The ubiquitin-like protein BAG-6 protects cells from newly synthesized misfolded proteins by tethering them to the proteasome.
Abstract
BAG-6/Scythe/BAT3 is a ubiquitin-like protein that was originally reported to be the product of a novel gene located within the human major histocompatibility complex, although the mechanisms of its function remain largely obscure. Here, we demonstrate the involvement of BAG-6 in the degradation of a CL1 model defective protein substrate in mammalian cells. We show that BAG-6 is essential for not only model substrate degradation but also the ubiquitin-mediated metabolism of newly synthesized defective polypeptides. Furthermore, our in vivo and in vitro analysis shows that BAG-6 interacts physically with puromycin-labeled nascent chain polypeptides and regulates their proteasome-mediated degradation. Finally, we show that knockdown of BAG-6 results in the suppressed presentation of MHC class I on the cell surface, a procedure known to be affected by the efficiency of metabolism of defective ribosomal products. Therefore, we propose that BAG-6 is necessary for ubiquitin-mediated degradation of newly synthesized defective polypeptides.
Introduction
The significance of protein quality control is demonstrated by a number of human disorders, such as cystic fibrosis and Parkinson’s disease, which can result from the accumulation of misfolded proteins (Ward et al., 1995; Meacham et al., 1999; Ardley et al., 2003; Olzmann et al., 2007; Metzger et al., 2008; Nakamura and Lipton, 2009). Therefore, understanding the mechanisms of misfolded protein metabolism, as well as cotranslational degradation of defective proteins in mammalian cells (Qian et al., 2006), is of primary importance. Hydrophobic residues that are normally buried in a variety of natively folded proteins may be exposed by protein misfolding and act as degrons to deliver the misfolded protein mainly to the ubiquitin–proteasome system for degradation (Metzger et al., 2008). Thus, ubiquitin-mediated protein quality control is essential for monitoring protein folding and endorsing misfolded and/or defective proteins for degradation/aggregation (Johnston et al., 1998; Schubert et al., 2000; Lelouard et al., 2004). In the ubiquitin system, recognition and recruitment of defective proteins to the degradation machinery is a key step in the selective elimination of aberrant proteins (Finley et al., 2004; Hartmann-Petersen and Gordon, 2004; Elsasser and Finley, 2005). Some cytosolic cochaperone proteins, such as CHIP, are known to be protein quality-control ubiquitin ligases that selectively target aberrant proteins for proteasomal degradation by promoting their ubiquitination (Meacham et al., 2001). In addition, targeting of polyubiquitinated substrates to the 26S proteasome is important because this process is thought to determine the final fate of defective proteins involved in various physiological and pathological reactions (Voges et al., 1999; Hershko et al., 2000; Madura, 2004; Hoeller et al., 2006). Previous studies have shown that the Rpn10 subunit of the 26S proteasome can recognize polyubiquitinated substrates, and it is thought to play a role as a ubiquitin receptor for the 26S proteasome (Deveraux et al., 1994; van Nocker et al., 1996; Kawahara et al., 2000; Wilkinson et al., 2001; Elsasser et al., 2004; Verma et al., 2004). Although it was believed that substrate recognition is an essential step in the proteasome-mediated degradation process, the role of Rpn10 remained obscure after the observation that its deletion in yeast and nematodes does not influence the viability of cells and causes only mild or highly specific phenotypes (van Nocker et al., 1996; Shimada et al., 2006). These observations suggested the existence of functionally redundant routes in the substrate recruitment system. Indeed, several proteins, such as Rad23p and Dsk2p, are involved in the delivery of polyubiquitinated substrates to the proteasome because they can bind simultaneously with polyubiquitinated substrates and the 26S proteasome (Kleijnen et al., 2000; Wilkinson et al., 2001; Chen and Madura, 2002; Elsasser et al., 2004). Accordingly, these gene products in the substrate delivery pathway may play a largely redundant role (Elsasser et al., 2004; Madura, 2004; Elsasser and Finley, 2005; Shimada et al., 2006). However, owing to this complex redundancy, a complete picture of substrate recognition and recruitment to the 26S proteasome in the ubiquitin system has not been adequately presented to date.
BAG-6–Scythe–BAT3 was originally identified as a novel gene located within the human major histocompatibility complex (Banerji et al., 1990) that encodes the anti-apoptotic ubiquitin-like protein (Thress et al., 1998, 1999; Desmots et al., 2005; Kikukawa et al., 2005; Sasaki et al., 2007), although its function and mechanism of action remain largely obscure. Our previous analysis revealed that Scythe, a BAG-6–homologous protein expressed in Xenopus embryos (Thress et al., 1998), binds to the proteasomal Rpn10c subunit via the N-terminal 436 residues and that this region is required for apoptotic control in Xenopus embryos (Kikukawa et al., 2005; Minami et al., 2007). Furthermore, our recent analysis indicated that the N terminus of Scythe regulates ubiquitin-mediated proteolysis of a pro-apoptotic protein (Minami et al., 2007), although how BAG-6 is involved in the ubiquitin-mediated pathway remains a mystery to be clarified. In this study, we provide a set of evidence that BAG-6 is a novel proteasomal substrates-associated protein. Importantly, BAG-6–associated polyubiquitinated substrates are newly synthesized defective ribosomal products (DRiPs) and BAG-6 plays a crucial role in their metabolism. BAG-6 offers a protective role against cell death induced by the accumulation of aberrant proteins. Furthermore, we suggest that BAG-6 collaborates with immunoproteasomes to generate MHC class I presented antigenic peptides via targeted degradation of DRiPs and might play crucial roles in antigen presentation. These findings support the hypothesis that BAG-6 is a novel tethering factor that mediates selective elimination of defective nascent chain polypeptides in mammalian cells.
Results
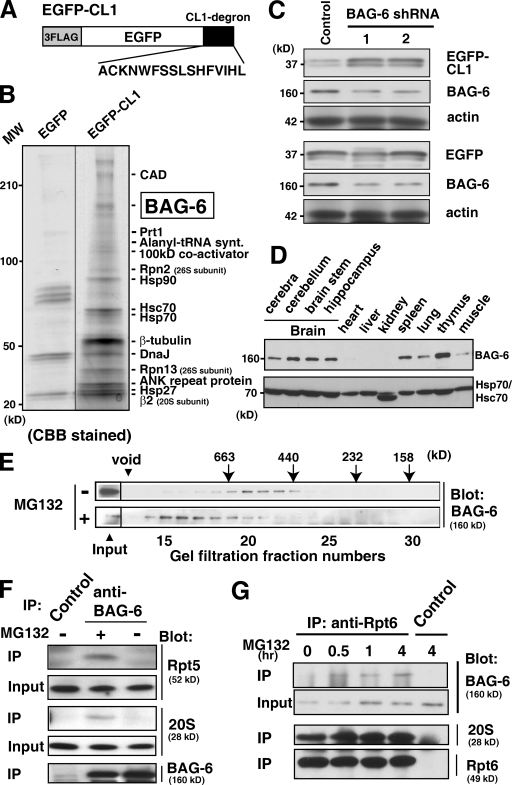
BAG-6 is essential for the CL1 degron–dependent degradation pathway
The hydrophobic CL1 peptide is known to function as an unstable degron that can be targeted to the 26S proteasome in a ubiquitin-dependent manner in yeast (Gilon et al., 1998; Metzger et al., 2008). We prepared a chimeric protein that fused the CL1 degron to the C terminus of EGFP (designated EGFP-CL1) (Fig. 1 A) and confirmed that this fusion protein is highly unstable in HeLa cells and stabilized by proteasome inhibitors, such as MG132 (unpublished data), indicating that EGFP-CL1 is indeed metabolized by the ubiquitin–proteasome-dependent degradation pathway in mammalian cells. Our peptide MS fingerprint analysis of EGFP-CL1 immunoprecipitates from MG132-treated HeLa cell extracts revealed a series of CL1-associated proteins including various chaperones, translation-related proteins, subunits of the 26S proteasome, and BAG-6 (Fig. 1 B), suggesting that they might cooperatively participate in CL1 peptide metabolism. Among these proteins, we focused on BAG-6 as a novel key molecule for protein degradation because the Xenopus homologue of mammalian BAG-6 (called Scythe) has been reported to regulate apoptosis via the control of XEF1AO proteolysis in Xenopus embryos (Minami et al., 2007). To examine the function of mammalian BAG-6 in CL1 metabolism, we suppressed endogenous BAG-6 expression by shRNA. As shown in Fig. 1 C, only a partial knockdown of BAG-6 blocked EGFP-CL1 degradation, indicating that BAG-6 is essential for the ubiquitin–proteasome-mediated metabolism of EGFP-CL1 in HeLa cells (see also Fig. S1, A and B).
Figure 1.
BAG-6 is essential for CL1 degron-dependent proteasomal degradation. (A) Schematic representation of the 3xFlag-tagged EGFP protein fused with CL1 degron used in this study. (B) CL1 degron-associated proteins identified in this study. After transfection of a 3xFlag-tagged EGFP-CL1 expression vector, HeLa cells were treated with 5 µM MG132 for 4.5 h. Proteins immunoprecipitated with antibody against Flag were subjected to SDS-PAGE and PMF analysis. 3xFlag-tagged EGFP immunoprecipitates were used as a negative control. (C) Knockdown of BAG-6 suppressed the degradation of the CL1 degron substrate. 3xFlag-tagged EGFP-CL1 was expressed in HeLa cells with two distinct shRNA vectors for BAG-6 (siRNA-1 and siRNA-2) or control siRNA. After 60 h of shRNA treatment, whole-cell extracts were prepared and subjected to immunoblot analysis with antibodies against Flag, BAG-6, and actin. (D) Expression patterns of endogenous BAG-6 protein in various adult mouse tissues (top). The anti-Hsp70/Hsc70 blot confirmed equal protein loading (bottom). (E) MG132 treatment stimulated the formation of a larger BAG-6 complex. Extracts of NIH3T3 cells were subjected to gel filtration with Superose 6, and the fractions were subjected to Western blotting with specific antibodies against BAG-6. Cells were cultured with (+) or without (−) 20 µM MG132 before harvesting. (F and G) BAG-6 was associated with the 26S proteasome in HeLa cells. (F) An antibody against BAG-6 was used to immunoprecipitate endogenous BAG-6 protein from extracts of HeLa cells cultured with (+) or without (−) 20 µM MG132 before harvesting. The precipitates were immunoblotted with antibodies to the 19S complex (Rpt5 subunit), 20S proteasome (α5 subunit), and BAG-6. (G) Using an antibody against the 26S proteasome subunit Rpt6, endogenous 26S proteasomes were immunoprecipitated from HeLa cell extracts. HeLa cells were treated with 20 µM MG132 for indicated periods. Immunoglobulin derived from nonimmune mouse serum was used as a negative control. The precipitates were immunoblotted with antibodies against BAG-6, 20S, and 19S complex (Rpt6 subunit).
BAG-6 is a novel 26S proteasome-associated protein
To detect the endogenous BAG-6 protein, we prepared a specific antibody and found that mammalian BAG-6 was mainly expressed in the brain and lymphoid tissues (Fig. 1 D). With this antibody, we examined whether BAG-6 forms complexes with other cellular proteins. We found that the endogenous BAG-6 protein was eluted from gel filtration at a molecular weight of ∼450 kD (Fig. 1 E, top). Unexpectedly, we found that treatment with MG132 stimulated the formation of a soluble, but much larger BAG-6 complex in HeLa cells (Fig. 1 E, bottom), and it apparently comigrated with 26S proteasomes, as determined by both gel filtration (Fig. S1 C) and glycerol density gradient fractionation (Fig. S1 D). Because we previously found that the BAG-6 homologue in Xenopus interacted with a proteasomal subunit, we further examined whether endogenous BAG-6 protein interacted with the 26S proteasomal complex. Our results clearly showed that the 26S proteasome associated with anti–BAG-6 immunoprecipitates (Fig. 1 F). Conversely, endogenous BAG-6 protein could be coimmunoprecipitated by antibody against the Rpt6 subunit of the 26S proteasome (Fig. 1 G). In both precipitation experiments, the association between BAG-6 and the 26S proteasome was strengthened by treatment with MG132 (Fig. 1, F and G). These observations suggest that mammalian BAG-6 is a novel member of 26S proteasome-associated proteins and that BAG-6 might be closely linked with ubiquitin-mediated protein degradation events (see also Fig. S1, E and F).
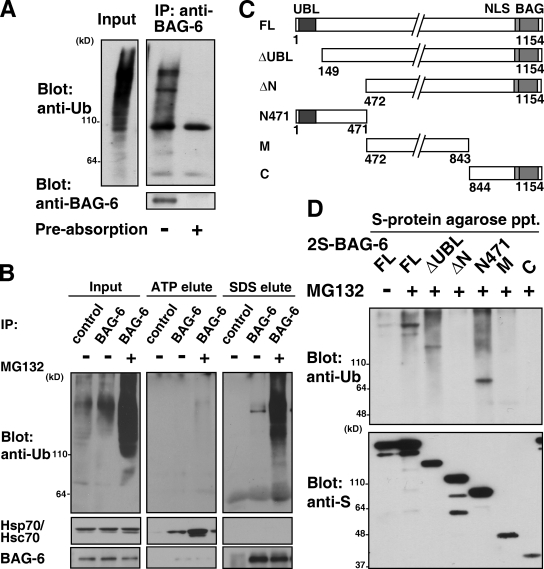
BAG-6 associates with polyubiquitinated proteasomal substrates
To examine the function of BAG-6 in the ubiquitin-mediated protein degradation pathway, we next immunoprecipitated endogenous BAG-6 protein from HeLa cell extracts and blotted it with an antibody against ubiquitin. We found that a much larger amount of polyubiquitinated proteins coimmunoprecipitated with endogenous BAG-6 protein from extracts of MG132-treated cells (Fig. 2 A and Fig. S2 A). The precipitation of polyubiquitinated proteins was specific because preabsorption with an excess of antigen completely abolished the coimmunoprecipitation (Fig. 2 A). Immunoprecipitation of BAG-6 after denaturation with SDS never coprecipitated polyubiquitinated moieties (Fig. S2 B), revealing polyubiquitinated identity as BAG-6–bound proteins but not BAG-6 itself. Thus, BAG-6 appears not to be directly polyubiquitylated. Indeed, BAG-6 itself was stable in HeLa cells (Fig. S2, C–F). It has been reported that proteins of the BAG family interact with heat-shock proteins (HSPs) via the C-terminal BAG domain (Demand et al., 2001; Takayama and Reed, 2001). It is conceivable that HSP substrate proteins might be modified by ubiquitin (Jiang et al., 2001) and thus coprecipitated with BAG-6. To determine whether polyubiquitinated species that coimmunoprecipitated with BAG-6 were proteins that bound with HSP, BAG-6 immunocomplexes were eluted by ATP (a procedure that stimulated the quantitative dissociation of Hsp70 from BAG-6; see Fig. 2 B, ATP elute). We found that polyubiquitinated proteins remained associated with BAG-6 after ATP elution but were released in the presence of SDS (Fig. 2 B, SDS elute). These results indicate that the majority of precipitated high molecular mass polyubiquitinated polypeptides are not Hsp70-associated client proteins or HSP itself. The results also suggest that a region other than the BAG domain is responsible for polyubiquitin coprecipitation.
Figure 2.
BAG-6 associates with polyubiquitinated proteasomal substrates. (A) Endogenous BAG-6 protein was affinity purified from extracts of MG132-treated HeLa cells with an antibody against BAG-6, and the precipitates were immunoblotted with antibodies against ubiquitin and BAG-6. Ubiquitin coprecipitation of BAG-6 was abolished by preabsorption with an excess of BAG-6 antigen. (B) Endogenous BAG-6 protein precipitated from HeLa cell extracts was treated with 5 mM ATP (ATP-elute) before boiling in 1% SDS (SDS-elute). Each eluate was immunoblotted with antibodies against ubiquitin, Hsp70/Hsc70, and BAG-6. Cells were treated with (+) or without (−) 20 µM MG132 for 6 h before being harvested as indicated. Immunoglobulin derived from nonimmune rabbit serum was used in immunoprecipitation as a negative control. (C) Schematic representation of the BAG-6 deletion mutant proteins used in this study. The numbers denote corresponding amino acid numbers. (D) N-terminal 471 amino acids of BAG-6 were essential for the binding with polyubiquitinated proteins. The full-length (FL) form of 2S-tagged BAG-6 and its truncated derivatives were expressed in HeLa cells as indicated. Before harvesting, cells were treated with or without 10 µM MG132 for 12 h. Each form of BAG-6 was affinity purified with S-protein agarose, and the bound materials were blotted with antibodies against ubiquitin and S peptide.
We next determined the region of BAG-6 required for interaction with polyubiquitinated proteins (Fig. 2, C and D). A mutant form of BAG-6 lacking either the N-terminal UBL domain (by deletion of the N-terminal 148 residues, designated ΔUBL, Fig. 2 C) or the C-terminal BAG domain bound polyubiquitinated proteins as efficiently as did full-length (FL) BAG-6 (Fig. 2 D and unpublished data), indicating that neither the UBL domain nor the BAG domain is essential for binding polyubiquitinated proteins. In contrast, a mutant BAG-6 (designated ΔN) with truncation of the N-terminal 471 amino acids (the region corresponding to the N-terminal 436 amino acids of Xenopus Scythe; Minami et al., 2007) did not bind polyubiquitinated proteins (Fig. 2 D). Consistent with this result, a fragment that exclusively encodes the N-terminal 471 amino acids (N471) coprecipitated polyubiquitinated proteins as efficiently as did the full-length form of BAG-6 (Fig. 2 D). These results indicate that the N-terminal 471 residues of BAG-6 are necessary and sufficient for its binding to polyubiquitinated substrates.
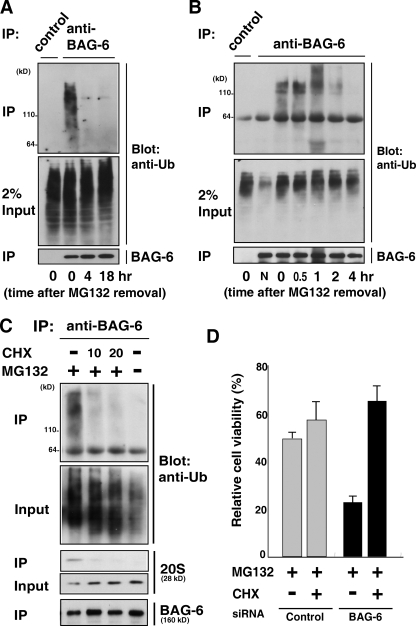
We then attempted to determine the fate of polyubiquitinated proteins associated with BAG-6 by “chasing” experiments. HeLa cells were treated with MG132 (5 µM) for 4.5 h, the inhibitor was washed out (this time point being defined as time zero), and the cells were cultured in fresh medium without proteasome inhibitors (Fig. S3 A). Although BAG-6–associated polyubiquitinated proteins were evident at time zero (Fig. 3, A and B), they gradually disappeared after the removal of MG132. Within 4 h after chasing, no detectable polyubiquitinated proteins coprecipitated with BAG-6 (Fig. 3, A and B). The disappearance of polyubiquitinated substrates on BAG-6 does not necessarily mean their degradation but instead their dissociation from BAG-6. To determine whether BAG-6 was involved in protein degradation, we established a semi-vitro degradation assay system. In this experiment, 3xFlag-tagged EGFP-CL1 and 2S-tagged BAG-6 were coexpressed in HeLa cells with the proteasome inhibitor MG132. EGFP-CL1 was immunoprecipitated by an anti-Flag antibody from cell extracts (first IP), and the eluted proteins were then affinity purified using S-protein agarose beads (second precipitation). The isolated materials (containing EGFP-CL1 and the 26S proteasome as a BAG-6 complex) were then incubated at 37°C in the presence or absence of ATP and/or MG132 in vitro and the stability of BAG-6–associated EGFP-CL1 during the incubation periods was evaluated. We found that EGFP-CL1 was degraded after incubation with ATP, which stabilizes/supports 26S proteasome function (Fig. S3 B). In contrast, polyubiquitinated EGFP-CL1 was stabilized when ATP was absent. When the active site within the 20S core particle was blocked by MG132, EGFP-CL1 degradation was inhibited, even in the presence of ATP (Fig. S3 B). Thus, these findings support the hypothesis that BAG-6 provides a platform that connects the 26S proteasome and its ubiquitinated substrates to promote their efficient degradation.
Figure 3.
BAG-6 associates with polyubiquitinated newly synthesized defective proteins. (A and B) Polyubiquitinated proteins associated with BAG-6 were degraded in vivo. HeLa cells were treated with MG132 (5 µM) for 4.5 h, the inhibitor was then washed out (this time point being defined as time zero), and the cells were cultured in a fresh medium (without MG132) for the indicated times. After harvesting the cells, endogenous BAG-6 was immunoprecipitated and the coprecipitated materials were probed with an antibody against ubiquitin. Normal rabbit immunoglobulin was used as a negative control for the immunoprecipitation procedures. “N” indicates no addition of MG132 to the cell culture. (C) Polyubiquitinated proteins associated with BAG-6 were abolished by addition of the translation inhibitor cycloheximide (CHX). HeLa cells were treated with 5 µM MG132 with or without 10 µg/ml or 20 µg/ml CHX as indicated for 1 h and then subjected to immunoprecipitation using antibody against BAG-6. The immunoprecipitates were probed with antibodies against ubiquitin, 20S proteasome, and BAG-6. (D) Depletion of BAG-6 made HeLa cells more sensitive to proteasome inhibitor-induced cell death, but the effect of BAG-6 knockdown was abrogated by adding CHX. 48 h after treatment with siRNA, cells were treated with 5 µM MG132 for 24 h with or without 10 µg/ml CHX, and cell viability was analyzed using an MTT cell counting system. The percent viability was normalized to the cells transfected with control siRNA without MG132 treatment (arbitrarily assigned a value of 100%). Error bars represent SD calculated from three experiments. P < 0.01, by t test.
BAG-6 is essential for metabolism of newly synthesized defective polypeptides
Identification of the client proteins of BAG-6 is critical to understanding the function of BAG-6. We observed only a slow degradation of ubiquitin conjugates in the lysate (not bound with BAG-6) after the removal of MG-132 (Fig. 3 A, input panel), in contrast to the rapid disappearance of polyubiquitinated substrates on BAG-6 (Fig. 3 A, top IP panel). These observations indicate that polyubiquitinated substrates that associate with BAG-6 might be only a part of the proteasomal substrates and that BAG-6 substrates degrade much faster than do those existing as free from BAG-6. To elucidate the endogenous client proteins of BAG-6, we analyzed the characteristics of polyubiquitin conjugates associated with BAG-6. We found that treatment with cycloheximide abolished most of the BAG-6–associated polyubiquitinated species in MG132-treated cells (Fig. 3 C; top IP panel), whereas there was only a small reduction of ubiquitin conjugates in the lysate (existing as free from BAG-6) after addition of cycloheximide (Fig. 3 C, input panel). These observations suggest that a significant part of BAG-6–associated polyubiquitinated species is cycloheximide-sensitive (and thus newly synthesized) polypeptides that are stabilized with MG132 treatment in cells. It has been reported that more than 30% of newly synthesized polypeptides are recognized as defective proteins by the ubiquitin-dependent degradation pathway (Schubert et al., 2000). Thus, we suspect that the primary target of BAG-6 might be defective ribosomal protein products (DRiPs) and that BAG-6 has a crucial function to support their efficient removal.
Inhibition of the proteasome-dependent proteolytic pathway by treatment with MG132 induced gradual cell death (Fig. 3 D) with accumulation of polyubiquitinated defective proteins. Although BAG-6 knockdown itself (up to 80%) did not significantly affect cell viability and growth (Fig. S3, C–E; and unpublished data), we found that knockdown of BAG-6 significantly enhanced the cytotoxicity of MG132 (and any other proteasome inhibitors; Fig. 3 D). This result indicates that BAG-6 knockdown renders cells more susceptible to MG132-induced death and that BAG-6 has an essential protective function against cell death induced by the accumulation of proteasomal substrates. Importantly, we found that the cytotoxicity enhanced by BAG-6 siRNA was also drastically suppressed by treatment with cycloheximide (Fig. 3 D). This observation further supports our conclusion that the effect of BAG-6 siRNA was mainly due to the excess accumulation of newly synthesized defective proteins. Thus, our data indicate that BAG-6 has a crucial function to metabolize defective ribosomal polypeptides.
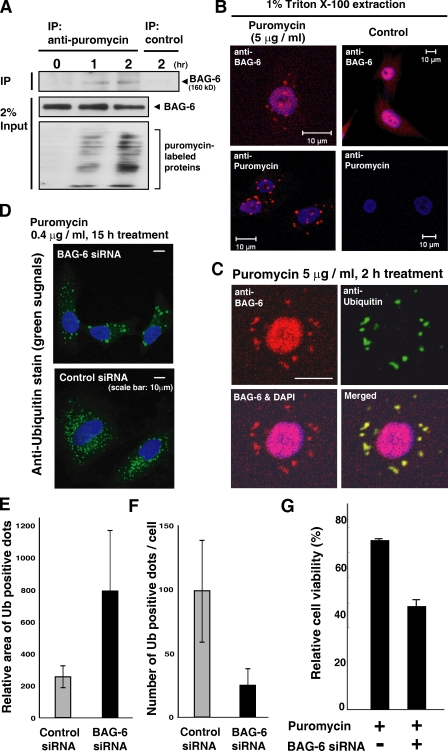
BAG-6 interacts with puromycin-labeled defective polypeptides
To investigate the relationship between BAG-6 and defective ribosomal products more directly, we examined whether BAG-6 recognized newly synthesized defective polypeptides. Puromycin is an analogue of the 3′ terminus of aminoacyl-tRNA that can be incorporated into newly synthesized polypeptides at their C termini, blocking further peptide elongation and thus producing truncated translational products (Fig. 4 A, bottom; Zhang et al., 1997; Lelouard et al., 2004). We found that BAG-6 could be coimmunoprecipitated by antibody against puromycin (Fig. 4 A, top IP panel). This result indicates that BAG-6 physically interacts with puromycin-labeled nascent chain polypeptides. Results of our immunocytochemical analyses of BAG-6 after puromycin treatment further support this notion. It has been reported that puromycin-labeled truncated proteins tend to form ubiquitin-positive cytoplasmic aggregates called ALIS (Lelouard et al., 2004). Interestingly, we also found that BAG-6 could be detected as cytoplasmic dots (Fig. 4 B, top panels) exclusively in puromycin-treated cells, and these dots looked similar to puromycin-labeled protein aggregates (Fig. 4 B, bottom panels). Indeed, we confirmed that the BAG-6–positive aggregates formed by puromycin treatment were co-ubiquitin stained (Fig. 4 C). These observations indicate that BAG-6 is intimately associated with cytoplasmic aggregates that are composed of polyubiquitinated nascent chain polypeptides in puromycin-treated cells.
Figure 4.
BAG-6 is involved in the metabolism of puromycin-induced nascent chain polypeptides. (A) BAG-6 bound to truncated puromycin-labeled proteins. After treatment with 5 µg/ml puromycin for the indicated times, HeLa cells were harvested, the puromycin-labeled polypeptides were immunoprecipitated from cell extracts with antibody against puromycin, and the precipitates were probed with antibody against BAG-6. (B and C) BAG-6 localized on cytoplasmic dots after puromycin treatment. HeLa cells were treated with 5 µg/ml puromycin for 2 h and extracted with 1% Triton X-100. Fixed cells were stained with antibodies against BAG-6, puromycin (B), and ubiquitin (C). Note that a part of BAG-6 also localized in the nucleus. (D–F) BAG-6 controlled the dynamics of puromycin-induced aggregate formation. Puromycin-treated HeLa cells with BAG-6 siRNA contained enlarged but reduced numbers of ubiquitin-positive cytoplasmic aggregates. Error bars represent SD calculated from three experiments. (G) 48 h after siRNA treatment, cells were treated with puromycin for 24 h and cell viability was analyzed with a cell counting kit. The percent viability was normalized to the cells without puromycin treatment.
To estimate the impact of BAG-6 knockdown on formation of puromycin-induced aggregates, we compared aggregates with or without BAG-6 siRNA (Fig. 4, D–F). We found that cells with BAG-6 knockdown contain enlarged cytoplasmic aggregates compared with control siRNA cells (Fig. 4, D and E). In contrast, the number of ubiquitin-positive aggregates was decreased with BAG-6 siRNA (Fig. 4 F). These results indicate that BAG-6 controls the dynamics of aggregate metabolism. In addition, we found that BAG-6 knockdown accelerated puromycin-induced cell death (Fig. 4 G), indicating that BAG-6 has protective roles against nascent chain polypeptide-induced cell toxicity. Collectively, the results indicate that BAG-6 might be an essential element for the metabolism of nascent chain polypeptides.
BAG-6–mediated degradation of nascent chain polypeptides in vitro
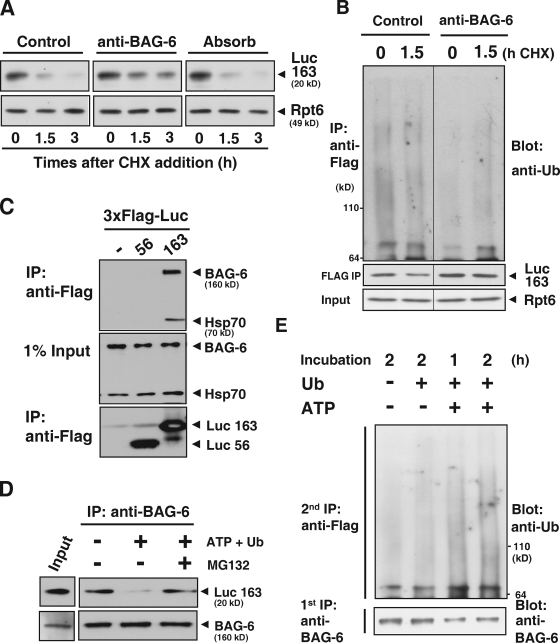
We then established a reticulocyte lysate-based in vitro degradation assay for artificial DRiPs. In this system, we used a puromycin-labeled, truncated luciferase (Luc163) that had been synthesized de novo as a model nascent chain polypeptide (Fig. S4 A). The Luc163 substrate, which was newly synthesized in a rabbit reticulocyte lysate, was unstable in the extract (Fig. S4 B) and was stabilized by the addition of proteasome inhibitors (Fig. S4 C), indicating that Luc163 was metabolized via a ubiquitin-dependent pathway. In accordance with this, we detected the accumulation of polyubiquitinated forms of Luc163 in a MG132-treated rabbit reticulocyte lysate (Fig. S4, D and G). With this system, we showed that addition of antibody against the N terminus of BAG-6 to the extracts blocked both degradation (Fig. 5 A) and polyubiquitin modification (Fig. 5 B and Fig. S4 E) of Luc163. In the reticulocyte lysate, Luc163 was associated with BAG-6 (Fig. 5 C). After isolation of the BAG-6 complex from lysates by anti–BAG-6 immunoprecipitation, the immunocomplex was incubated at 37°C in the presence or absence of MG132 and/or ATP in vitro and the stability of BAG-6–associated Luc163 during the incubation periods was evaluated (Fig. 5 D). We found that isolated Luc163 in the complex was degraded after incubation with ATP (which stabilizes/supports 26S proteasome function) and ubiquitin. In contrast, Luc163 was stabilized when ATP was absent. When the 26S proteasome was blocked by MG132, Luc163 degradation was inhibited, even in the presence of ATP (Fig. 5 D). The situation is similar in polyubiquitination of Luc163. Incubation of the BAG-6–Luc163 immunocomplex in vitro with ATP, ubiquitin, and MG132 enhanced polyubiquitin modification of Luc163 on BAG-6 (Fig. 5 E and Fig. S4 F). Thus, these findings support the idea that BAG-6 provides a platform that connects degradation machinery and nascent chain polypeptides to promote their efficient degradation.
Figure 5.
BAG-6 provides a platform that is necessary for linking puromycin-labeled defective protein with degradation machinery. (A) Addition of antibody against BAG-6 inhibited the degradation of puromycin-labeled, truncated luciferase in vitro. A messenger RNA encoding the 3xFlag-tagged N-terminal 163 residues of luciferase (Luc163) was incubated in a rabbit reticulocyte lysate chasing with 2 mM puromycin addition. After addition of 50 µg/ml anti–BAG-6 antibody, cycloheximide (CHX) chase analysis was performed, and lysates were harvested at the indicated times and probed with antibodies against Flag and Rpt6 to evaluate the stability of puromycin-labeled Luc163 (Luc163). Non-immune rabbit IgG or antibody against BAG-6 that was preabsorbed with an excess amount of recombinant antigen was used as a negative control. (B) Addition of antibody against BAG-6 inhibited ubiquitination of puromycin-labeled, truncated luciferase. Blot with Flag-Luc163 and Rpt6 are indicated as loading controls. (C) Immunoprecipitation of in vitro translated Luc163 with anti-Flag M2 beads coprecipitated endogenous BAG-6 and Hsp70 from lysates. (D) BAG-6 provided a platform for the targeted degradation of Luc163. Endogenous BAG-6 was immunoprecipitated from a rabbit reticulocyte lysate that was translating 3xFlag-tagged Luc163. The precipitated immunocomplex was further incubated in the presence (+) or absence (−) of 5 mM ATP, ubiquitin, and 25 µM MG132 at 37°C for 2 h as indicated. After incubation, the precipitated complexes were subjected to Western blot analysis with an antibody against Flag to examine the stability of Luc163 on BAG-6 during the incubation periods. (E) Endogenous BAG-6 was immunoprecipitated (first IP) as in D, and the precipitated complex was incubated with MG132 under the conditions indicated. After incubation, the complexes were denatured by SDS and diluted samples were further subjected to precipitation with anti-Flag M2 agarose (second IP). Precipitated, Flag-tagged Luc163 was blotted with antibody against ubiquitin to estimate the extent of its modification with ubiquitin.
BAG-6 knockdown blocks the cell surface presentation of MHC class I
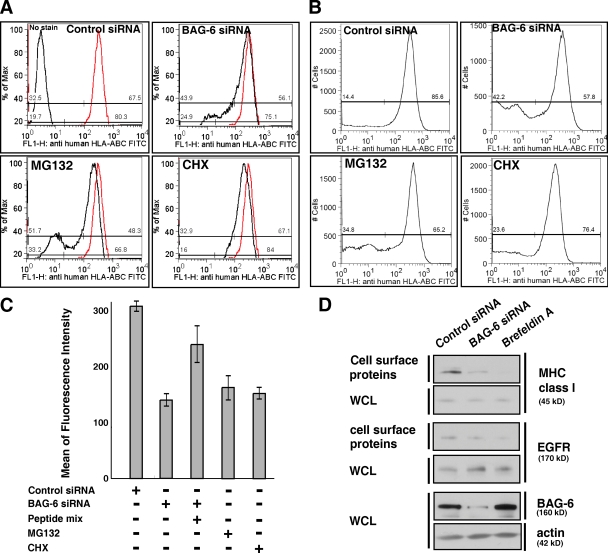
All of our observations support the idea that BAG-6 is essential for selective elimination of defective ribosomal products. It is reported that degradation products of DRiPs are the major source of peptide ligands presented on the cell surface MHC class I molecules of the major histocompatibility complex (Reits et al., 2000; Schubert et al., 2000; Khan et al., 2001; Yewdell and Bennink, 2001; Yewdell et al., 2003). If BAG-6–mediated processing of DRiPs is a major source of peptide ligands for MHC class I molecules, then knockdown of BAG-6 function in vivo should rapidly decrease the peptide supply and should block the cell surface presentation of MHC class I molecules because peptide binding is required for the rapid export of class I molecules. To directly examine whether BAG-6 knockdown affected the presentation of MHC class I molecules, we performed live-cell flow cytometric analysis with FITC-labeled anti–HLA-ABC antibody (W6/32). As our positive controls, we confirmed that treatment of HeLa cells with 10 µM MG132 increased the populations of FITC-negative cells (Fig. 6, A–C). Treatment with 10 µg/ml cycloheximide (CHX) also reduced the mean of fluorescence intensity of FITC-HLA-ABC on the surface of HeLa cells (Fig. 6, A and C). These results support a previous hypothesis that proteasome-mediated degradation of newly synthesized protein is essential for MHC class I presentation (Yewdell et al., 1996; Rock and Goldberg, 1999; Reits et al., 2000; Khan et al., 2001). In the case of BAG-6 knockdown, we found that the populations of FITC-negative cells increased significantly (Fig. 6, A and B). A quantitative evaluation of the mean of fluorescence intensity of FITC-HLA-ABC on the surface of HeLa cells indicated that the fluorescence decreased less than half in BAG-6 knockdown samples compared with the negative control (Fig. 6 C). These results suggest that knockdown of BAG-6 results in the suppression of MHC class I cell surface presentation. In addition, we found that an excess of exogenously supplied antigenic peptides partially rescued MHC class I cell surface expression (Fig. 6 C), suggesting that peptide production by the proteasome is limited after BAG-6 down-regulation.
Figure 6.
BAG-6 modulates expression of MHC class I molecules on the cell surface. (A and B) Live-cell flow cytometric analysis of HeLa cells with FITC-labeled antibody against MHC class I that recognizes cell surface HLA-ABC antigen. As positive controls, the results of treatment with 10 µM MG132 (A and B) and 10 µg/ml CHX (A) are shown. The data were obtained by linear scale analysis (A) and by log scale analysis (B). The flow cytometric pattern of negative control siRNA is indicated as a red line, and those after treatment with BAG-6 siRNA, MG132, and CHX are indicated with black lines (A). BAG-6 knockdown was performed with two independent duplex siRNAs as described in Materials and methods, and both siRNA gave a similar result. Representatives are results with duplex siRNAs of BAG-6-2 (5′-ATGATGCACATGAACATTC-3′). (C) Quantitative evaluations of the mean of fluorescence intensity of FITC-HLA-ABC on the surface of HeLa cells. Antigenic peptide mixture was pulsed with cells at 50 µM for 16 h. The data shown are the results of at least three independent experiments. (D) Biotin-labeling experiments. Cell surface proteins were biotinylated and affinity purified with avidin beads and blotted with antibody against MHC class I. Knockdown of BAG-6 reduced the expression of MHC class I on the cell surface, whereas the amount of EGF receptor (EGFR) on the cell surface was not affected. Brefeldin A treatment was used as a positive control for general suppression of transport of MHC class I and EGFR to the cell surface. Actin blots for whole-cell lysates (WCL) were used as a loading control for whole cellular proteins.
To further verify the expression of MHC class I with another independent method, we performed an experiment to label the cell surface with biotin. In this experiment, cell surface proteins were biotinylated after siRNA, affinity purified with avidin beads, and blotted with antibody against MHC class I. As shown in Fig. 6 D, the abundance of biotinylated MHC class I molecules was clearly reduced with BAG-6 siRNA. The reduction of cell surface MHC molecules is not caused by simple defects in the peptide transport system inside cells, because the amount of EGF receptor protein (EGFR) was not affected by BAG-6 knockdown but was reduced by brefeldin A, a drug that is known to block anterograde protein transport from the ER to the Golgi. Thus, all of our experiments showed that BAG-6 is essential for supplying antigenic peptides to the immune system.
Finally, we also provided evidence that BAG-6 could be associated with immunoproteasomes, the principal cytosolic proteases used for generating MHC class I peptide ligands (Monaco and Nandi, 1995; Tanaka and Kasahara, 1998; Rock and Goldberg, 1999), after induction by treatment with interferon-γ (Fig. S5 A). Although our results showed that BAG-6 could be associated with constitutive 26S proteasome as well (Fig. 1, F and G), these observations suggest that BAG-6 collaborates with immunoproteasomes to generate antigenic peptides via targeted degradation of defective ribosomal products and might play a crucial role for antigen presentation in immune response.
Discussion
The CL1 degron used in this study was originally identified in a screen for a genomic sequence that destabilized the cytosolic protein Ura3p by targeting it for degradation via Ubc6p/Ubc7p in the yeast Saccharomyces cerevisiae (Gilon et al., 1998). Metzger et al. (2008) reported that CL1 represents a frame-shifted region of the yeast PMD1 gene and that it contains a strongly hydrophobic region and thus may resemble a misfolded protein when it is exposed. Therefore, a study using a CL1 degron might reveal mechanisms for the targeted removal of improper translational products. A previous study indicated that Ura3p-CL1 degradation in yeast is dependent on the molecular chaperones Ydj1p (a DnaJ homologue) and Ssa1p (Hsp70 homologue) as well as the proteasome (Metzger et al., 2008). In accordance with the results of that study, we found that the CL1 degron could interact with human homologues of DnaJ, Hsp70, and 26S proteasome subunits in HeLa cells (Fig. 1 B). Furthermore, we identified BAG-6 as a novel mammalian CL1-associated protein. This observation suggested that BAG-6 may participate in the metabolism of misfolded proteins for proteasomal degradation. Indeed, we found in this study that BAG-6 recognized a CL1 degron substrate and supported its proteasomal degradation. We showed evidence that not only a CL1 model substrate but also newly synthesized polyubiquitinated polypeptides were associated with BAG-6 after proteasome inhibition (Fig. 3 C). All of our data suggest that BAG-6 provides a transient platform that is necessary for linking the 26S proteasome and its defective substrates for targeted degradation.
It has been reported that ∼25% of rapidly degraded polypeptides (RDPs) lose their solubility within an hour of blocking proteasome activity (Qian et al., 2006). The expression of BAG-6 was diffuse and soluble in normal HeLa cells, whereas treatment with MG132 significantly increased the amount of BAG-6 in the detergent-insoluble fraction (Fig. S5 B). These results support the notion that the BAG-6 protein in the soluble fraction moves to and accumulates in the insoluble aggregates associated with ubiquitinated RDPs during proteasomal dysfunction. It has been reported that polyubiquitinated, aggregation-prone misfolded proteins are transported on microtubules to the MTOC and then form large perinuclear insoluble aggregates/aggresomes (Johnston et al., 1998; Bence et al., 2001; Ardley et al., 2003). Because we found that BAG-6 interacted with polyubiquitinated proteins (Fig. 2; and Fig. 3) and that a part of BAG-6 moved to the insoluble fractions after treatment with MG132 (Fig. S5 B), we investigated whether BAG-6 colocalized with an aggresome. As shown in Fig. S5 C, confocal microscopic observation revealed that the treatment of MG132 resulted in the formation of a large perinuclear structure and that BAG-6 accumulated on this aggregate. Importantly, we found that BAG-6–positive aggregates were coimmunostained with either polyubiquitin or the intermediate filament protein vimentin, markers for aggresome formation (Johnston et al., 1998), in the presence of MG132 (Fig. S5 C). Our observations suggest that BAG-6 is functionally linked with the aggresome at the time of proteasome inhibition (Fig. S5 D).
Schubert et al. (2000) reported that 30% or more of newly synthesized proteins are destroyed by proteasomes of their synthesis. These unstable nascent polypeptides that emerge from the ribosome into the cytosol were designated as defective ribosomal products (Yewdell et al., 1996). DRiPs are polypeptides that fail to attain a stable conformation because of errors in translation, folding, and post-translational modification as well as errors in transcription and mRNA processing. Puromycin is mistakenly inserted during protein synthesis by the ribosome in place of normal amino acids, resulting in truncated DRiPs containing the drug at their C termini (Vazquez, 1974; Lelouard et al., 2004). Our immunocytologic and immunoprecipitation experiments provided evidence that BAG-6 could associate with puromycin-labeled defective nascent polypeptides in vivo. We also showed that BAG-6 controlled in vivo formation of puromycin-induced aggregation structures. Furthermore, our in vitro analysis clearly showed that puromycin-labeled truncated luciferase, a model DRiP substrate, was ubiquitinated and degraded on BAG-6 in rabbit reticulocyte lysates. All of these observations strongly support our conclusion that mammalian BAG-6 is essential for selective elimination of defective nascent chain polypeptides.
It has been shown that newly synthesized proteins are the major source of peptide ligands presented by MHC class I molecules of the major histocompatibility complex on the cell surface (Townsend et al., 1986; Anton et al., 1997; Khan et al., 2001). It has been reported that blocking protein synthesis for 30 min is sufficient to deplete cells of most TAP-transported antigenic peptides (Reits et al., 2000). Because a similar degree of depletion is reported to be achieved within 15 min of blocking proteasomes, the primary source of peptides is from proteins in the first 15 min of their synthesis (Reits et al., 2000). To date, there is considerable evidence that a significant source of self and viral peptides is DRiPs, which consist of prematurely terminated and/or misfolded polypeptides (Yewdell et al., 1996, 2003; Yewdell, 2002; Princiotta et al., 2003). If BAG-6–mediated processing of DRiPs is a major source of peptide ligands for MHC class I molecules, then blocking BAG-6 function should rapidly decrease the peptide supply and should subsequently slow the cell surface presentation of MHC class I molecules because peptide binding is required for the rapid export of class I molecules. That is the reason why we examined the cell surface presentation of MHC molecules in this study (Fig. 6). Both our FACS and biochemical analyses clearly showed that knockdown of BAG-6 resulted in the suppression of MHC class I cell surface presentation without affecting the intracellular protein transport system. These results also support our hypothesis that BAG-6 is essential for supplying antigenic peptides to the immune system. In good agreement with this observation, we found that BAG-6 was associated with immunoproteasome complexes after treatment with interferon-γ (Fig. S5 A). The initial evidence implicating immunoproteasomes in antigen processing was the discovery that the MHC region contains genes that encode two proteasome subunits and that their expression is controlled by cytokines released by activated T cells (Michalek et al., 1993; Monaco and Nandi, 1995; Tanaka and Kasahara, 1998). When induced, these subunits replace constitutively expressed subunits in newly assembled proteasomes to create immunoproteasomes, which appear to be better at producing peptides favored by MHC class I molecules (Tanaka and Kasahara, 1998). It is worth recalling that BAG-6 was originally described as an MHC-encoded gene product (Banerji et al., 1990) and that its expression is apparently enriched in lymphoid tissues (this paper; Fig. 1 D). Although we have not yet detected obvious induction of BAG-6 by interferon-γ, we present here an interesting possibility that BAG-6 is a novel factor that modulates immune responses via DRiP-mediated antigen presentations.
The role of BAG-6 in apoptotic cell death appears to be an area of controversy. In several previous studies, BAG-6 has been shown to be required for the induction of apoptosis in response to a variety of stimuli, and the loss of BAG-6 is associated with protection against apoptosis induced by calcium overloading in the ER as well as by menadione and thapsigargin (Desmots et al., 2008). Currently, we do not exactly know how the reported apoptotic function of BAG-6 is linked to the proteolytic function identified in this study. However, the region that is required for apoptotic control (N-terminal 436 residues in Xenopus Scythe–BAG-6; Minami et al., 2007) superficially overlaps with the region of substrate recognition in mammalian BAG-6. In addition, our previous study indicated that the N-terminal region of Scythe–BAG-6 interacts with XEF1AO, a maternal form of polypeptide elongation factor that was suggested to be a potential inducer of apoptosis in vertebrates (Minami et al., 2007). The binding stimulates polyubiquitin modification and subsequent degradation of XEF1AO in Xenopus embryos. In addition, we found in this study that BAG-6 provided protection against cell death induced by MG132 and puromycin treatment in mammalian cells. These observations imply that BAG-6–mediated modification of protein degradation is, at least in part, important for apoptotic control caused by the accumulation of aggregation-prone defective proteins. Because aggregated proteins with polyubiquitin have been proposed to be central to the pathology of a number of neurological diseases (Bence et al., 2001; Taylor et al., 2002; Ardley et al., 2003), we are also interested in the possibility that BAG-6 is involved in protein quality control in the neural system.
In the ubiquitin-dependent protein degradation pathway, the substrate sorting process depends on the cooperation of chaperone machineries and ubiquitin chain recognition factors (Hartmann-Petersen and Gordon, 2004; Verma et al., 2004; Richly et al., 2005; Westhoff et al., 2005). These factors sequentially support the process through protein–protein interactions and thereby escort substrate recognition, ubiquitination, and ubiquitin–protein conjugate presentation to the proteasome (Richly et al., 2005). Although we do not fully know what kinds of ubiquitin chain recognition factors and ubiquitination machinery are associated with BAG-6 at present, we favor the idea that BAG-6 may possess roles in ubiquitin modification of tethered substrates. We observed that there was an inexplicable increase in BAG-6–bound ubiquitin conjugates for 1 h after MG132 removal (Fig. 4 B). In our previous study, we reported that Scythe–BAG-6 expression stimulates polyubiquitin modification of XEF1AO substrate (Minami et al., 2007). These observations suggest that the BAG-6–Scythe complex plays a role in modification of polyubiquitin chains. We suggest that BAG-6 provides a transient platform that links the ubiquitinating machinery, the 26S proteasome, and its newly synthesized substrates to promote their efficient destruction. In any case, we have presented the first evidence that BAG-6–Scythe–BAT3 is a novel polyubiquitinated substrate-associated protein, and our results shed light on the importance of BAG-6 in the degradation of defective proteasomal substrates. Elucidation of the involvement of BAG-6 in the regulation of viral infections and/or onset of neurodegeneration caused by defects in the metabolism of DRiPs should be the next big challenge in understanding the significance of BAG-6 in the development of various human diseases.
Materials and methods
Plasmid construction
The cDNAs of BAG-6 were amplified by PCR from the library prepared from NIH3T3 and HeLa cells. The PCR products of BAG-6 were digested with SalI–NotI and inserted into the pCI-neo-3xFlag or pCI-neo-2S vectors for expression in cultured cells. It should be noted that pCI-neo-3xFlag and pCI-neo-2S expression vectors contain three repeats of a Flag tag or two repeats of an S peptide sequences, respectively, at the N-terminal regions of their products. The truncated and mutated versions of BAG-6 were constructed by PCR and cloned into appropriate pCI-neo vectors (Promega). The synthetic oligonucleotide encoding the CL1 peptide (ACKNWFSSLSHFVIHL; Gilon et al., 1998) was inserted into the EcoRI site of pCI-neo-3xFlag-EGFP expression vector. Each vector was used for experiments after verification of the sequence of inserted DNA.
Mammalian cell culture and transfection
HeLa, Neuro2a, and NIH3T3 cells were cultured in DME (Sigma-Aldrich) supplemented with 10% heat-inactivated calf serum at 37°C under a 5% CO2 atmosphere. Transfection to HeLa cells was performed according to the standard calcium phosphate precipitation protocol. The total amount of plasmid DNA was adjusted to 2 µg with an empty vector. At 24 h after transfection, the cells were harvested and subjected to immunological analysis unless otherwise noted.
Immunological analysis
An anti–BAG-6 antibody was prepared as follows. 200 μg of bacterially produced Xenopus BAG-6 (N-terminal 436 amino acids) was mixed and emulsified with an equal amount of TiterMaxGold (TiterMax USA, Inc.) and then inoculated into a rabbit. The antibody was obtained after three rounds of immunization at 1-wk intervals and used after affinity purification. We confirmed that this antibody can efficiently recognize mammalian BAG-6.
For immunoprecipitation analysis, cultured cells were washed with ice-cold phosphate-buffered saline and lysed with immunoprecipitation (IP) buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Tween 20, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride (PMSF), 3 µg/ml pepstatin A, 5 µg/ml aprotinin, 10 mM N-ethylmaleimide, and 25 µM MG132. The lysate was sonicated for 1 s and centrifuged at 20,000 g for 20 min at 4°C, and the resulting supernatant was incubated with 3 µl of anti-Flag M2-agarose beads (Sigma-Aldrich), 5 µl of S-protein agarose beads (EMD), or anti–BAG-6 antibody stabilized to Protein A–Sepharose beads (GE Healthcare) for 2 h at 4°C. After the beads had been washed four times with the IP buffer, the precipitated immunocomplexes were subjected to SDS-PAGE. ATP elution was performed as described previously (Demand et al., 2001).
For Western blotting, the whole-cell lysate and immunoprecipitates were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad Laboratories). The membranes were immunoblotted with specific antibodies as indicated and then incubated with horseradish peroxidase–conjugated antibody against mouse or rabbit immunoglobulin (GE Healthcare), followed by detection with ECL Western blotting detection reagents (GE Healthcare).
The following antibodies were used for immunological analyses in this study: anti-19S complex (Rpt1, Rpt5, and Rpt6 subunits; Thermo Fisher Scientific and Enzo Life Sciences, Inc.), anti-20S proteasome (α5 and β1i subunit; Thermo Fisher Scientific and Enzo Life Sciences, Inc.), anti-S peptide (Santa Cruz Biotechnology, Inc.), anti-Flag tag (Sigma-Aldrich), anti-polyubiquitin FK2 (Nippon Bio-Test Laboratories Inc.), anti-Hsp70/Hsc70 (MBL), anti-tubulin (ICN), anti-Chk2 (Santa Cruz Biotechnology, Inc.), anti-MHC class I (Santa Cruz Biotechnology, Inc.), anti-EGFR (Santa Cruz Biotechnology, Inc.), anti-GST (Santa Cruz Biotechnology, Inc.), anti-vimentin (Santa Cruz Biotechnology, Inc.), and anti–human HLA-ABC monoclonal antibody W6/32 (Bay Bioscience Co., Ltd.). Anti-puromycin antibody is a gift from Peter Walter at University of California, San Francisco (San Francisco, CA).
Identification of CL1-associated proteins
An expression vector encoding 3xFlag-tagged EGFP-CL1 was transfected into HeLa cells. After 4.5 h treatment with 5 µM MG132, cells were harvested and crushed with IP buffer. Using anti-Flag M2 agarose beads (Sigma-Aldrich), EGFP-CL1 were immunopurified and subjected to SDS-PAGE analysis. 3xFlag-tagged EGFP was used as a negative control. PMF analysis was performed with Voyager Biospectrometry Workstation DE-PRO (PerSeptive Biosystems) as described previously (Minami et al., 2007).
Gene knockdown experiments
For knockdown analysis of BAG-6 in Fig. 1 C, the oligonucleotides encoding two independent targeted sequences of BAG-6 (targeted sequence of siRNA-1: 5′-TGGGTCCCTATTATCCAGC-3′, siRNA-2: 5′-TTTCTCCAAGAGCAGTTTA-3′) were inserted into the pSUPER vector (Oligo Engine). The plasmids for siRNA were transfected to HeLa cells. 60 h after treatment of siRNA, cells were subjected to Western blot analyses. For other knockdown experiments, two independent duplex siRNAs encoding the targeted sequences of BAG-6 (BAG-6-1: 5′-TTTCTCCAAGAGCAGTTTA-3′, BAG-6-2: 5′-ATGATGCACATGAACATTC-3′) were synthesized (SIGMA Genosys) and used with Lipofectamine 2000 (Invitrogen) according to the protocol provided by the manufacturer. Negative control siRNA was purchased from Invitrogen. The efficacies of each siRNA were verified by immunoblot and RT-PCR (Fig. S3, D and E) or immunocytochemical observations with anti-BAG-6 antibody.
Gel filtration analysis of the cell extracts of NIH3T3
Extracts of NIH3T3 cells were subjected to gel filtration on a column of Superose 6 HR 10/30 (GE Healthcare) in buffer A (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 1% Tween 20, 10% glycerol, 2 mM ATP, 1 mM dithiothreitol, 10% glycerol, 1 mM PMSF, 3 µg/ml pepstatin A, 5 µg/ml aprotinin, 10 mM N-ethylmaleimide, and 25 µM MG132) at a flow rate of 0.25 ml/min. Fractions of 0.5 ml were collected and subjected to SDS-PAGE, followed by Western blotting. Gel Filtration LMW/HMW Marker kits (GE Healthcare) were used as molecular mass standard proteins for gel filtration.
Microscopic observations
For immunocytochemical observations of cultured cells, HeLa cells were grown on micro coverglass (Matsunami), fixed by incubating in 4% paraformaldehyde, and were then permeabilized with 0.1% Triton X-100. Fixed cells were blocked with 1% bovine serum albumin in PBS and reacted with a series of primary antibodies at room temperature for 1 h. Alexa 594–conjugated anti–rabbit IgG antibody (Invitrogen) and FITC-conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies at dilutions of 1:800. To observe the nucleus, cells were stained with 2.5 µg/ml DAPI in PBS at the time of antibody staining. Immunofluorescent images were obtained with an invert confocal microscopy system (LSM510; Carl Zeiss, Inc.).
Cell viability assay
HeLa cells were treated with BAG-6–specific siRNA or negative control siRNA (Invitrogen). After 12 h, cells were plated in 96-well plates at a density of 104 cells/well and cultured for a further 72 h with or without treatment of 5 µM MG132 or 5 µg/ml puromycin for the indicated time. Cell viability was determined using a cell counting kit 8 (Dojindo) according to the protocol provided by manufacturer. The absorbance of 450 nm was measured by a densitometer (CS-9300PC; Shimadzu).
Rabbit reticulocyte lysate-based preparation of model DRiP substrate
Flag-tagged luciferase gene in mammalian expression vector (pCI-neo; Promega) was digested with restriction enzyme Bsp1407I. Bsp1407I exclusively cut the middle of luciferase ORF at the position of 163 residue of its encoding luciferase protein. Using this linearized DNA as a template, mRNA was synthesized in vitro using mMESSAGE smMACHINE (Applied Biosystems). The synthesized mRNAs was added to rabbit reticulocyte lysate and incubated at 26°C for 40 min, followed by an additional 20-min incubation with 2 mM puromycin, resulting in a production of puromycin-labeled truncated luciferase as a model nascent chain polypeptide. Flag-tagged full-length luciferase without puromycin treatment was used as a non-DRiP control polypeptide.
Assay for presentation of MHC class I on the cell surface
For FACS analysis, knockdown experiments were performed with two independent duplex siRNAs encoding the targeted sequences of BAG-6 (BAG-6 siRNA-1: 5′-TTTCTCCAAGAGCAGTTTA-3′, BAG-6 siRNA-2: 5′-ATGATGCACATGAACATTC-3′) with Lipofectamine 2000 (Invitrogen). After 72 h of siRNA transfection, 106 HeLa cells were harvested and probed with FITC-labeled anti–human HLA-ABC monoclonal antibody W6/32 (Bay Bioscience Co., Ltd.) in PBS (containing 10% FBS) at 4°C for 30 min. As positive controls, HeLa cells were treated with 10 µM MG132 or 10 µg/ml cycloheximide for 22 h (MG132) or 8 h (CHX), respectively, before harvesting. Exogenously supplied peptides (YVIKVSARV for HLA-A*6802, VQRLNATGY for HLA-B*1503, CCFHCQVC for HLA-Cw*1203) were synthesized by SIGMA Genosys and were pulsed with cells at 50 µM for 16 h before harvesting. Negative control siRNA was purchased from Invitrogen. Living-cell FACS analyses were performed with a FACSCalibur flow cytometer (Becton Dickinson) by ReproCELL, Inc.
Biochemical labeling experiments were performed as follows. After 72 h of siRNA transfection, HeLa cells were harvested and the cell surface proteins were biotinylated. Immediately after cell lysis with biotin lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% Triton X-100), cell surface–biotinylated proteins were affinity purified with avidin beads. The bound cell surface proteins were detected by antibodies against MHC class I and EGF receptor (EGFR; control of cell surface proteins). Brefeldin A treatment at 1 µg/ml for 4 h was used as a positive control for the suppression of the transport of MHC class I and EGFR to the cell surface. Anti-actin blot was used as a loading control.
Online supplemental material
Fig. S1 shows our additional data that BAG-6 is essential for CL1 degron-dependent proteasomal degradation. Fig. S2 demonstrates that BAG-6 is a stable protein that associates with polyubiquitinated substrates. Fig. S3 shows that polyubiquitinated proteins associated with BAG-6 are destined for degradation. Fig. S3 also shows the efficacies of BAG-6 knockdown. Fig. S4 describes our proteasome-dependent nascent polypeptide chain degradation system. Fig. S5 demonstrates that BAG-6 interacts with immunoproteasome and with aggresome. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908092/DC1.
Acknowledgments
We thank Dr. Peter Walter for his generous gift of anti-puromycin antibody. We also thank Drs. K. Tanaka (Rinshoken Institute); K. Iwai (Osaka University); F. Inagaki, T. Suzuki, H. Ariga, and N. Noda (Hokkaido University); S. Hisanaga and T. Saito (Tokyo Metropolitan University); and members in laboratory for many of constructive suggestions and discussions.
This work was supported in part by grants from the Ministry of Education, Culture, Science and Technology of Japan.
Footnotes
Abbreviations used in this paper:
- ALIS
- aggresome-like induced structures
- BAG
- bcl-2-associated athanogene
- CHX
- cycloheximide
- DRiP
- defective ribosomal product
- EGFP
- enhanced green fluorescent protein
- HSP
- heat shock protein
- IP
- immunoprecipitation
- Luc163
- puromycin-labeled truncated luciferase with N-terminal 163 residues
- MG132
- benzoyl-Leu-Leu-Leu-aldehyde
- MHC
- major histocompatibility complex
- MTOC
- microtubule-organizing center
References
- Anton L.C., Yewdell J.W., Bennink J.R. 1997. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J. Immunol. 158:2535–2542 [PubMed] [Google Scholar]
- Ardley H.C., Scott G.B., Rose S.A., Tan N.G., Markham A.F., Robinson P.A. 2003. Inhibition of proteasomal activity causes inclusion formation in neuronal and non-neuronal cells overexpressing Parkin. Mol. Biol. Cell. 14:4541–4556 10.1091/mbc.E03-02-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Sands J., Strominger J.L., Spies T. 1990. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. USA. 87:2374–2378 10.1073/pnas.87.6.2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Chen L., Madura K. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22:4902–4913 10.1128/MCB.22.13.4902-4913.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand J., Alberti S., Patterson C., Höhfeld J. 2001. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11:1569–1577 10.1016/S0960-9822(01)00487-0 [DOI] [PubMed] [Google Scholar]
- Desmots F., Russell H.R., Lee Y., Boyd K., McKinnon P.J. 2005. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25:10329–10337 10.1128/MCB.25.23.10329-10337.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F., Russell H.R., Michel D., McKinnon P.J. 2008. Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 283:3264–3271 10.1074/jbc.M706419200 [DOI] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. 1994. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269:7059–7061 [PubMed] [Google Scholar]
- Elsasser S., Finley D. 2005. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 7:742–749 10.1038/ncb0805-742 [DOI] [PubMed] [Google Scholar]
- Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. 2004. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279:26817–26822 10.1074/jbc.M404020200 [DOI] [PubMed] [Google Scholar]
- Finley D., Ciechanover A., Varshavsky A. 2004. Ubiquitin as a central cellular regulator. Cell. 116(2, Suppl):S29–S32: 2: S32 10.1016/S0092-8674(03)00971-1 [DOI] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., Kulka R.G. 1998. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17:2759–2766 10.1093/emboj/17.10.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R., Gordon C. 2004. Protein degradation: recognition of ubiquitinylated substrates. Curr. Biol. 14:R754–R756 10.1016/j.cub.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. 2000. The ubiquitin system. Nat. Med. 6:1073–1081 10.1038/80384 [DOI] [PubMed] [Google Scholar]
- Hoeller D., Hecker C.M., Dikic I. 2006. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat. Rev. Cancer. 6:776–788 10.1038/nrc1994 [DOI] [PubMed] [Google Scholar]
- Jiang J., Ballinger C.A., Wu Y., Dai Q., Cyr D.M., Höhfeld J., Patterson C. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938–42944 10.1074/jbc.M101968200 [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H., Kasahara M., Nishiyama A., Ohsumi K., Goto T., Kishimoto T., Saeki Y., Yokosawa H., Shimbara N., Murata S., et al. 2000. Developmentally regulated, alternative splicing of the Rpn10 gene generates multiple forms of 26S proteasomes. EMBO J. 19:4144–4153 10.1093/emboj/19.15.4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., de Giuli R., Schmidtke G., Bruns M., Buchmeier M., van den Broek M., Groettrup M. 2001. Cutting edge: neosynthesis is required for the presentation of a T cell epitope from a long-lived viral protein. J. Immunol. 167:4801–4804 [DOI] [PubMed] [Google Scholar]
- Kikukawa Y., Minami R., Shimada M., Kobayashi M., Tanaka K., Yokosawa H., Kawahara H. 2005. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J. 272:6373–6386 10.1111/j.1742-4658.2005.05032.x [DOI] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih A.H., Zhou P., Kumar S., Soccio R.E., Kedersha N.L., Gill G., Howley P.M. 2000. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 6:409–419 10.1016/S1097-2765(00)00040-X [DOI] [PubMed] [Google Scholar]
- Lelouard H., Ferrand V., Marguet D., Bania J., Camosseto V., David A., Gatti E., Pierre P. 2004. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 164:667–675 10.1083/jcb.200312073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K. 2004. Rad23 and Rpn10: perennial wallflowers join the melee. Trends Biochem. Sci. 29:637–640 10.1016/j.tibs.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Meacham G.C., Lu Z., King S., Sorscher E., Tousson A., Cyr D.M. 1999. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18:1492–1505 10.1093/emboj/18.6.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Patterson C., Zhang W., Younger J.M., Cyr D.M. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100–105 10.1038/35050509 [DOI] [PubMed] [Google Scholar]
- Metzger M.B., Maurer M.J., Dancy B.M., Michaelis S. 2008. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283:32302–32316 10.1074/jbc.M806424200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M.T., Grant E.P., Gramm C., Goldberg A.L., Rock K.L. 1993. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 363:552–554 10.1038/363552a0 [DOI] [PubMed] [Google Scholar]
- Minami R., Shimada M., Yokosawa H., Kawahara H. 2007. Scythe regulates apoptosis through modulating ubiquitin-mediated proteolysis of the Xenopus elongation factor XEF1AO. Biochem. J. 405:495–501 10.1042/BJ20061886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco J.J., Nandi D. 1995. The genetics of proteasomes and antigen processing. Annu. Rev. Genet. 29:729–754 10.1146/annurev.ge.29.120195.003501 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Lipton S.A. 2009. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 14:455–468 10.1007/s10495-008-0301-y [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Li L., Chudaev M.V., Chen J., Perez F.A., Palmiter R.D., Chin L.-S. 2007. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178:1025–1038 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princiotta M.F., Finzi D., Qian S.-B., Gibbs J., Schuchmann S., Buttgereit F., Bennink J.R., Yewdell J.W. 2003. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 18:343–354 10.1016/S1074-7613(03)00051-7 [DOI] [PubMed] [Google Scholar]
- Qian S.-B., Princiotta M.F., Bennink J.R., Yewdell J.W. 2006. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 281:392–400 10.1074/jbc.M509126200 [DOI] [PubMed] [Google Scholar]
- Reits E.A., Vos J.C., Grommé M., Neefjes J. 2000. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 404:774–778 10.1038/35008103 [DOI] [PubMed] [Google Scholar]
- Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 120:73–84 10.1016/j.cell.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Rock K.L., Goldberg A.L. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739–779 10.1146/annurev.immunol.17.1.739 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Gan E.C., Wakeham A., Kornbluth S., Mak T.W., Okada H. 2007. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21:848–861 10.1101/gad.1534107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U., Antón L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 404:770–774 10.1038/35004754 [DOI] [PubMed] [Google Scholar]
- Shimada M., Kanematsu K., Tanaka K., Yokosawa H., Kawahara H. 2006. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol. Biol. Cell. 17:5356–5371 10.1091/mbc.E06-05-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Reed J.C. 2001. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3:E237–E241 10.1038/ncb1001-e237 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kasahara M. 1998. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-γ-inducible proteasome activator PA28. Immunol. Rev. 163:161–176 10.1111/j.1600-065X.1998.tb01195.x [DOI] [PubMed] [Google Scholar]
- Taylor J.P., Hardy J., Fischbeck K.H. 2002. Toxic proteins in neurodegenerative disease. Science. 296:1991–1995 10.1126/science.1067122 [DOI] [PubMed] [Google Scholar]
- Thress K., Henzel W., Shillinglaw W., Kornbluth S. 1998. Scythe: a novel reaper-binding apoptotic regulator. EMBO J. 17:6135–6143 10.1093/emboj/17.21.6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K., Evans E.K., Kornbluth S. 1999. Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 18:5486–5493 10.1093/emboj/18.20.5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A.R.M., Rothbard J., Gotch F.M., Bahadur G., Wraith D., McMichael A.J. 1986. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 44:959–968 10.1016/0092-8674(86)90019-X [DOI] [PubMed] [Google Scholar]
- van Nocker S., Sadis S., Rubin D.M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R.D. 1996. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16:6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D. 1974. Inhibitors of protein synthesis. FEBS Lett. 40:S63–S84 10.1016/0014-5793(74)80689-7 [DOI] [PubMed] [Google Scholar]
- Verma R., Oania R., Graumann J., Deshaies R.J. 2004. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 118:99–110 10.1016/j.cell.2004.06.014 [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015–1068 10.1146/annurev.biochem.68.1.1015 [DOI] [PubMed] [Google Scholar]
- Ward C.L., Omura S., Kopito R.R. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 83:121–127 10.1016/0092-8674(95)90240-6 [DOI] [PubMed] [Google Scholar]
- Westhoff B., Chapple J.P., van der Spuy J., Höhfeld J., Cheetham M.E. 2005. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol. 15:1058–1064 10.1016/j.cub.2005.04.058 [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939–943 10.1038/ncb1001-939 [DOI] [PubMed] [Google Scholar]
- Yewdell J. 2002. To DRiP or not to DRiP: generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol. Immunol. 39:139–146 10.1016/S0161-5890(02)00097-4 [DOI] [PubMed] [Google Scholar]
- Yewdell J.W., Bennink J.R. 2001. Cut and trim: generating MHC class I peptide ligands. Curr. Opin. Immunol. 13:13–18 10.1016/S0952-7915(00)00175-8 [DOI] [PubMed] [Google Scholar]
- Yewdell J.W., Antón L.C., Bennink J.R. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157:1823–1826 [PubMed] [Google Scholar]
- Yewdell J.W., Reits E., Neefjes J. 2003. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3:952–961 10.1038/nri1250 [DOI] [PubMed] [Google Scholar]
- Zhang J.X., Braakman I., Matlack K.E., Helenius A. 1997. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol. Biol. Cell. 8:1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]