Generation of PI3P in the normally PI3P-deficient ER membrane makes the organelle a platform for autophagosome formation.
Abstract
Autophagy is a catabolic process that allows cells to digest their cytoplasmic constituents via autophagosome formation and lysosomal degradation. Recently, an autophagy-specific phosphatidylinositol 3-kinase (PI3-kinase) complex, consisting of hVps34, hVps15, Beclin-1, and Atg14L, has been identified in mammalian cells. Atg14L is specific to this autophagy complex and localizes to the endoplasmic reticulum (ER). Knockdown of Atg14L leads to the disappearance of the DFCP1-positive omegasome, which is a membranous structure closely associated with both the autophagosome and the ER. A point mutation in Atg14L resulting in defective ER localization was also defective in the induction of autophagy. The addition of the ER-targeting motif of DFCP1 to this mutant fully complemented the autophagic defect in Atg14L knockout embryonic stem cells. Thus, Atg14L recruits a subset of class III PI3-kinase to the ER, where otherwise phosphatidylinositol 3-phosphate (PI3P) is essentially absent. The Atg14L-dependent appearance of PI3P in the ER makes this organelle the platform for autophagosome formation.
Introduction
Autophagy is an intracellular degradation process that allows cells to digest cytoplasmic constituents by delivering them into the lysosome via the autophagosome. The autophagosome sequesters substrates to be degraded within a double membrane structure. The molecules involved in autophagosome formation are well conserved throughout eukaryotes, from yeast to mammals (He and Klionsky, 2009; Nakatogawa et al., 2009). Suppression of TOR (target of rapamycin) kinase activity has been linked to autophagy induced by nutrient starvation (Noda and Ohsumi, 1998). Although several physiological processes, including immunity and pathology, have been shown to be directly linked to autophagy (Mizushima et al., 2008), much of the basic mechanism remains unclear (Yoshimori and Noda, 2008).
A longstanding question in the field of autophagy is what is the origin of the autophagosome membrane (Reggiori, 2006)? Despite a variety of hypotheses regarding this question, only recently was an innovative finding reported: autophagosome formation is preceded by the formation of the omegasome, which is a specialized subdomain of the ER, and autophagosome formation takes place inside the omegasome (Axe et al., 2008). Omegasome formation was visualized using GFP-tagged DFCP1, a double FYVE (Fab-1, YGL023, Vps27, and EEA1)–containing protein, also called Taff1. Although the function of DFCP1 in autophagy is unclear, GFP-DFCP1 has been shown to localize to the omegasome. In nutrient-rich conditions, DFCP1 localizes to the ER membrane ubiquitously and to the Golgi apparatus, but autophagy is not induced (Cheung et al., 2001; Axe et al., 2008). However, upon the induction of autophagy, GFP-DFCP1 has a ringlike signal representing the omegasome. This localization depends on the FYVE domain, which mediates phosphatidylinositol 3-phosphate (PI3P) binding capacity. Electron microscopic tomography analysis revealed that there is a direct connection between the forming autophagosome (isolation membrane/phagophore) and the omegasome (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009). Therefore, the ER is the best candidate for the source of the autophagosome membrane. However, localization of PI3P is thought to be restricted to endocytic organelles such as the early endosome and is absent in the ER (Gillooly et al., 2000). Thus, some unknown mechanism must underlie the connection between the ER and PI3P.
PI3P is one of the key mediators of autophagosome membrane regulation (Simonsen and Tooze, 2009). In the case of yeast, phosphatidylinositol 3-kinase (PI3-kinase) activity and the PI3P-binding protein Atg18 have been shown to be necessary for autophagy (Obara et al., 2008a, b). A previous study in yeast established the existence of an autophagy-specific PI3-kinase complex consisting of Vps34, Vps15, Atg6, and Atg14 (Kihara et al., 2001b). However, whether a similar model is applicable to mammalian cells has been unclear, although hVps34 forms a complex with Beclin-1, the mammalian homologue of yeast Atg6/Vps30 (Liang et al., 1999; Kihara et al., 2001a). Recently, four research groups, including our own, independently identified an autophagy-specific PI3-kinase complex in mammalian cells. It consists of hVps34, hVps15, Beclin-1, and Atg14L/Atg14/Barkor (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Together, hVps34 and hVps15 form heteromeric core complexes that are involved in diverse cellular processes as class III PI3-kinase and its regulatory protein kinase (Lindmo and Stenmark, 2006; Yan et al., 2009). Additionally, Beclin-1 functions in other processes, including endocytosis and autophagosome and lysosome fusion steps, together with the other subcomplexes composed of UVRAG (UV radiation resistance–associated gene) and Rubicon (Liang et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Therefore, Atg14L is the sole specific subunit in the autophagy-specific PI3-kinase complex in mammalian cells. In yeast, the absence of Atg14 leads to dislocation of Vps34 from the PAS (preautophagosomal structure), a mysterious structure potentially specialized for autophagosome formation (Obara et al., 2006). To date, the detailed function of Atg14L in mammalian autophagy has not yet been characterized.
In this study, we reveal the role of Atg14L in autophagosome formation. It mediates the localization of the autophagy-specific PI3-kinase complex to the ER, and its recruitment is a critical determinant of the identity of the autophagic membrane structure.
Results and discussion
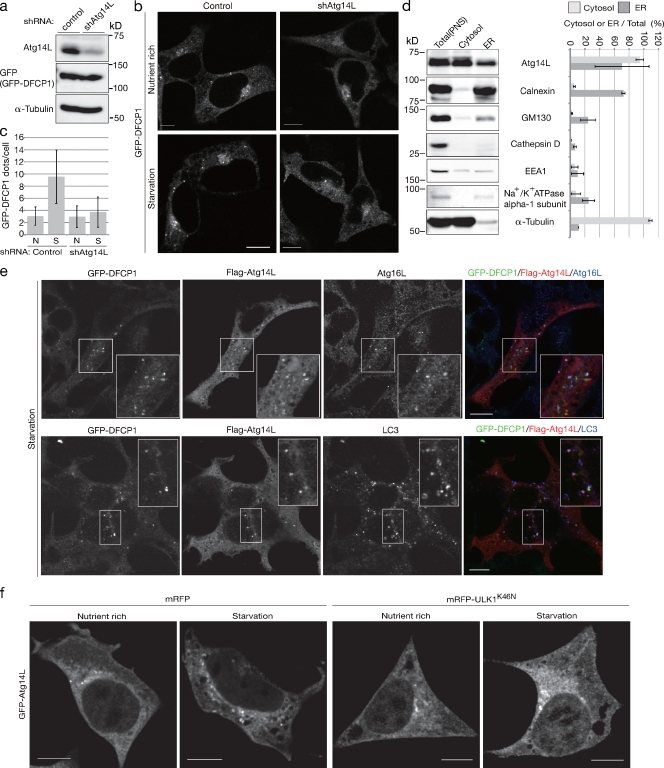
Atg14L is the sole specific subunit of the autophagy-specific PI3-kinase complex in mammalian cells, but how it is involved in the autophagic process is almost unknown (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). The process of autophagosome formation consists of three main phases, initiation of biogenesis of the membrane, elongation of the isolation membrane, and final closure of the autophagosomal mouth to form the sealed autophagosome. Some of the autophagy-executing molecules (Atgs) have been suggested to function in the later phase (Fujita et al., 2008a; Noda et al., 2009). To determine whether Atg14L is involved in the initial phase of autophagosome formation or at a later phase, we observed the GFP-DFCP1 distribution pattern before and after autophagy-inducing nutrient starvation conditions. In the control cells, GFP-DFCP1 formed puncta upon nutrient starvation treatment (Fig. 1 b). Short hairpin RNA (shRNA)–mediated Atg14L knockdown clearly impaired GFP-DFCP1–positive omegasome formation, characterized by puncta formation and nutrient starvation treatment (Fig. 1, a–c). These data indicated that Atg14L is critical for the initiation of autophagic membrane biogenesis. This conclusion is further supported by the localization of Atg14L. We previously reported that GFP-Atg14L shows both a punctate pattern and an ER localization pattern (Matsunaga et al., 2009). In this study, we show that endogenous Atg14L is enriched in the purified ER fraction by subcellular fractionation from nonstarved cells, corroborating the idea that part of Atg14L usually localizes to the ER (Fig. 1 d). Besides the ER pattern, GFP-Atg14L–positive puncta partially colocalized with Atg16L, a marker for the isolation membrane/phagophore, and with LC3, a marker for both the completed autophagosome and isolation membrane/phagophore in starved cells (Kabeya et al., 2000; Mizushima et al., 2003; Matsunaga et al., 2009). As reported, some Atg16L- and LC3-positive puncta are negative for Atg14L (Fig. 1 e; Matsunaga et al., 2009). Atg14L puncta and GFP-DFCP1 puncta overlapped substantially (Fig. 1 e). ULK1, a mammalian homologue of Atg1 protein kinase, functions in the initial step of autophagosome formation, and the kinase-dead mutant ULK1 (ULK1K46N) functions in a dominant-negative manner (Hara et al., 2008). GFP-Atg14L puncta but not the ER distribution disappeared in conditions to inhibit autophagy by overexpression of dominant-negative ULK1, both under nutrient-rich and starvation conditions (Fig. 1 f). We also observed that colocalization of Atg14L and DFCP-1 was induced by lithium or etoposide treatment, which induced autophagy independent of starvation (Fig. S1 a; Sarkar et al., 2005; Nishida et al., 2009). These results clearly indicated the role of Atg14L in the early phase involving omegasome formation, although they do not necessarily exclude roles in later phases. It is very likely that upon input of autophagy induction signals, part of Atg14L that constitutively localizes to the entire ER moves to specific points in the ER to form omegasomes.
Figure 1.
Involvement of Atg14L in the initial phase of autophagosome formation. (a) HEK293 cells stably expressing GFP-DFCP1 were transfected with or without shRNA against Atg14L. The cell lysates were subjected to immunoblotting with the indicated antibodies. (b) GFP signals were observed in these cells before or after starvation. (c) The number of GFP-positive puncta per cell was counted; mean ± SD values are presented. N, nutrient rich; S, starvation. (d) HEK293 cells were lysed and subjected to subcellular fractionation as described in Materials and methods. Equivalent amounts of total, cytosolic, and ER fractions were subjected to immunoblotting with each organelle marker antibody. The percentage of recovery in each fraction is presented as mean ± SD values. (e) HEK293 cells stably expressing GFP-DFCP1 were transiently transformed with Flag-tagged Atg14L and subjected to starvation. The cells were fixed and stained with the indicated antibodies. Insets show higher magnification of the boxed areas. (f) HEK293 cells were transfected with GFP-Atg14L with or without mCherry-linked dominant-negative ULK1, and these cells were cultured in complete or starvation medium for 60 min. Bars, 10 µm.
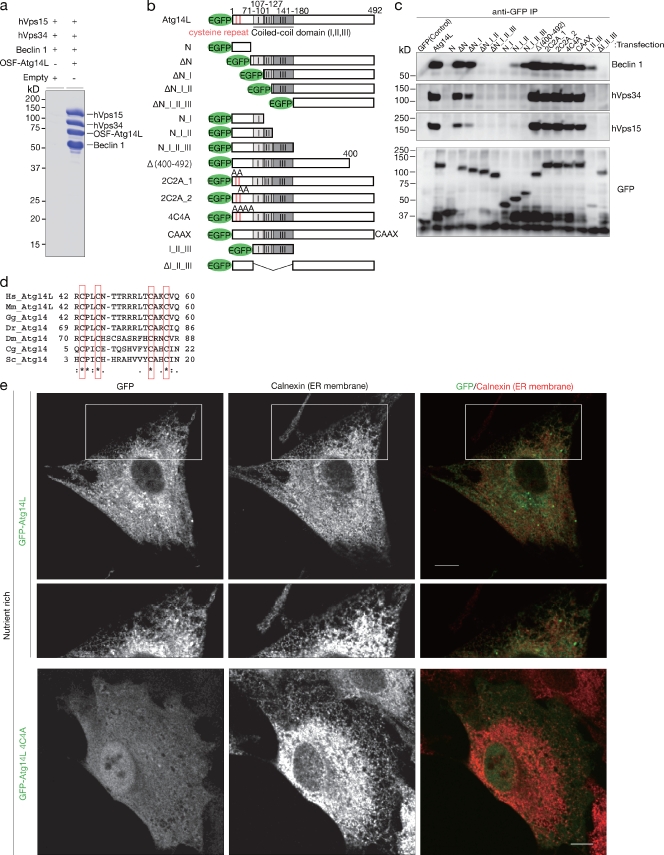
Although direct interaction between Atg14L and Beclin-1 has been suggested from yeast two-hybrid analysis, the possible interaction between Atg14L–Beclin-1 and hVps34–hVps15 has remained unclear (Matsunaga et al., 2009). To elucidate this, we transfected HEK293T cells simultaneously with four plasmids harboring Beclin-1, Atg14L, hVps34, and hVps15. The lysates were pulled down using the Strep-Tactin tag connected to Atg14L, and resulting precipitates were subjected to SDS-PAGE and Coomassie brilliant blue staining. As a result, there were no bands other than Beclin-1, Atg14L, hVps34, and hVps15 equimolar to these four subunits (Fig. 2 a). This strongly suggests that a subunit binds directly to another subunit. We then analyzed the structural and functional relationship of Atg14L. Initially, we analyzed the Atg14L domain architecture in terms of binding to Beclin-1 and hVps34–hVps15. Coimmunoprecipitation experiments were performed using a series of Atg14L deletion constructs as bait. Among the three coiled-coil domains at the N terminus of Atg14L, the central domain, termed II, is indispensable for binding to Beclin-1 and hVps34–hVps15 (Fig. 2, b and c; ΔN_I and ΔN_I_II). Even without the coiled-coil domain III, Atg14L binds to Beclin-1, but with lower efficiency (Fig. 2, b and c; N_I_II and N_I_II_III), suggesting that the coiled-coil domain III is involved in binding to Beclin-1. In contrast, the noncharacterized portion (181–400 aa) at the C terminus is indispensable for binding to hVps34–hVps15 but dispensable for binding to Beclin-1 (Fig. 2, b and c; N_I_II_III and Δ400–492). This undefined region (181–400 aa) plays a role in the interaction with hVps34–hVps15, together with coiled-coil domain II (and potentially with domain III). Therefore, whether hVps34–hVps15 binds to Atg14L directly or indirectly through Beclin-1 is still an open question.
Figure 2.
Domain analysis of Atg14L structure. (a) HEK293 cells were simultaneously transfected with four plasmids harboring One-Strep-Flag (OSF)–tagged Atg14L, Beclin-1, hVps34, and hVps15, and Atg14L was pulled down using Strep-Tactin Sepharose beads. Empty plasmid was used as control instead of Atg14L. The precipitates were subjected to SDS-PAGE and Coomassie brilliant blue staining. (b) Diagram of Atg14L deletion mutant construction. (c) HEK293 cells transiently expressing each construct were subjected to coimmunoprecipitation analysis with the anti-GFP antibody. Each precipitant was subjected to immunoblotting with the indicated antibodies. IP, immunoprecipitation. (d) Alignment of N-terminal conserved region of Atg14 orthologues. Conserved cysteine repeats are boxed. The degree of sequence conservation is denoted: asterisks indicate a perfect match among interspecies, periods indicate mostly conserved, and colons indicate more conserved than the periods. Homo sapiens (Hs), Mus musculus (Mm), Gallus gallus (Gg), Danio rerio (Dr), Drosophila melanogaster (Dm), Candida glabrata (Cg), and Saccharomyces cerevisiae (Sc) are shown. (e) NRK cells were transiently transfected with adenovirus expressing GFP-Atg14L or GFP-Atg14L4C4A and fixed under nutrient-rich conditions. The cells were stained with anticalnexin antibody as an ER membrane marker. Higher magnification of the boxed areas is shown below. Bars, 10 µm.
Constructs consisting of only coiled-coil domains of Atg14L failed to localize to the ER (Fig. S1 b). However, Atg14L lacking the whole coiled-coil domain restored the ER localization pattern (Fig. S1 c). We found that neither the N-terminal portion alone nor the C-terminal portion alone was sufficient for ER localization (unpublished data). Thus, the N-terminal domain together with the C-terminal portion is important for ER localization.
Although overall homology is quite low between yeast and mammalian Atg14, the N-terminal region is most conserved among species over the whole Atg14L sequence (Matsunaga et al., 2009). Moreover, the N-terminal region contains characteristic cysteine repeats that are completely conserved from yeast to mammals (Fig. 2 d). To explore the function of these cysteine repeats, either two of four or all four cysteine residues were replaced with alanine residues. As expected, these mutations did not affect binding to Beclin-1 and hVps34–hVps15 (Fig. 2, b and c; Atg14L2C2A and Atg14L4C4A). However, GFP-Atg14L4C4A failed to show any reticular pattern but rather had a dispersed pattern (Fig. 2 e). These results suggest that the localization of Atg14L to the ER is determined by Atg14 itself through these cysteine repeats and does not require interactions with Beclin-1, hVps34, or hVps15.
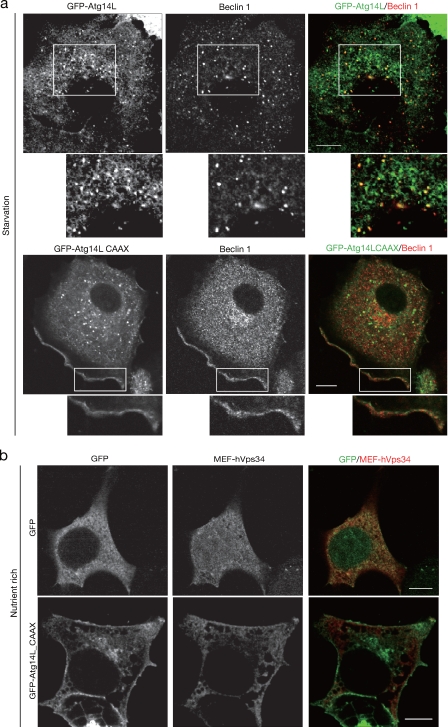
To support the suggestion that Atg14L determines the localization of the autophagy-specific PI3-kinase complex, the effect of artificially altering the localization of Atg14L was examined. The CAAX motif containing 17 aa (KDGKKKKKKSKTKCVIM) of K-ras, which is sufficient for heterologous protein targeting to the plasma membrane (Hancock et al., 1991), was added to Atg14L. This modification did not affect the binding to Beclin-1, hVps34, and hVps15 (Fig. 2, b and c; CAAX). As expected, GFP-Atg14LCAAX demonstrated a plasma membrane localization pattern significantly different from the wild-type pattern (Fig. 3 a and Fig. S1 d), although some populations still remained in intracellular structures. These intracellular structures seem not to have been bona fide Atg14L localization because addition of the CAAX motif to GFP-Atg14L4C4A gave a similar pattern (Fig. S1 d). By expression of GFP-Atg14LCAAX, Beclin-1 and hVps34 molecules were also targeted to the plasma membrane (Fig. 3, a and b). These data indicate that Atg14L is the determining component for membrane localization of the autophagy-specific PI3-kinase complex, Beclin-1 and hVps34.
Figure 3.
Ectopic expression of Atg14L in the plasma membrane. (a) A549 cells were transiently transfected with adenovirus expressing GFP-Atg14L or GFP-Atg14LCAAX and subjected to starvation. The cells were fixed and stained with anti–Beclin-1 antibody. Higher magnification of the boxed areas is shown below. (b) HEK293 cells were cotransfected with MEF-hVps34 and GFP or GFP-Atg14LCAAX and stained with anti-Flag antibody. Bars, 10 µm.
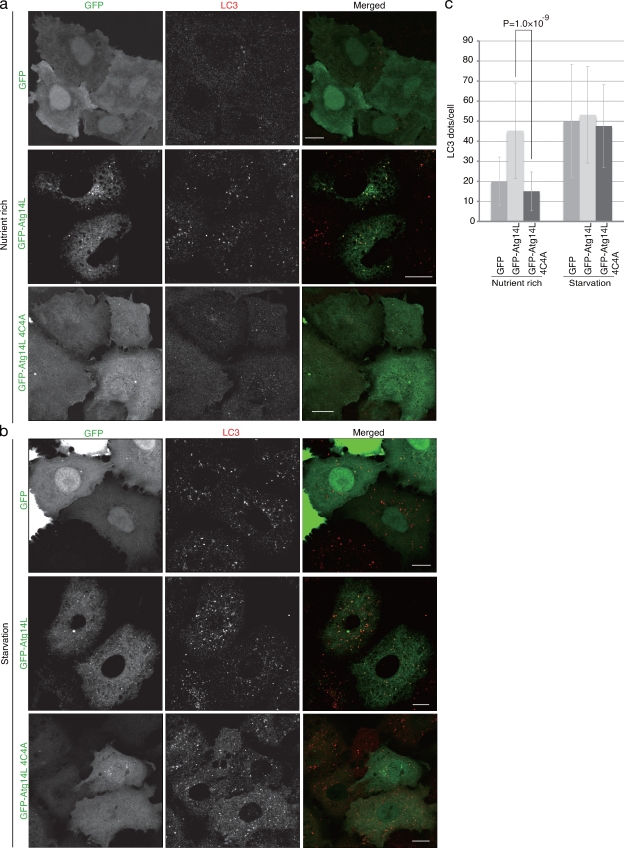
Next we investigated how the localization of Atg14L is related to its function. A549 cells were transfected with an adenovirus harboring GFP-Atg14L. Interestingly, the amount of PI3P-positive puncta found by immunostaining of the GST-FYVE domain was increased by overexpression of GFP-Atg14L, indicating that extra PI3P is generated in response to the overexpression of Atg14L (Fig. S2, a and b). This transient overexpression resulted in the appearance of both GFP-Atg14L–positive puncta, in addition to ER localization, and a significant increase of LC3-positive puncta even under nutrient-rich conditions (Fig. 4, a and c). The GFP-Atg14L–positive puncta were colocalized with the LC3 puncta (Fig. 4, a and b). Conditions of starvation did not further increase the number of LC3-positive puncta (Fig. 4, b and c). These results suggest that ectopic expression of cellular Atg14L, at least partially, bypassed the requirement for an autophagy induction signal triggered by starvation. Next, we tested the effect of overexpression of an Atg14L mutant defective in ER localization. This mutant induced neither the formation of GFP-Atg14L–positive puncta nor LC3-positive puncta (Fig. 4, a and c). These results suggest that ER localization is associated with Atg14L function in response to autophagy induction caused by its overexpression.
Figure 4.
Autophagy induction by overexpressed Atg14L. A549 cells were transiently transfected with adenovirus harboring GFP-Atg14L or GFP-Atg14L4C4A. (a and b) The cells before starvation (a) or after starvation (b) were fixed and stained with anti-LC3 antibody. (c) The number of LC3-positive puncta per cell was counted, and mean ± SD values are presented. Bars, 10 µm.
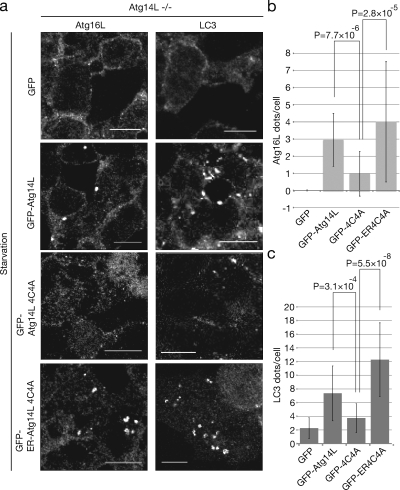
However, Atg14L function under native autophagy-inducing conditions, namely nutrient starvation, is still unclear. Therefore, we extended our analysis using Atg14L knockout embryonic stem (ES) cells (Matsunaga et al., 2009). Control GFP, wild-type GFP-Atg14L, and GFP-Atg14L4C4A were transfected into Atg14L knockout ES cells, which were then subjected to nutrient starvation (Fig. 5). As already reported, in the control GFP-expressing cells, Atg16L puncta were not detected, and very few LC3 puncta, which were assumed to be aberrant or forming intermediates of autophagosomes, were detected (Matsunaga et al., 2009). The expression of wild-type GFP-Atg14L reverted this autophagy-defective phenotype; however, GFP-Atg14L4C4A scarcely reverted the defective phenotype (Fig. 5). Although this result strongly supports the critical role of Atg14L localization to the ER, there still remains the possibility that the Atg14L cysteine repeats are required not only for Atg14L localization but also for another essential function of Atg14L. We tried to circumvent this problem by adding an extra ER-targeting motif to GFP-Atg14L4C4A. We added the ER-targeting motif of DFCP-1, which is necessary and sufficient for ER targeting of DFCP1 but insufficient for omegasome targeting (Axe et al., 2008). This construct was still able to associate with Beclin-1, hVps34, and hVps15 (Fig. S3 a). We confirmed that the construct (GFP-ER-Atg14L4C4A) showed an ER localization pattern that overlapped with the ER-resident protein, calnexin, and puncta overlapped with LC3 puncta in HEK293 cells (ER localization is difficult to visualize with ES cells because of their compact cytoplasmic spaces; Fig. S3, b and d). Then we transfected Atg14L knockout cells with this construct and found that it fully complemented the defect in LC3 puncta formation, similar to that of wild-type Atg14L (Fig. 5; GFP-ER-Atg14L4C4A). We also confirmed that autophagic flow is completed depending on ER localization of Atg14L using a tandem fluorescent-tagged LC3 system (Kimura et al., 2007). Overexpression of GFP-ER-Atg14L4C4A but not GFP-Atg14L4C4A can induce formation of the puncta with an RFP signal and without a GFP signal, which represent completed autophagic flow (Fig. S3 c). Therefore, we concluded that recruitment of the autophagy-specific PI3-kinase complex to the ER is the critical function of Atg14L in autophagy.
Figure 5.
The effect of Atg14L localization on its function. (a) Atg14L knockout mouse ES cells were transfected with GFP, GFP-Atg14L, GFP-Atg14L4C4A, or GFP-ER-Atg14L4C4A of the pCAG vector and subjected to starvation. The cells were fixed and immunostained with anti-Atg16L or anti-LC3 antibodies. (b and c) The number of Atg16L-positive puncta (b) and LC3-positive puncta (c) per cell was counted in at least 10 GFP-positive cells, and mean ± SD values are presented. Bars, 10 µm.
In this study, we have elucidated the molecular mechanism by which the autophagy-specific PI3-kinase complex is recruited to the ER. This explains the enigma raised by the study of Axe et al. (2008) regarding the existence of PI3P in the ER. Atg14L needs to be targeted to the ER to fulfill its function in autophagy, and PI3-kinase is recruited accordingly. We did not see an apparent difference in the efficiency of ER recruitment of GFP-Atg14L in nutrient-rich versus nutrient starvation conditions. Furthermore, our previous subcellular fractionation analysis showed that ∼20–30% of endogenous Atg14L protein is recovered in the pelletable fraction, and this percentage was largely unchanged in response to starvation (Matsunaga et al., 2009; unpublished data). Therefore, ER recruitment of the PI3-kinase complex is critical to autophagosome formation but is not an autophagosome formation trigger downstream of starvation.
This is a very interesting strategy of the cells to use PI3P as a secondary signal in the ER, where it is otherwise essentially absent, because it will provide a very specific localization message. Potential downstream effectors of PI3P are WIPI-1 family proteins and/or DFCP-1, and these will lead to subsequent events required to complete autophagosome formation (Proikas-Cezanne et al., 2007).
PI3P is largely absent over the surface of the general ER membrane, even if a certain amount of PI3-kinase is recruited by Atg14L because a FYVE domain does not lead to ER localization of DFCP-1 (Axe et al., 2008). The PI3-phosphatase Jumpy and MTMR3 may contribute to keeping PI3P out of the ER (Vergne et al., 2009; Taguchi-Atarashi et al., 2010). The GFP-Atg14L puncta signal associated with the omegasome is apparently much brighter than the signal on the ER (Fig. 1 e). The next key question is how the PI3-kinase complex is nucleated to the omegasome on the ER. The density of the PI3-kinase complex on the ER membrane may be a critical factor because overexpression of Atg14L alone induces autophagy. It is possible that once the density exceeds some threshold, through its artificial overexpression, Atg14L may become concentrated at some specific points. In contrast, the starvation signal directs, via ULK1, Atg14L to be concentrated at points without an increment in its density using unknown mechanisms. GFP-Atg14L lacking the whole coiled-coil domain, which does not bind to Beclin-1, hVps34–hVps15, also makes GFP-positive puncta (Fig. 2, b and c; and Fig. S1 c), suggesting that Atg14L itself possesses the ability for concentration. The mechanism may be critical in autophagy initiation downstream of ULK1 signaling (Fig. 1 f).
To our knowledge, this is the first study showing the mechanism of recruitment of the PI3-kinase to the ER and its necessity in autophagosome formation. This is not only strong supportive evidence for recent challenging of a model of omegasome involvement in autophagy but also an excellent example of specification of intracellular membranes mediated by phosphoinositide turnover. It is interesting that a specific phosphoinositide in the ER plays an unexpected role in autophagy, especially in a manner quite different from those discussed in a past study (Hayashi-Nishino et al., 2009). We hope our study provides some advances in the longstanding debate as to the origin of the autophagosomal membrane.
Materials and methods
Cell culture
HEK293 stably expressing GFP-DFCP1 (clone 201; Axe et al., 2008), HEK293, NRK, and A549 cells were cultured in DME D6546 (Sigma-Aldrich) containing 10% fetal bovine serum supplemented with 4 mM l-glutamine in 5% CO2 at 37°C. The clone 201 cell line was cultured in medium containing 500 µg ml−1 geneticin (Invitrogen). Mouse ES cells were cultured in DME D6546 containing 0.1 mM nonessential amino acids (Invitrogen), 1 µM 2-mercapthoethanol (Wako Chemicals USA, Inc.), 2 mM l-glutamine (Sigma-Aldrich), 20% fetal bovine serum (Invitrogen), and 1,000 U ml−1 Leukemia inhibitory factor (EMD) on plastic plates coated with 0.1% gelatin (Wako Chemicals USA, Inc.). For amino acid and serum starvation, cells were cultured in Earle’s balanced salt solution (Sigma-Aldrich) for 3–4 h. For starvation-independent autophagy induction, LiCl (Wako Chemicals USA, Inc.) was added to a final concentration of 10 mM, and etoposide (Sigma-Aldrich) dissolved in DMSO was added to a final concentration of 10 µM.
Plasmid construction and transfection
The pcDNA3.1/3xFlag-His(-)A-Atg14L plasmid has been previously described (Matsunaga et al., 2009). Truncation and deletion mutants of Atg14L (residues 1–64, 65–492, 103–492, 132–492, 181–492, 1–101, 1–127, 1–180, and 1–400) were cloned into the EcoRI–KpnI site of the pEGFP-C2 vector using standard techniques. Site-directed mutagenesis to generate point mutants within Atg14L was performed using standard PCR techniques, and these constructs were verified by DNA sequencing. All point mutants of Atg14L were cloned into the EcoRI–KpnI site of the pEGFP-C2 vector. To generate the GFP-Atg14LCAAX construct and GFP-Atg14L4C4ACAAX, the CAAX motif sequence was amplified by PCR as described previously (Fujita et al., 2008b) and cloned into the pEGFP-C2-Atg14L vector. To generate the GFP-ER-Atg14L4C4A construct, the ER-targeting sequence of DFCP1 (Axe et al., 2008) was amplified by PCR and cloned into the pEGFP-C2-Atg14L4C4A vector. The pCAG vector was described previously (Morita et al., 2007). mCherry-ULK1K46N was subcloned into the pEGFP vector. GFP, GFP-Atg14L, GFP-Atg14L4C4A, and GFP-ER-Atg14L4C4A were subcloned into site NheI–BglII of the pCAG vector. MEF-hVps34 was subcloned into site EcoRI–NotI of the pcDNA-MEF vector (Ichimura et al., 2005). The pGEM-1-GST-2×FYVE was a gift from H. Stenmark (Norwegian Radium Hospital, Oslo University Hospital, Oslo, Norway; Gillooly et al., 2000). The tandem fluorescent-tagged LC3 (tfLC3) plasmid has been previously described (Kimura et al., 2007). Subconfluent cells were transfected with plasmids using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
Antibodies
Polyclonal antibodies against Atg14L, Beclin-1, and hVps34 have been previously described (Matsunaga et al., 2009). Other antibodies and reagents were as follows: rabbit anti-Flag (Sigma-Aldrich), rabbit anticalnexin (Stressgen), rabbit anti-GFP (MBL), mouse monoclonal anti-GFP (Roche), rabbit anti–Beclin-1 (MBL), rabbit anti-Atg16L (MBL), rabbit anti-LC3 (MBL), rabbit anti-GST (Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-p150 (1G12; Abnova), and mouse monoclonal anti–α-tubulin (Sigma-Aldrich) antibodies.
Adenoviral expression system
The pENTR-1A-EGFPC2 and pENTR-1A-EGFPC2-Atg14L vectors have been previously described (Matsunaga et al., 2009). GFP-Atg14L4C4A, GFP-ER-Atg14L4C4A, and GFP-Atg14LCAAX cDNAs were subcloned into the DraI–EcoRV sites of the pENTR-1A vector and transferred into the pAd/CMV vector (Invitrogen) by performing the Clonase LR recombination reaction (Invitrogen). The production of adenovirus and adenoviral infection were performed according to the manufacturer’s protocol.
Isolation of ER fraction by differential sucrose gradient centrifugation
In a 140-mm plastic plate, 293A cells were trypsinized and resuspended in 2 ml MTE solution (270 mM d-mannitol, 10 mM Tris-HCl, 0.1 mM EDTA, and 1 mM PMSF, pH 7.4). The cell suspension was homogenized by repeatedly shearing 15 times through a 25-gauge needle mounted on a 1-ml syringe, as described previously (Matsunaga et al., 2009), and centrifuged (1,400 g for 10 min at 4°C). The supernatant was centrifuged for 10 min at 15,000 g and 4°C. For ER purification, the supernatant was layered onto a sucrose density gradient (2.0-, 1.5-, and 1.3-M discontinuous sucrose gradient into ultracentrifuge tube) and ultracentrifuged for 70 min at 152,000 g and 4°C. The large band was collected at the interface of the 1.3-M sucrose gradient layer (Bozidis et al., 2007). An equivalent amount of each fraction was subjected to immunoblotting. The band intensity was quantified from three independent experiments using the Image Gauge (Fujifilm) image analyzer.
Atg14L complex purification
HEK293T cells were cotransfected with Atg14L complex expression plasmids (pCAG-OSF.Atg14L, pCAG-NT.hVps15, pCAG-NT.hVps34, and pCAG-NT.Beclin-1). Cells were harvested 48 h after transfection by incubation in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, proteinase inhibitor cocktail, and 1% Triton X-100). Lysates were clarified by centrifugation (18,000 g for 10 min at 4°C) and incubated with Strep-Tactin Sepharose (IBA GmbH; for 2 h at 4°C). The matrix was washed four times in wash buffer (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100), and purified complexes were eluted with 2× SDS-PAGE sample buffer.
Vector-based small interference RNA expression
The mammalian expression vector pSUPER.retro.puro (OligoEngine) was used for expression of shRNA in HEK293 stably expressing GFP-DFCP1 cells (clone 201). pSUPER.retro.puro empty vector was used as a control vector. The gene-specific insert specified a 19-nt sequence 523–541 of human Atg14L (5′-AGAAGATTCAGAGGCATAA-3′). This sequence was inserted into the pSUPER.retro.puro after digestion with BglII and HindIII. Subconfluent cells in 35-mm dishes were transfected with pSUPER.retro.puro-shAtg14L and pSUPER.retro.puro–empty vector and transferred to 60-mm dishes 24 h later. After an additional 24 h, the cells were transfected again with the vector. The medium was changed to DME containing 1 µg ml−1 puromycin after an additional 24 h. After a further 72 h, the cells were transferred to 35-mm dishes with coverslips. After an additional 24 h, cells were cultured in Earle’s balanced salt solution for 45 min.
GST-2×FYVE probe assay
GST-2×FYVE recombinant protein expression and purification in Escherichia coli BL21 were performed according to the manufacturer’s protocols (GE Healthcare). GFP-Atg14L– or GFP-Atg14L4C4A–transfected cells on coverslips were permeabilized with 0.05% saponin in PBS for 5 min, then fixed with 3% PFA for 10 min and incubated 20 µg/ml GST-2×FYVE for 60 min, and then stained with anti-GST antibody (Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor 594–conjugated secondary antibody (Invitrogen) for 60 min. Samples were mounted using Slow Fade Gold (Invitrogen) and observed using a laser confocal microscope (FV1000; Olympus).
Immunoprecipitation and Western blotting
For immunoprecipitation, cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [wt/vol] glycerol, 100 mM NaF, 10 mM EGTA, 1 mM Na3VO4, 1% Triton X-100, 5 µM ZnCl2, 1 mM PMSF, and complete protease inhibitor cocktail [Roche]). Lysates were centrifuged for 10 min at 15,000 g and 4°C. The supernatants were subjected to immunoprecipitation using primary antibody and protein G–Sepharose 4FF (GE Healthcare). Immunocomplexes were washed five times with wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton X-100), subjected to SDS-PAGE, and transferred to polyvinylidenefluoride membranes. The membranes were blocked with TBST (TBS and 0.1% Tween 20) containing 1% nonfat dried milk and were then incubated overnight at room temperature with primary antibodies diluted in Can Get Signal solution I (TOYOBO). Membranes were washed three times with TBST, incubated for 1 h at room temperature with 5,000× dilutions of HRP-conjugated secondary antibodies (GE Healthcare) in TBST containing 1% nonfat dried milk, and washed five times. Immunoreactive bands were then detected using ECL plus (GE Healthcare) and a chemiluminescence detector (LAS-3000; Fujifilm; Matsunaga et al., 2009).
Immunofluorescence and microscopy
Cells were cultured on coverslips, fixed with 3% PFA in PBS for 10 min, and permeabilized with 50 µg ml−1 digitonin in PBS for 5 min. Cells were then treated with 50 mM NH4Cl−PBS for 10 min at room temperature and blocked with PBS containing 3% BSA for 15 min. Primary antibodies were diluted 1:100 or 1:200, and Alexa Fluor–conjugated secondary antibodies (Invitrogen) were diluted 1:500 in PBS containing 3% BSA. Coverslips were incubated with primary antibodies for 60 min, washed six times with PBS, and incubated with secondary antibodies for 60 min. Samples were mounted using Slow Fade Gold and observed with an FV1000 laser confocal microscope (Matsunaga et al., 2009). The microscope images were taken using an FV1000 confocal laser-scanning microscope system equipped with 100× NA 1.40 oil immersion objective lens and an A1 (Nikon) equipped with 100× NA 1.49 oil immersion objective lens. Image acquisition software used was Fluoview (Olympus) with NIS elements (Nikon) integrated in the system. Fluorochromes associated with the secondary antibodies were Alexa Fluor 405 or 594. The images were adjusted using the software Photoshop CS4 (Adobe). We counted the number of puncta per cell in at least 10 cells. Fluorescence intensity of GFP-Atg14LCAAX or GFP-Atg14L4C4ACAAX was quantified using ImageJ 1.43 (National Institutes of Health). The plasma membrane GFP signal intensity of the cell was acquired by drawing a 3-pixel-wide line around the cell using the segmented line selection tool, and the total GFP signal intensity of the cell was acquired by surrounding the cell using the polygon selection tool.
Statistical analysis
Statistical analyses were performed using a two-tailed unpaired t test. P-values <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows the localization of Atg14L treated with nonconventional autophagy inducers and Atg14L mutants. Fig. S2 shows the effect of overexpression of Atg14L or Atg14L4C4A mutant on extra PI3P generation. Fig. S3 shows the character of Atg14L4C4A added with the ER-targeting motif of DFCP-1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911141/DC1.
Acknowledgments
The work described in this report was supported in part by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Takeda Science Foundation.
Footnotes
Abbreviations used in this paper:
- ES
- embryonic stem
- PI3-kinase
- phosphatidylinositol 3-kinase
- PI3P
- phosphatidylinositol 3-phosphate
- shRNA
- short hairpin RNA
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozidis P., Williamson C.D., Colberg-Poley A.M. 2007. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.27 10.1002/0471143030.cb0327s37 [DOI] [PubMed] [Google Scholar]
- Cheung P.C., Trinkle-Mulcahy L., Cohen P., Lucocq J.M. 2001. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 355:113–121 10.1042/0264-6021:3550113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. 2008a. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 19:4651–4659 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. 2008b. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 19:2092–2100 10.1091/mbc.E07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.F., Cadwallader K., Paterson H., Marshall C.J. 1991. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10:4033–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D.J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., Isobe T. 2005. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 280:13187–13194 10.1074/jbc.M412884200 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. 2001a. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2:330–335 10.1093/embo-reports/kve061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. 2001b. Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119:605–614 10.1242/jcs.02855 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11:385–396 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116:1679–1688 10.1242/jcs.00381 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Alam S.L., Eckert D.M., Gygi S.P., Sundquist W.I. 2007. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2:41–53 10.1016/j.chom.2007.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 461:654–658 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- Noda T., Fujita N., Yoshimori T. 2009. The late stages of autophagy: how does the end begin? Cell Death Differ. 16:984–990 10.1038/cdd.2009.54 [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Ohsumi Y. 2006. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:1527–1539 10.1091/mbc.E05-09-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K., Noda T., Niimi K., Ohsumi Y. 2008a. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 13:537–547 10.1111/j.1365-2443.2008.01188.x [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Niimi K., Ohsumi Y. 2008b. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283:23972–23980 10.1074/jbc.M803180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne T., Ruckerbauer S., Stierhof Y.D., Berg C., Nordheim A. 2007. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 581:3396–3404 10.1016/j.febslet.2007.06.040 [DOI] [PubMed] [Google Scholar]
- Reggiori F. 2006. 1. Membrane origin for autophagy. Curr. Top. Dev. Biol. 74:1–30 10.1016/S0070-2153(06)74001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L.J., Rubinsztein D.C. 2005. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170:1101–1111 10.1083/jcb.200504035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Tooze S.A. 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186:773–782 10.1083/jcb.200907014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 105:19211–19216 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Atarashi N., Hamasaki M., Matsunaga K., Omori H., Ktistakis N.T., Yoshimori T., Noda T. 2010. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 11:468–478 10.1111/j.1600-0854.2010.01034.x [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., Deretic V. 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 28:2244–2258 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Flinn R.J., Wu H., Schnur R.S., Backer J.M. 2009. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem. J. 417:747–755 10.1042/BJ20081865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Noda T. 2008. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 20:401–407 10.1016/j.ceb.2008.03.010 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., Yue Z. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11:468–476 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]