacidic structures such as autophagosomes, and is red only in the autolysosomes due to quenching of GFP in these acidic structures. Upon expression of GFP-mCherry-DrAtg8a in the germline, several GFP-mCherry-DrAtg8a yellow puncta were detected in the cytoplasm of nurse cells during early stage 12 (Fig. 1 A). After the completion of transport of the majority of the nurse cell cytoplasm to the growing oocyte during late stage 12, GFP-mCherry-DrAtg8a yellow puncta remained in nurse cell cytoplasm in close proximity to the nurse cell nuclei (Fig. 1 B). In agreement with previous studies (Velentzas et al., 2007; Bass et al., 2009), ultrastructural analysis of the nurse cells at the same developmental stage also revealed the presence of autophagosomes in the remaining nurse cell cytoplasm (Fig. 1 D). Interestingly, during late stage 13 when the majority of nurse cells have degenerated, we observed a large number of red structures, indicating that the majority of the autophagosomes became autolysosomes (Fig. 1 C). We further confirmed this by ultrastructural analysis through detection of large autolysosomes associated with the condensed and fragmented nurse cell nucleus (Fig. 1 E; Velentzas et al., 2007; Bass et al., 2009). These autolysosomes often contained condensed material resembling the material of the fragmented nurse cell nucleus, suggesting that the nurse cell nuclear remnants are removed by autophagy. Indeed, nurse cells of late stage 13 egg chambers expressing UASp-mCherry-DrAtg8a (Nezis et al., 2009) exhibited mCherry-DrAtg8a puncta that are located either adjacent to or attached to the fragmented nucleus, indicative of nuclear autophagy (Fig. 1 F). To further examine the presence of autophagy during late oogenesis in Drosophila, we used protein trap lines that express GFP-tagged Atg5 and Atg8a (Kelso et al., 2004). Atg5-GFP and Atg8a-GFP were detected as punctae around the nurse cell nuclei during late oogenesis, revealing the presence of autophagic compartments (Fig. S1, A and B). These findings indicate that autophagy occurs during nurse cell death and degradation in late oogenesis in D. melanogaster.
Figure 1.
Autophagy is activated in dying nurse cells during late oogenesis in D. melanogaster. (A–C) Confocal micrographs of egg chambers of flies expressing the UASp-GFP-mCherry-DrAtg8a transgene exclusively in the germline (genotype: UASp-GFP-mCherry-DrAtg8a/+; nanos-VP-16 Gal4/+). Expression of UASp-GFP-mCherry-DrAtg8a exhibits punctate staining pattern (arrows). Note that during developmental stages early 12 (A) and late 12 (B), the punctate dots are yellow (merge). In late stage 13 (C), large red dots are evident, indicating the presence of acidic compartments (autolysosomes; arrows). The arrowheads in A point to the physical connection between the oocyte and the nurse cells during early stage 12 (not observed during late stage 12). (D and E) Transmission electron micrographs of nurse cells in stage 12 (D) and in stage 13 (E) egg chambers. Autophagosomes (inset in D) and autolysosomes (arrow in E) are evident in the remaining nurse cell cytoplasm (arrow in D indicates actin bundles). Note that the autolysosome shown in E contains condensed material resembling the material of the condensed and fragmented nurse cell nucleus (NCN), providing evidence for nuclear autophagy. (F) Nurse cell of a late stage 13 egg chamber expressing UASp-mCherry-DrAtg8a exclusively in the germline (genotype: UASp-mCherry-DrAtg8a/+; nanos-VP-16 Gal4/+). mCherry-DrAtg8a puncta are attached to or located adjacent to the fragmented nucleus (arrows and insets). Hoechst staining (blue) was performed to visualize the nuclei. Nurse cells are outlined with a white line. FC, follicle cell; FCN, follicle cell nucleus; OC, oocyte. Bars: (A–C) 10 µm; (D–F) 1 µm.
Autophagy controls DNA fragmentation and expression of cleaved caspase-3 in the nurse cells
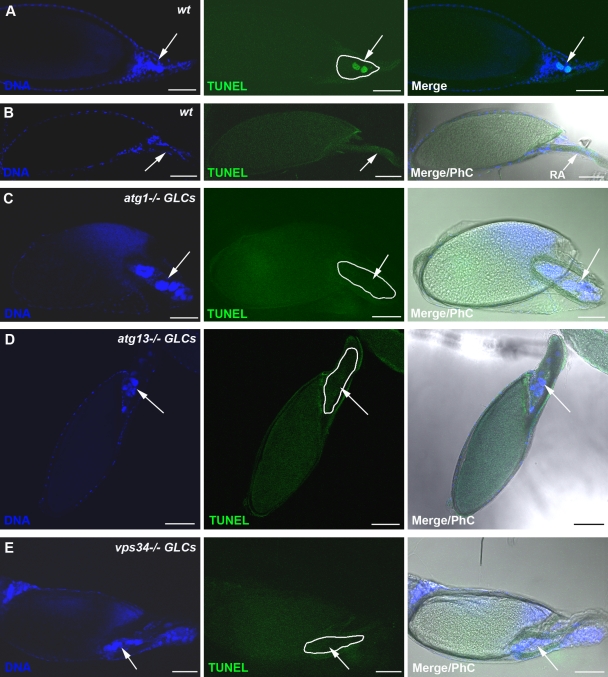
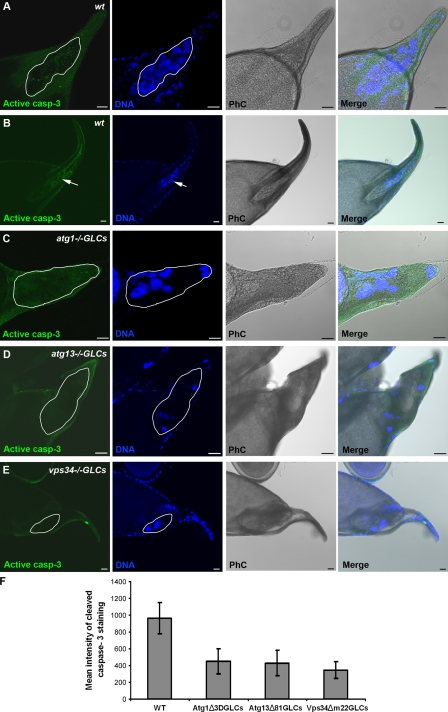
To explore the potential role of autophagy in nurse cell death during late oogenesis, we generated germline mutant cells for the core Drosophila autophagy genes atg1 and atg13 (Scott et al., 2004; Chang and Neufeld, 2009) and examined for cell death using the TUNEL assay to detect fragmented DNA. Interestingly, we observed that in either atg1 or atg13 germline mutants, there was a significant increase in the number of stage 14 egg chambers that had persisting TUNEL-negative nurse cell nuclei (Fig. 2, C and D; and Table S1). This phenotype differs from wild-type stage 14 egg chambers, in which nurse cell nuclei can rarely be detected (Fig. 2 B and Table S1; Bass et al., 2009), and those few that remain are exclusively TUNEL positive. TUNEL-positive nurse cell nuclei can be detected in the wild-type egg chambers in earlier developmental stages (Fig. 2 A; Nezis et al., 2000; Pritchett et al., 2009) but not in autophagy germline mutants (Fig. S1, C and D). To further examine the role of autophagy in nurse cell degeneration, we generated germline mutants for vps34, a member of the class III PI3-kinase complex that is responsible for the production of phosphatidylinositol 3-phosphate, a phosphoinositide required for autophagy (Juhász et al., 2008). Like the other autophagy mutants, the vps34 germline mutant egg chambers displayed significant increase in the number of egg chambers that had persisting TUNEL-negative nurse cell nuclei during late oogenesis (Fig. 2 E, Fig. S1 E, and Table S1). All autophagy germline mutants exhibited accumulation of Ref(2)P, a marker for autophagic flux (Klionsky et al., 2008; Nezis et al., 2008), in the nurse cell cytoplasm compared with the wild type, further confirming that autophagy was inhibited (Fig. S2, A–D). Interestingly, in all the autophagy germline mutants, the persisting nurse cell nuclei exhibited condensed nuclear staining (Fig. 2, C–E; and Fig. S2, C–E). To examine whether proteolytic processing of caspase-3 was affected by inhibition of autophagy, we performed immunolabeling for cleaved caspase-3 in the atg1, atg13, and vps34 germline mutant egg chambers. Cleaved caspase-3 levels were markedly attenuated in autophagy germline mutants compared with the wild type, with 92% cleaved caspase-3 labeling in w1118 late stage 12–14 egg chambers (n = 135) versus 35% in atg1−/− germline clones (GLCs; n = 212), 38% in atg13−/− GLCs (n = 225), and 33% in vps34−/− GLCs (n = 162) late stage 12–14 egg chambers (Fig. 3, A–F; and Fig. S1, F–I). Together, these data demonstrate that autophagy functions upstream of caspase processing and DNA fragmentation during late oogenesis in D. melanogaster.
Figure 2.
Genetic inhibition of autophagy in the germline prevents DNA fragmentation. Confocal micrographs of stage 13/14 egg chambers after TUNEL staining. (A) Wild-type (wt) stage 13 egg chamber contains nurse cell nuclei exhibiting fragmented DNA (positive TUNEL staining; arrows). (B) Wild-type stage 14 egg chambers do not contain nurse cells. Arrows point to the respiratory appendages (RA). (C–E) Stage 14 atg1−/− (C), atg13−/− (D), and vps34−/− (E) germline mutant chambers have persisting nurse cell nuclei that do not contain fragmented DNA (negative TUNEL staining; arrows). Nurse cells are outlined with a white line. Draq5/Hoechst staining (blue) was performed to visualize the nuclei. PhC, phase contrast. Bars, 50 µm.
Figure 3.
Germline autophagy mutant egg chambers exhibit reduced expression of cleaved caspase-3. Confocal micrographs of stage 13/14 egg chambers stained for cleaved caspase-3 (casp-3). (A) Wild-type (wt) late stage 13 egg chamber showing cleaved caspase-3 staining in the degenerating nurse cell cluster. (B) In wild-type stage 14 egg chamber, nurse cells are completely degraded (arrows). (C–E) atg1−/− (C), atg13−/− (D), and vps34−/− (E) germline mutant stage 14 egg chambers exhibit significantly reduced staining for cleaved caspase-3 and contain persisting nurse cell nuclei. Nurse cells are outlined with a white line. Draq5/Hoechst staining (blue) was performed to visualize the nuclei. PhC, phase contrast. (F) Quantification of mean intensity of cleaved caspase-3 staining observed in the nurse cells of stage 12–14 egg chambers in wild-type and germline autophagy mutant egg chambers. Wild type (WT): three independent experiments, n = 30 egg chambers; atg1−/− GLCs: four independent experiments, n = 30 egg chambers; atg13−/− GLCs: four independent experiments, n = 30 egg chambers; vps34−/− GLCs: three independent experiments, n = 30 egg chambers. Data are presented as mean ± SD. Difference was significant with P < 0.001 for all values versus wild type. Bars, 20 µm.
Drosophila IAP dBruce is degraded by autophagy in the nurse cells
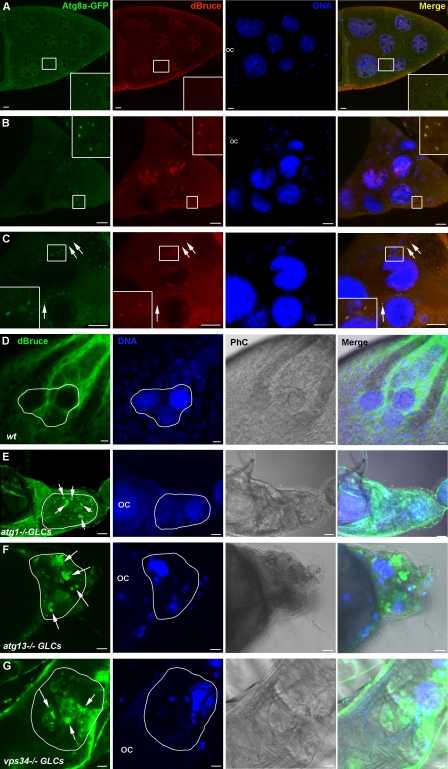
How can autophagy promote caspase activity, DNA fragmentation, and cell death in the same cell? We hypothesized that proteins crucial for cell survival could be degraded by autophagy, thus promoting cell death. To test our hypothesis, we investigated the localization of Drosophila IAPs in the nurse cells during late oogenesis and their relationship to the autophagic marker GFP-Atg8a. We tested three of four known Drosophila IAPs, DIAP1, DIAP2, and dBruce (Xu et al., 2009). DIAP1 and DIAP2 exhibit a rather diffuse cytoplasmic staining that did not colocalize with GFP-Atg8a (unpublished data). In contrast, dBruce exhibited an interesting localization pattern. We could not detect dBruce in stage 10B egg chambers (n = 112; Fig. 4 A). Interestingly, during early stage 12, we observed colocalization of dBruce and Atg8a-GFP in structures 0.5–1.5 µm in diameter resembling autophagosomes. A similar pattern of colocalization was observed during late stage 12 (n = 155; Fig. 4, B and C). In contrast, in later stages when nurse cell cytoplasm was completely transferred to the oocyte, dBruce exhibited a diffuse localization pattern mainly in the follicle cells surrounding the nurse cells remnants (Fig. 4 D). Our data suggest that dBruce might be degraded by autophagy. To test this hypothesis, we investigated the localization of dBruce in atg1, atg13, and vps34 germline mutants. Significantly, dBruce accumulated in the remaining cytoplasm of the nurse cells of all of these autophagy mutants and formed large aggregates 5–10 µm in diameter (Fig. 4, E–G; and Fig. S2 M). Western blot analyses showed that autophagy germline mutant egg chambers contain higher levels of dBruce protein than wild-type egg chambers (Fig. S2 N). These observations support our hypothesis and indicate that dBruce is degraded by autophagy in the nurse cells during late oogenesis.
Figure 4.
dBruce colocalizes with the autophagic marker Atg8a-GFP in the nurse cells during late oogenesis and is accumulated in autophagy germline mutants. Confocal micrographs of late stage egg chambers expressing Atg8a-GFP and stained for dBruce. (A–C) Stage 10 (A), early stage 12 (B), and late stage 12 (C). dBruce colocalizes with Atg8a-GFP in punctuate structures during stages 12 and 13 (insets in B and insets and arrows in C) and not during stage 10 (insets in A). (D–G) Confocal micrographs of stage 14 wild-type (D) and autophagy mutant egg chambers (E–G) stained for dBruce. dBruce is accumulated in atg1, atg13, and vps34 mutant nurse cells (arrows). Draq5/Hoechst staining (blue) was performed to visualize the nuclei. Nurse cells are outlined with a white line. OC, oocyte; PhC, phase contrast. Bars, 10 µm.
We next asked the question of how dBruce might be targeted for autophagy. p62 is a known adaptor protein that targets substrates for autophagic degradation (Pankiv et al., 2007). We investigated whether the Drosophila orthologue of p62, Ref(2)P (Nezis et al., 2008), may target dBruce for autophagy. Immunofluorescence analysis demonstrated that Ref(2)P staining in the nurse cells of late stage egg chambers has no correlation with the autophagic marker Atg8a-GFP (Fig. S2, F–H). Additionally, Ref(2)P mutant egg chambers exhibited a normal pattern of DNA fragmentation, cell death, and degradation in the nurse cells during late oogenesis (Fig. S2, I–L), which suggests that targeting dBruce for autophagy does not depend on Ref(2)P function.
Autophagic degradation of dBruce controls DNA fragmentation in the nurse cells
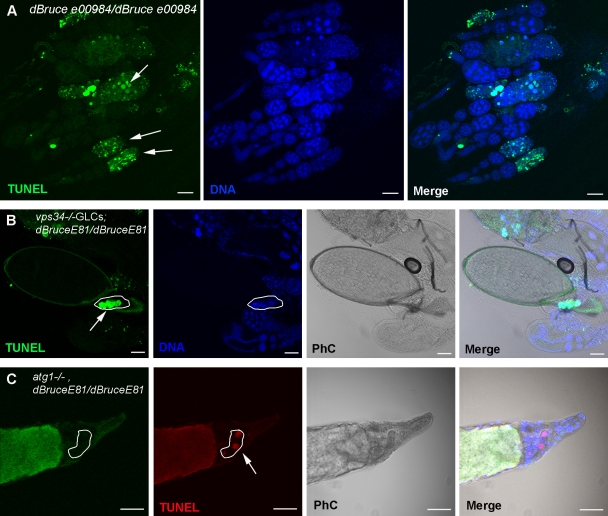
dBruce belongs to the IAP protein family. It contains both BIR (baculoviral IAP repeat, which is responsible for caspase inhibition) and UBC (responsible for ubiquitin conjugation) domains in the N and C termini, respectively (Vernooy et al., 2002). We tested the function of three different dBruce mutant alleles that result in truncated proteins with deletions either in the BIR or UBC domains. Two of them (dBruceE16 and dBrucee00984) have a deletion in the UBC domain (Vernooy et al., 2002; Sathyanarayanan et al., 2008), and one of them (dBruceE81) has a deletion in the BIR domain (Arama et al., 2003). All dBruce mutant alleles displayed a significant increase in the number of degenerating egg chambers during mid-oogenesis compared with the wild type, as previously shown (percentage of the number of degenerating mid-stage egg chambers: dBruceE81/+, 9.8 ± 1.1%; dBruceE81/dBruceE81, 53.2 ± 3.2%; dBrucee00984/dBrucee00984, 55.1 ± 4.7%; and dBruceE16/dBruceE16, 51.9 ± 1.2%; Fig. 5 A and Table S2; Hou et al., 2008). To further investigate the role of autophagic degradation of dBruce in nurse cell death, we constructed double mutants for either atg1 and dBruceE81 or vps34 and dBruceE81. Both double mutant egg chambers contained persistent nurse cell nuclei that were TUNEL positive (Fig. 5, B and C; and Table S1). These data indicate that autophagic degradation of dBruce controls DNA fragmentation in the nurse cells during oogenesis in D. melanogaster.
Figure 5.
dBruce is required for nurse cell survival by controlling DNA fragmentation during oogenesis. (A) Confocal micrographs of dBrucee00984 mutant ovarioles stained for TUNEL and DNA. Arrows point to degenerating stage 8/9 egg chambers that are TUNEL positive. (B and C) Confocal micrographs of vps34/dBruceE81 and atg, dBruceE81 double mutant egg chambers stained for TUNEL (green and red, respectively) and DNA. (B) vps34−/−GLCs; dBruceE81/dBruceE81 stage 14 mutant egg chamber contain persisting TUNEL-positive nurse cell nuclei (outlined; arrow). (C) atg1−/−GLCs, dBruceE81/dBruceE81 stage 14 mutant egg chamber contain persisting TUNEL-positive nurse cell nuclei. atg1 mutant nurse cells are identified by the lack of GFP staining (outlined; arrow). Draq5/Hoechst staining (blue) was performed to visualize the nuclei. PhC, phase contrast. Bars, 50 µm.
The role of autophagy in cell death has been controversial. Previous studies have shown that autophagy promotes cell death in Drosophila larval salivary glands, midgut, and embryonic serosal membrane (Berry and Baehrecke, 2007; Denton et al., 2009; Mohseni et al., 2009). However, the precise mechanism by which autophagy executes the death of these cells is not clear. In this study, we have shown that autophagic degradation of the IAP dBruce controls DNA fragmentation in nurse cells during Drosophila late oogenesis. Our data also demonstrate that autophagy acts genetically upstream of caspase activation and DNA fragmentation in this developmental context and indicate that autophagy directly contributes to the activation of cell death. This agrees with recent evidence from cultured mammalian cells in which autophagy appears to act upstream of caspase-3 activation under specific experimental settings (Laane et al., 2009; Zalckvar et al., 2010).
dBruce has been previously shown to suppress cell death in the Drosophila eye (Vernooy et al., 2002) and also has a crucial function in nuclear degeneration during sperm differentiation in Drosophila (Arama et al., 2003). Interestingly, dBruce was recently shown to regulate autophagy and cell death during early and mid-oogenesis in Drosophila (Hou et al., 2008). In this case, dBruce and caspase activity were shown to influence autophagy. In contrast, this is the first evidence for a mechanism by which autophagy regulates dBruce and cell death. In this study, we provide genetic evidence that dBruce is degraded by autophagy in the degenerating nurse cells during late oogenesis and that it regulates DNA fragmentation. The fact that chromatin condensation is not affected in autophagy mutants indicates that this process is regulated independently from DNA fragmentation, as shown previously (Nezis et al., 2006).
Degradation of proteins that are crucial for cell survival is one of the mechanisms by which a cell can trigger its own death (Yu et al., 2008). For instance, selective depletion of catalase by autophagy has been shown to promote cell death in mammalian cells in vitro (Yu et al., 2006). Furthermore, it was recently shown that chaperone-mediated autophagy modulates the neuronal survival machinery by regulating the neuronal survival factor MEF2D, and dysregulation of this pathway is associated with Parkinson’s disease (Yang et al., 2009). In a recent study, it was also demonstrated that autophagy promotes synaptogenesis in Drosophila neuromuscular junction by degrading Highwire, an E3 ubiquitin ligase which limits neuromuscular junction growth (Shen and Ganetzky, 2009). Our in vivo data further support the idea that autophagic degradation of survival factors can promote cell death and indicate that IAPs can be degraded by autophagy, thereby causing cell death. Autophagy not only functions during late oogenesis as the cause of cell death, but can also function to efficiently degrade the nurse cell nuclei remnants, as previously shown in salivary glands (Berry and Baehrecke, 2007). It was recently reported that dying nurse cells during late oogenesis exhibit characteristics of programmed necrosis and that the lysosomal genes dor, spinster, and cathepsin D are required for this process (Bass et al., 2009), showing that autophagy and necrosis participate in nurse cell death and degradation during late oogenesis. In conclusion, our findings indicate that autophagy plays an important role in nurse cell death during late oogenesis in Drosophila, first by acting upstream of DNA fragmentation, thereby causing cell death, and then by scavenging nurse cell remnants.
Materials and methods
Drosophila culturing conditions
Drosophila insects were reared at 25°C and fed on standard diet supplemented with wet yeast paste. 15–20 adult insects (equal numbers of males and females) were kept in single vials for 3–4 d before dissection. Dissections were performed in cold Ringer’s solution or PBS, and ovaries were separated into single ovarioles. Staging of egg chambers during late oogenesis was determined by the size and shape of the respiratory appendages.
Fly strains and crosses
The w1118 strain was used as wild-type control. The atg1Δ3DFRT80B, FTR82Batg13Δ81 and FRT42Dvps34Δm22 fly lines were a gift from T.P. Neufeld (University of Minnesota, Minneapolis, MN). Germline mutant cells were generated by using the FLP-FRT recombinase-dominant female sterile technique (Chou and Perrimon, 1996), using the following ovoD1 strains: (a) yw,hsflp; ovoD1FRT80B/TM6B/MKRS, (b) yw,hsflp; FRT42DovoD1/CyO (gifts from P. Rørth, Institute of Molecular and Cell Biology, Singapore), and (c) w*;FRT82BovoD1/st1, Tub85D,ss1,e/TM3 (Bloomington Stock Center). Females of the genotype yw,hsflp; atg1Δ3DFRT80B/TM6B, yw,hsflp; FRT82Batg13Δ81/TM6B, and yw,hsflp; FRT42Dvps34Δm22/SM6-TM6B were crossed to yw,hsflp; ovoD1FRT80B/TM6B, yw,hsflp; FRT42DovoD1/CyO, and w; FRT82BovoD1/TM3 males, respectively, and were allowed to lay eggs for 1 d. Larval progeny were heat shocked on day 4 and day 5 (L2 and L3 of larvae development) for 1.5 h at 37°C in a circulating water bath, to induce the generation of mitotic clones in the developing ovary. For vps34d/BruceE81 double mutant egg chambers yw/Y;FRT42DovoD1/CyO;dBruceE81/TM3 males were crossed to yw,hsflp; FRT42Dvps34Δm22/CyO;dBruceE81/TM3 females. Adult females carrying germline mutants were dissected and analyzed as described in Drosophila culturing conditions. For atg1Δ3D/dBruceE81 double mutant egg chambers, atg1Δ3DFRT80BdBruceE81 and UbiGFP[w+]FRT80B,dBruceE81 lines were generated by recombination.
For generation of atg1Δ3D/dBruceE81 double mutant flies, w;+; atg1Δ3DFRT80BdBruceE81/TM6B males were crossed to yw,hsflp;+; UbiGFPFTR80B dBruceE81/TM6B females. Adult females with genotype ywhsflp;+; atg1Δ3D FRT80B dBruceE81/UbiGFPFTR80B dBruceE81 were fed for 1–2 d with yeast paste, and then they were heat shocked in a water bath at 37°C for 1.5 h. After the first heat shock, they were recovered for 6–7 h, and then they were heat shocked again for 1.5 h. Then they were fed up to 4 d with food supplemented with yeast paste. Ovaries were dissected between 72 and 96 h after the first heat shock. The protein trap lines for Atg8a (CG32672 and CA07477) and for Atg5 (CG1643 and YD0603) were obtained from the FlyTrap collection (http://flytrap.med.yale.edu; Kelso et al., 2004).
Construction of the GFP-mCherry-DrAtg8a transgene
The pUASp vector for the GFP-mCherry-DrAtg8a (CG32672) double tag fusion, pPGW-mCherry-DrAtg8a, was made by a Gateway LR reaction between pENTR-mCherry-DrAtg8a (Nezis et al., 2009) and the destination vector pPGW (obtained from the Drosophila Gateway Vector Collection; Pankiv et al., 2007).
Immunofluorescence labeling and confocal microscopy
Drosophila egg chambers were processed for TUNEL staining and immunofluorescence labeling as previously described (Nezis and Papassideri, 2008; Nezis et al., 2009). The primary antibodies used in this study were the following: (a) rabbit antibody against cleaved caspase-3 (Cell Signaling Technology) used at 1:500, (b) rabbit anti-Ref(2)P antibody used at 1:1,000 (Nezis et al., 2008), (c) anti–rabbit anti-GFP antibody used at 1:1,000 (raised against EGFP purified as a GST fusion from Escherichia coli; before immunization, the GST tag was removed using precision protease; the antibody was produced by Eurogentec), and (d) anti–rat anti-dBruce antibody (gift from Y. Kaplan, E. Arama [Weizmann Institute of Science, Rehovot, Israel], and H. Steller [The Rockefeller University, New York, NY]; Arama et al., 2007) used at 1:100. The secondary antibodies, conjugated with either Cy2 or Cy3, were purchased from Jackson ImmunoResearch Laboratories, Inc. Draq5 (Biostatus Limited) or Hoechst (Merck) was used to stain DNA at a dilution of 1:1,000. Finally, the egg chambers were mounted in antifading mounting medium (Prolong Antifade; Invitrogen) and observed under a confocal laser-scanning microscope (LSM 510 META; Carl Zeiss, Inc.) equipped with a Neo-Fluar 16× NA 0.50, 63× NA 1.4, and 100× NA 1.45 oil immersion objectives at 20°C. Image processing and analysis were performed with LSM 510 software version 3.2 (Carl Zeiss, Inc.) and Photoshop CS2 (Adobe). For the quantification of cleaved caspase-3 and dBruce staining, confocal images of nurse cells were scanned at the same pinhole, offset gain, and amplifier values below pixel value saturation. Mean intensity of each staining was calculated with the histogram function in the LSM 510 software. Colocalization line plots were created with the histogram/colocalization function in the LSM 510 software.
Conventional transmission electron microscopy and immunoblotting
Dissected egg chambers were processed for transmission electron microscopy as previously described (Nezis et al., 2000). To determine the levels of dBruce in wild-type and autophagy mutants, stage 12–14 egg chambers from wild-type and autophagy mutant flies were processed for Western blot analysis as previously described (Nezis et al., 2008). Band intensities were quantified using Quantity One imaging and analysis software (Bio-Rad Laboratories).
Online supplemental material
Fig. S1 shows that genetic inhibition of autophagy in the germline prevents DNA fragmentation and results in reduced expression of cleaved caspase-3. Fig. S2 shows that genetic inhibition of autophagy in the germline causes accumulation of Ref(2)P and dBruce in the nurse cells. Tables S1 and S2 show quantification of nurse cell death during late and mid-oogenesis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002035/DC1.
Acknowledgments
I.P. Nezis would like to dedicate this work to the memory of his lovely sister Maria, who passed away unexpectedly the 15th of July 2009.
We are very grateful to Yossef Kaplan, Eli Arama, Thomas P. Neufeld, Hermann Steller, Bruce Hay, Pascal Meier, Pernille Rørth, Ruth Lehmann, and the Bloomington Stock Center for fly lines and antibodies.
This work was supported by grants from the Norwegian Cancer Society (to H. Stenmark), the Hartmann Family foundation (to H. Stenmark), the Stem Cell Research Programme of the Research Council of Norway (to I.P. Nezis), and the National Institutes of Health (GM079431 to E.H. Baehrecke).
Footnotes
Abbreviations used in this paper:
- GLC
- germline clone
- IAP
- inhibitor of apoptosis
References
- Arama E., Agapite J., Steller H. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell. 4:687–697 10.1016/S1534-5807(03)00120-5 [DOI] [PubMed] [Google Scholar]
- Arama E., Bader M., Rieckhof G.E., Steller H. 2007. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 5:e251 10.1371/journal.pbio.0050251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B.P., Tanner E.A., Mateos San Martín D., Blute T., Kinser R.D., Dolph P.J., McCall K. 2009. Cell-autonomous requirement for DNaseII in nonapoptotic cell death. Cell Death Differ. 16:1362–1371 10.1038/cdd.2009.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.L., Baehrecke E.H. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere V., Taddei C., Gargiulo G. 1998. Apoptosis of nurse cells at the late stages of oogenesis of Drosophila melanogaster. Dev. Genes Evol. 208:106–112 10.1007/s004270050160 [DOI] [PubMed] [Google Scholar]
- Chang Y.Y., Neufeld T.P. 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 20:2004–2014 10.1091/mbc.E08-12-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.B., Perrimon N. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D., Shravage B., Simin R., Mills K., Berry D.L., Baehrecke E.H., Kumar S. 2009. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 19:1741–1746 10.1016/j.cub.2009.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K., Cooley L. 1998. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 125:1075–1082 [DOI] [PubMed] [Google Scholar]
- Gozuacik D., Kimchi A. 2007. Autophagy and cell death. Curr. Top. Dev. Biol. 78:217–245 10.1016/S0070-2153(06)78006-1 [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D.J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.C., Chittaranjan S., Barbosa S.G., McCall K., Gorski S.M. 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182:1127–1139 10.1083/jcb.200712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J.H., Yan Y., Sass M., Baehrecke E.H., Backer J.M., Neufeld T.P. 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181:655–666 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso R.J., Buszczak M., Quiñones A.T., Castiblanco C., Mazzalupo S., Cooley L. 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32:D418–D420 10.1093/nar/gkh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laane E., Tamm K.P., Buentke E., Ito K., Kharaziha P., Khahariza P., Oscarsson J., Corcoran M., Björklund A.C., Hultenby K., et al. 2009. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 16:1018–1029 10.1038/cdd.2009.46 [DOI] [PubMed] [Google Scholar]
- McCall K., Steller H. 1998. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 279:230–234 10.1126/science.279.5348.230 [DOI] [PubMed] [Google Scholar]
- McPhee C.K., Baehrecke E.H. 2009. Autophagy in Drosophila melanogaster. Biochim. Biophys. Acta. 1793:1452–1460 10.1016/j.bbamcr.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez A., Neufeld T.P. 2008. The cell biology of autophagy in metazoans: a developing story. Development. 135:2347–2360 10.1242/dev.016105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohseni N., McMillan S.C., Chaudhary R., Mok J., Reed B.H. 2009. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 5:329–338 10.4161/auto.5.3.7444 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Papassideri I. 2008. Monitoring autophagy in insect eggs. Methods Enzymol. 451:669–683 10.1016/S0076-6879(08)03237-0 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Stravopodis D.J., Papassideri I., Robert-Nicoud M., Margaritis L.H. 2000. Stage-specific apoptotic patterns during Drosophila oogenesis. Eur. J. Cell Biol. 79:610–620 10.1078/0171-9335-00088 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Stravopodis D.J., Margaritis L.H., Papassideri I.S. 2006. Chromatin condensation of ovarian nurse and follicle cells is regulated independently from DNA fragmentation during Drosophila late oogenesis. Differentiation. 74:293–304 10.1111/j.1432-0436.2006.00076.x [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D., Rusten T.E., Stenmark H., Brech A. 2008. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180:1065–1071 10.1083/jcb.200711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis I.P., Lamark T., Velentzas A.D., Rusten T.E., Bjørkøy G., Johansen T., Papassideri I.S., Stravopodis D.J., Margaritis L.H., Stenmark H., Brech A. 2009. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 5:298–302 10.4161/auto.5.3.7454 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Peterson J.S., Barkett M., McCall K. 2003. Stage-specific regulation of caspase activity in Drosophila oogenesis. Dev. Biol. 260:113–123 10.1016/S0012-1606(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Pritchett T.L., Tanner E.A., McCall K. 2009. Cracking open cell death in the Drosophila ovary. Apoptosis. 14:969–979 10.1007/s10495-009-0369-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Kumar S., Chen C.H., Chen D., Hay B., Sehgal A. 2008. Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 22:1522–1533 10.1101/gad.1652308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.C., Schuldiner O., Neufeld T.P. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7:167–178 10.1016/j.devcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Shen W., Ganetzky B. 2009. Autophagy promotes synapse development in Drosophila. J. Cell Biol. 187:71–79 10.1083/jcb.200907109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velentzas A.D., Nezis I.P., Stravopodis D.J., Papassideri I.S., Margaritis L.H. 2007. Mechanisms of programmed cell death during oogenesis in Drosophila virilis. Cell Tissue Res. 327:399–414 10.1007/s00441-006-0298-x [DOI] [PubMed] [Google Scholar]
- Vernooy S.Y., Chow V., Su J., Verbrugghe K., Yang J., Cole S., Olson M.R., Hay B.A. 2002. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr. Biol. 12:1164–1168 10.1016/S0960-9822(02)00935-1 [DOI] [PubMed] [Google Scholar]
- Xu D., Woodfield S.E., Lee T.V., Fan Y., Antonio C., Bergmann A. 2009. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin). 3:78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J.J., Mao Z. 2009. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 323:124–127 10.1126/science.1166088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E.H., Lenardo M. 2006. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA. 103:4952–4957 10.1073/pnas.0511288103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Strandberg L., Lenardo M.J. 2008. The selectivity of autophagy and its role in cell death and survival. Autophagy. 4:567–573 [DOI] [PubMed] [Google Scholar]
- Zalckvar E., Yosef N., Reef S., Ber Y., Rubinstein A.D., Mor I., Sharan R., Ruppin E., Kimchi A. 2010. A systems level strategy for analyzing the cell death network: implication in exploring the apoptosis/autophagy connection. Cell Death Differ. 17:1244–1253 10.1038/cdd.2010.7 [DOI] [PubMed] [Google Scholar]