PKA puts the brakes on autophagy by inhibiting LC3 recruitment to autophagosomes.
Abstract
Macroautophagy is a major catabolic pathway that impacts cell survival, differentiation, tumorigenesis, and neurodegeneration. Although bulk degradation sustains carbon sources during starvation, autophagy contributes to shrinkage of differentiated neuronal processes. Identification of autophagy-related genes has spurred rapid advances in understanding the recruitment of microtubule-associated protein 1 light chain 3 (LC3) in autophagy induction, although braking mechanisms remain less understood. Using mass spectrometry, we identified a direct protein kinase A (PKA) phosphorylation site on LC3 that regulates its participation in autophagy. Both metabolic (rapamycin) and pathological (MPP+) inducers of autophagy caused dephosphorylation of endogenous LC3. The pseudophosphorylated LC3 mutant showed reduced recruitment to autophagosomes, whereas the nonphosphorylatable mutant exhibited enhanced puncta formation. Finally, autophagy-dependent neurite shortening induced by expression of a Parkinson disease–associated G2019S mutation in leucine-rich repeat kinase 2 was inhibited by dibutyryl–cyclic adenosine monophosphate, cytoplasmic expression of the PKA catalytic subunit, or the LC3 phosphorylation mimic. These data demonstrate a role for phosphorylation in regulating LC3 activity.
Introduction
The major regulatory pathways promoting induction of autophagy converge on the covalent lipidation of LC3 (microtubule-associated protein 1 light chain 3). Like its yeast homologue Atg8, LC3 is cleaved to liberate a C-terminal glycine needed for its conjugation to phospholipids upon autophagy induction. This processing and recruitment is essential for extension, curvature, and closure of the isolation membrane to form autophagosomes (He and Klionsky, 2009), also playing a role in cargo recognition (Pankiv et al., 2007). Lipidated LC3 is a useful marker of autophagic membranes, as it migrates to an apparently lower Mr position (LC3-II) by electrophoresis, and autophagosomes are visualized as bright GFP-LC3 puncta (Klionsky et al., 2008). Although much has been learned about the processing of LC3 necessary to initiate autophagy, regulatory mechanisms capable of modulating or fine tuning its participation in autophagy remain poorly understood.
It is recognized that a fine balance of stress-induced autophagy is important (Cuervo, 2004), particularly in neuronal cells (Boland and Nixon, 2006; Jaeger and Wyss-Coray, 2009), as autophagy functions to reduce protein aggregates but can also contribute to neurite degeneration (Yang et al., 2007; Plowey et al., 2008). Indeed, a current translational challenge is to harness beneficial effects of autophagy without eliciting harm (Zhang et al., 2007). We have previously shown that treatment with dibutyryl-cAMP (db-cAMP) protects against parkinsonian neurotoxins (Chalovich et al., 2006). Because PKA signaling can regulate autophagy in other cell types (Holen et al., 1996; Budovskaya et al., 2004), we investigated whether PKA regulation of autophagy protected against parkinsonian neurodegeneration.
Results and discussion
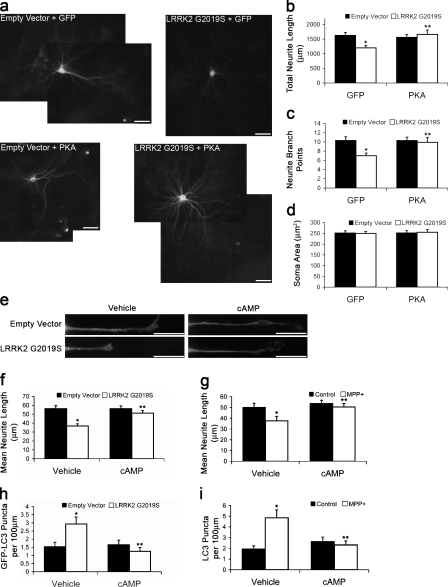
Neuritic/synaptic dysfunction or degeneration are observed in clinical studies of Parkinson disease (PD) patients and in genetic and toxic/environmental models of PD. For example, expression of the autosomal dominant PD-associated G2019S mutant form of LRRK2 (leucine-rich repeat kinase 2) causes shortening of neurite lengths and simplification of branch points as indices of neuritic injury (MacLeod et al., 2006; Plowey et al., 2008). In this study, we observed that treatment with db-cAMP or coexpression of a GFP-tagged PKA catalytic subunit prevented neurite injury caused by LRRK2 G2019S in cortical neurons (Fig. 1, a–c). As previously observed with LRRK2 (MacLeod et al., 2006), there were no effects on the mean soma area (Fig. 1 d). db-cAMP also protected against neuronal injury caused by LRRK2 G2019S and the parkinsonian neurotoxin MPP+ in differentiated SH-SY5Y cells (Fig. 1, e–g). Because both injuries are mediated by autophagy (Fig. S1 a; Zhu et al., 2007; Plowey et al., 2008), we investigated whether PKA activity regulated autophagy in neuronal cells. Treatment with db-cAMP reduced the number of autophagic vacuoles (AVs) induced by either LRRK2 G2019S or MPP+ (Fig. 1, e, h, and i).
Figure 1.
PKA signaling reduces neurite injury and autophagy. (a–d) Mouse cortical neurons coexpressing GFP or GFP-PKA and LRRK2 G2019S or vector 7 d after transfection. (b) Quantification of neurite lengths. *, P = 0.001 versus empty vector/GFP; **, P = 0.007 versus LRRK2 G2019S/GFP (n = 50–60 cells/condition). (c) Quantification of branch points. *, P = 0.001 versus empty vector/GFP; **, P = 0.015 versus LRRK2 G2019S/GFP (n = 50–60 cells/condition). (d) Quantification of soma area (n = 50–60 cells/condition). (e, f, and h) SH-SY5Y neurites expressing GFP-LC3 48 h after transfection with/without cAMP for 24 h. (f) Quantification of neurite lengths. *, P = 1.48 × 10−5 versus empty vector/vehicle; **, P = 9.21 × 10−5 versus LRRK2 G2019S/vehicle (n = 80–90 cells/condition). (g) Quantification of neurite lengths from MPP+-treated SH-SY5Y cells with/without cAMP for 48 h. *, P = 0.0298 versus control/vehicle; **, P = 0.0166 versus MPP+/vehicle (n = 50–60 cells/condition). (h) Quantification of GFP-LC3–labeled puncta with/without cAMP for 24 h. *, P = 0.006 versus empty vector/vehicle; **, P = 6.58 × 10−4 versus LRRK2 G2019S/vehicle (n = 80–90 cells/condition). (i) Quantification of GFP-LC3 puncta from MPP+-treated SH-SY5Y cells with/without cAMP. *, P = 6.06 × 10−4 versus control/vehicle; **, P = 0.004 versus MPP+/vehicle (n = 50–55 cells/condition). Error bars indicate mean ± SEM. Bars, 20 µm.
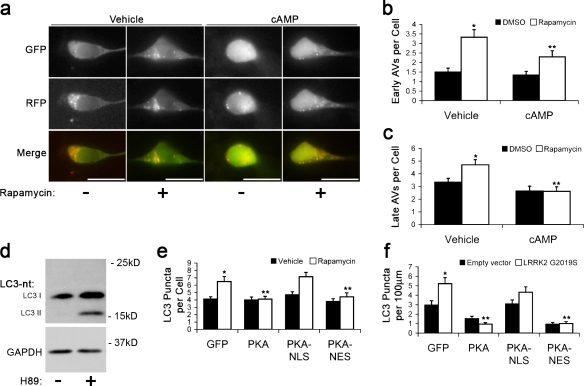
To determine whether db-cAMP reduced GFP-LC3 puncta by decreasing autophagy induction or by increasing AV maturation, we treated cells with rapamycin to induce autophagy and used the tandem reporter RFP-GFP-LC3 (Kimura et al., 2007), which labels early AVs with dual red and green fluorescence and late AVs with red only (Fig. 2 a). Rapamycin increased the number of early AVs at 1 h (Fig. S1 c). At 3 h, there was an increase in both early and late AVs (Fig. 2, b and c), which is consistent with maturation of rapamycin-induced AVs. Cotreatment with db-cAMP reduced the number of rapamycin-induced AVs at all time points (Fig. 2, a–c; and Fig. S1 c). Because db-cAMP reduced the number of rapamycin-induced early AVs without increasing late AVs, these data suggest that cAMP/PKA signaling suppresses autophagy induction. Treatment with the PKA inhibitor (E)-N-(2-(3-(4-bromophenyl)allylamino)ethyl)isoquinoline-5-sulfonamide (H89) led to an increase in lipidated LC3-II in SH-SY5Y, HeLa, and 293T cells (Fig. 2 d and Fig. S1 b), further indicating that PKA down-regulates autophagy induction.
Figure 2.
Cytoplasmic PKA inhibits autophagy. (a) RFP-GFP-LC3–labeled AVs in SH-SY5Y cells treated with rapamycin ± cAMP for 1 h. (b) Quantification of early AVs (GFP+ RFP+ puncta) in SH-SY5Y cells cotreated with rapamycin and cAMP for 3 h. *, P = 2.65 × 10−6 versus vehicle lacking rapamycin; **, P = 0.001 versus vehicle with rapamycin (n = 45–50 cells/condition). (c) Quantification of late AVs (RFP+-only puncta) in SH-SY5Y cells cotreated with rapamycin and cAMP for 3 h. *, P < 0.008 versus vehicle without rapamycin; **, P < 0.05 versus vehicle with rapamycin (n = 50–55 cells/condition). (d) LC3 immunoblot of SH-SY5Y cells treated with H89 or vehicle for 1 h. (e) Quantification of endogenous LC3-immunolabeled AVs in SH-SY5Y cells after 1-h treatment with rapamycin or DMSO (vehicle). *, P = 0.003 versus vehicle/GFP; **, P < 0.013 versus rapamycin/GFP (n = 35–45 cells/condition). (f) Quantification of endogenous LC3-immunolabeled AVs in SH-SY5Y cells 48 h after transfection. *, P = 3.52 × 10−4 versus empty vector/GFP group; **, P < 4.66 × 10−5 versus LRRK2 G2019S/GFP group (n = 80–90 cells/condition). Error bars indicate mean ± SEM. Bars, 20 µm.
Because PKA has both transcriptional (Chalovich et al., 2006) and posttranslational (Bok et al., 2003) neuroprotective effects, we used untargeted, nuclear-targeted (NLS), or cytoplasmic-targeted (nuclear export signal) GFP-tagged PKA (Bok et al., 2003) to study the effects of PKA subcellular localization on autophagy. Although the targeted PKA constructs elicited similar levels of cAMP response element binding protein phosphorylation (not depicted), nuclear localized PKA was unable to significantly reduce the number of AVs induced by rapamycin or LRRK2 G2019S (Fig. 2, e and f), whereas nuclear excluded PKA showed the same potency as untargeted PKA. These results suggest that the effect of PKA on autophagy is mediated by a cytoplasmic target.
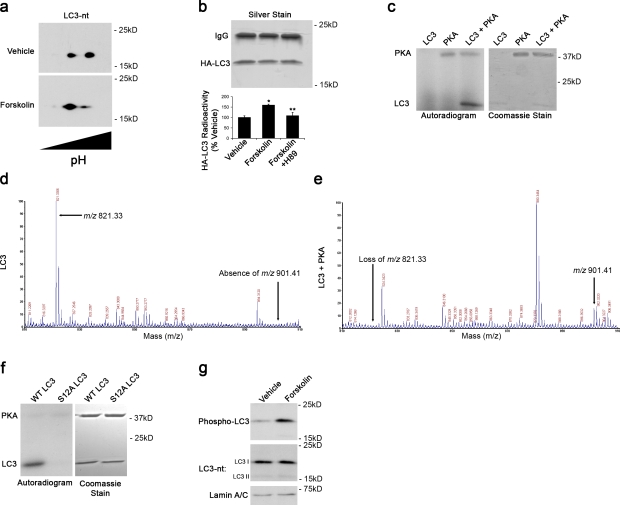
To identify potential autophagy-modulating cytoplasmic targets of PKA, we performed isoelectric focusing/SDS-PAGE 2D gel analysis on untreated cortical neurons (Fig. 3 a) and SH-SY5Y cells (not depicted). 2D immunoblots for LC3 revealed the presence of distinct species differing in isoelectric points, which is consistent with multiple phosphorylation states. Furthermore, treatment with forskolin, a known regulator of adenylate cyclase, increased the intensity of species migrating at more acidic isoelectric points, which is consistent with increased phosphorylation (Fig. 3 a). In living cells, we found that forskolin elicited increased 32P incorporation into HA-tagged LC3 (Fig. 3 b), which was prevented by cotreatment with the PKA inhibitor H89 (Fig. 3 b). Using in vitro kinase assays, we determined that PKA was capable of directly phosphorylating recombinant rat LC3 (Fig. 3 c).
Figure 3.
PKA directly phosphorylates LC3. (a) 2D immunoblot probed for LC3 shows distinct species differing by isoelectric points. Forskolin increases abundance of species with a more acidic isoelectric point. (b) Metabolic labeling reveals greater 32P incorporation in HA-LC3 immunoprecipitated from forskolin-treated cells compared with control cells. Error bars indicate mean ± SEM. *, P < 0.05 versus vehicle; **, P < 0.05 versus forskolin (n = 3 independent experiments). (c) In vitro kinase assay with recombinant PKA and purified rat LC3. Phosphorylation was detected by autoradiography. Coomassie stain shows migration of the recombinant proteins. (d and e) Recombinant LC3 was incubated with ATP in the presence and absence of PKA. In the presence of PKA and ATP, the m/z 821.33 ion is lost with the appearance of an m/z 901.41 ion. (f) Mutation of the LC3 phosphorylation site to S12A abolishes PKA phosphorylation of LC3. (g) Immunoblot of cortical neurons treated with forskolin or DMSO (vehicle) probed with phospho-specific LC3 antibody, total LC3 antibody (LC3-nt), and lamin antibody as a loading control.
We used matrix-assisted laser desorption/ionization (MALDI) time of flight (TOF) mass spectrometry (MS) to identify the PKA phosphorylation site on LC3. Analysis of the MS1 spectra revealed a singly charged peptide [M+H]+ ion signal at m/z 821.33 (Fig. 3 d) that was lost from samples treated with PKA, accompanied by the appearance of a new peptide [M+H]+ ion signal at m/z 901.41, corresponding to the predicted 80 D gain of a phosphate group (Fig. 3 e). Alkaline phosphatase treatment of PKA-phosphorylated LC3 eliminated the ion signal at m/z 901.41 (unpublished data). Tandem MS/MS analysis identified the m/z 901.41 ion as residues 8–13 containing a phosphorylation at serine 12 (Fig. S2 a). To confirm that serine 12 was the PKA phosphorylation site, mutagenesis of serine 12 to alanine (S12A) abolished LC3 phosphorylation by PKA (Fig. 3 f). Custom antibodies generated to the phosphorylated peptides effectively labeled PKA-phosphorylated recombinant LC3 (Fig. S2 b) and endogenously phosphorylated LC3 in SH-SY5Y and cortical neurons but not in the presence of the immunizing peptide (Fig. S2 c). Endogenously phosphorylated LC3 was further increased by treatment with forskolin to stimulate PKA (Fig. 3 g).
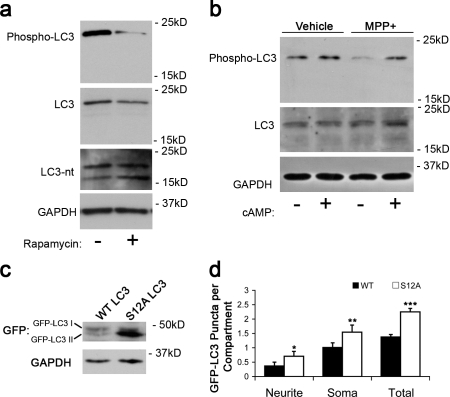
We investigated whether endogenous LC3 phosphorylation was regulated by rapamycin or MPP+, two stimuli that induce increased autophagic flux (Budovskaya et al., 2005; Zhu et al., 2007). Treatment with either rapamycin or MPP+ led to a reduction in phospho-LC3 as compared with vehicle-treated cells (Fig. 4, a and b) with no significant change in levels of LC3 probed using an antibody raised against a C-terminal region epitope of mature LC3 distant to the phosphorylation site (LC3). These results suggest that LC3 is dephosphorylated during autophagy induction. Blots reprobed using the N-terminal antibody for LC3 revealed the expected increase in LC3-II caused by rapamycin. Endogenously phosphorylated LC3 comigrated with nonlipidated forms of LC3 (Fig. 3 g and Fig. S2 c). To determine whether loss of phosphorylation was sufficient to increase the number of GFP-LC3–positive AVs, we transfected cells with a nonphosphorylatable mutant, GFP-LC3–S12A. Under basal conditions, GFP-LC3–S12A migrated predominantly as a lipidated LC3-II band, forming more puncta than GFP-LC3–wild type (WT; Fig. 4, c and d), indicating that phosphorylation of LC3 regulates its incorporation into AVs.
Figure 4.
LC3 dephosphorylation is associated with enhanced recruitment to autophagosomes. (a and b) Immunoblots of SH-SY5Y cells treated with rapamycin for 1 h (a) or MPP+ for 24 h (b) were probed using the phospho-specific antibody, a C-terminal LC3 antibody (LC3), an N-terminal LC3 antibody (LC3-nt), and GAPDH as loading control. (c) Immunoblotting of SH-SY5Y cells expressing GFP-LC3–WT or –S12A reveals that a greater fraction of GFP-LC3–S12A migrates as LC3-II. (d) Quantification of GFP-LC3 puncta in SH-SY5Y cells expressing GFP-LC3–WT or –S12A. *, P = 0.023 versus WT neurite; **, P = 0.014 versus WT soma; ***, P = 0.003 versus WT total (n = 100–115 cells/condition). Error bars indicate mean ± SEM.
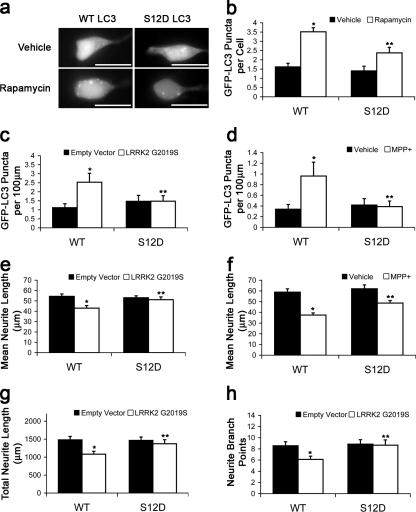
We determined whether dephosphorylation contributed to autophagy induction using the S12D phosphomimetic mutant in cells treated with rapamycin. The S12D mutation reduced the number of GFP-LC3 puncta in rapamycin-treated cells (Fig. 5, a and b). Similarly, the S12D mutant formed significantly fewer puncta in cells expressing LRRK2 G2019S or in cells treated with MPP+ (Fig. 5, c and d). Furthermore, the autophagy-related neurite shortening caused by LRRK2 G2019S or MPP+ was attenuated by the S12D mutation in SH-SY5Y cells and cortical neurons (Fig. 5, e–h). Collectively, the data indicate that LC3 phosphorylation by PKA suppresses induced autophagy and autophagic neurite degeneration elicited by LRRK2 G2019S and MPP+.
Figure 5.
LC3 phosphomimic shows reduced recruitment and suppresses neurite degeneration. (a and b) SH-SY5Y cells expressing GFP-LC3–WT or –S12D treated with rapamycin or vehicle for 1 h. Quantification of GFP-LC3 puncta. *, P = 2.31 × 10−8 versus vehicle/WT; **, P = 0.004 versus rapamycin/WT (n = 70–80 cells/condition). (c) Quantification of GFP-LC3 puncta in SH-SY5Y 48 h after transfection. *, P = 0.001 versus empty vector/GFP-LC3–WT; **, P = 0.025 versus LRRK2 G2019S/GFP-LC3–WT (n = 100–115 cells/condition). (d) Quantification of GFP-LC3 puncta in SH-SY5Y cells treated with MPP+ or vehicle for 48 h. *, P = 0.026 versus vehicle/WT; **, P = 0.043 versus MPP+/WT (n = 85–95 cells/condition). (e) Quantification of neurite lengths from SH-SY5Y cells 48 h after transfection. *, P = 6.37 × 10−4 versus empty vector/WT; **, P = 0.008 versus LRRK2 G2019S/WT (n = 100–115 cells/condition). (f) Quantification of neurite lengths from SH-SY5Y cells treated with MPP+ or vehicle for 48 h. *, P = 3.58 × 10−8 versus vehicle/WT; **, P = 3.74 × 10−4 versus MPP+/WT (n = 85–95 cells/condition). (g and h) Quantification of neurite length and branch points in cortical neurons coexpressing either GFP-LC3–WT or –S12A with empty vector or LRRK2 G2019S. *, P < 0.01 versus empty vector/WT; **, P < 0.036 versus LRRK2 G2019S/WT (n = 30–40 cells/condition). Error bars indicate mean ± SEM. Bars, 20 µm.
This study identifies a new phosphorylation site on LC3 that reduces its recruitment and participation in autophagy. To our knowledge, this represents the first example of phosphorylation-based regulation of this important autophagy effector, whose recruitment represents the convergence point for several pathways of autophagy induction (He and Klionsky, 2009). As autophagy has been implicated in a growing number of human diseases, it has become clear that a fine balance of autophagy underlies healthy mammalian physiology. Phosphorylation-based regulation of autophagy has thus far centered on the nutrient-sensing target of rapamycin–Atg1 regulatory axis, whereas loss of inhibitory mechanisms in the phosphoinositide 3-kinase–beclin 1 pathway have been implicated in hereditary myopathies and pathological cell death (Zhu et al., 2007; Scarlatti et al., 2008; Vergne et al., 2009). This study implicates components of the ubiquitin-like lipidation system itself as a third potential point of autophagy down-regulation capable of suppressing autophagy induced by metabolic or pathological stimuli. Other autophagy effector proteins may also be subject to phosphoregulation, linking autophagic responses to cellular injury and stress signals.
In yeast, recent studies indicate a nutrient-sensing role for PKA as a parallel pathway to target of rapamycin in suppressing autophagy, with phosphorylation of the kinase Atg1 as a convergence point (Yorimitsu et al., 2007; Stephan et al., 2009). There was no effect on Atg1 kinase activity, but phosphorylation inhibited localization to the preautophagosomal structure (Budovskaya et al., 2005). Although a corresponding preautophagosomal structure has not been identified in mammals, phosphorylation of LC3 reduces its recruitment to rapamycin or MPP+-induced autophagosomes.
Interestingly, yeast and Drosophila melanogaster Atg8 lack this particular PKA consensus site, although it is conserved in all mammalian forms of LC3 (Fig. S3) but not in the other mammalian Atg8 homologues, GABARAP and GATE16. The crystal structures of LC3, GABARAP, and GATE16 demonstrate significant charge differences in the N-terminal helices, which underlie the differences in their biological functions (Sugawara et al., 2004). Phosphorylation-related decreases in the net-positive charge of the LC3 N-terminal region may affect its interactions with phospholipids, proteins involved in target recognition, lipidation (Yamada et al., 2007), cytoskeleton, or other functions (Kouno et al., 2005; Wang et al., 2006).
Although PKA is a major nutrient-sensing pathway in yeast, it plays additional important roles in the mammalian nervous system. Among these is the ability of PKA to promote neuronal differentiation and neurite outgrowth, which has been predominantly studied in terms of transcriptional regulation. However, recent studies indicate that autophagy plays a direct role in mediating neurite retraction in neurons subjected to trophic factor withdrawal, physical axotomy, or toxicity because of chemical or genetic factors (Yang et al., 2007; Plowey et al., 2008). As neurite extension/retraction likely reflects a balance of biosynthetic and degradative processes, the ability of cytoplasmically expressed PKA catalytic subunit to phosphorylate LC3 and suppress autophagy-dependent neurite retraction represents a novel mechanism contributing to effects of PKA in maintaining neuronal function.
These data implicate LC3 phosphorylation as a novel switch that modulates its biological function in mammalian cells. Both neuronal and nonneuronal cells exhibit endogenous levels of phosphorylated LC3. These observations support the concept of a reserve pool of phosphorylated LC3 that can be rapidly recruited for autophagy in response to external stimuli such as nutrient deprivation or mitochondrial injury. Dephosphorylation of LC3 would allow cells to rapidly switch from basal to induced autophagy, allowing the compensatory expansion of the autophagic compartment that is observed after injury or stress in mammalian cells (Yue et al., 2009).
Materials and methods
Cell culture
293T, HeLa, and SH-SY5Y cells (American Type Culture Collection) were maintained in DME with 10% FBS (BioWhittaker) and 2 mM l-glutamine (BioWhittaker). SH-SY5Y cells were differentiated with retinoic acid 3 d before transfection as previously described (Plowey et al., 2008). Primary cortical neurons were maintained in Neurobasal (Invitrogen) supplemented with 2% B27 (Invitrogen) and 2 mM l-glutamine and transfected on 7 d in vitro. In some experiments, cells were treated with 250 µM db-cAMP (Sigma-Aldrich), 10 µM forskolin (Sigma-Aldrich), 10 nM FK506 (Enzo Life Sciences, Inc.), 10 µM H89 (EMD), 1 mM MPP+ (1-methyl-4-phenylpyridinium; Sigma-Aldrich), 250 nM okadaic acid (EMD), 2 µM rapamycin (LC Laboratories), or 10 µM rolipram (Sigma-Aldrich). Lipofectamine 2000 (Invitrogen) was used to transfect the following: pcDNA 3.1(+), pcDNA-HA-LRRK2-G2019S, pEGFP-C1, GFP-PKA, GFP-PKA-NLS, GFP-PKA–nuclear export signal, pEGFP-C1-LC3, GFP-RFP-LC3, and HA-LC3. Targeted PKA constructs were provided by S. Green (University of Iowa, Iowa City, IA). pEGFP-C1-LC3 and GFP-RFP-LC3 were provided by T. Yoshimori (Osaka University, Osaka, Japan). Site-directed mutagenesis of pEGFP-C1-LC3 was performed using QuickChange Mutagenesis II kit (Agilent Technologies).
Immunofluorescence and immunoblotting
Cells were fixed with 4% paraformaldehyde in PBS. Cells were permeabilized in PBS with 0.1% Triton X-100 and blocked in 2% BSA in PBS. Cells were incubated with antibodies against GFP (Invitrogen), LC3 (Abgent), and/or HA tag (Covance) followed by the appropriate Alexa Fluor 488– or 546–conjugated secondary antibodies (Invitrogen). Cells were imaged in PBS at 25°C using a microscope (IX71; Olympus) with 20× (air) or 40× (oil) objectives with numerical apertures of 0.45 or 1.30, respectively. Microsuite Five imaging software (Olympus) was used to capture images with a camera (DP70; Olympus), and data were analyzed with ImageJ (National Institutes of Health) as previously described (Chu et al., 2009). Representative images display the green channel, which was extracted as a grayscale image using ImageJ. GFP-LC3–labeled AVs (puncta) were defined as bright dots >1.5 SD above the mean cytosolic fluorescence. For mouse cortical neurons, montages were constructed to capture all projecting neurites, and the total neurite length was calculated as the summation of all processes projecting from the cell body and all subsequent branches. For SH-SY5Y cells, which primarily have one major neurite projecting from the cell body, the longest neurite from the cell body was measured. The means of both these parameters are presented in graphical format.
Immunoblots were performed as previously described (Plowey et al., 2008) with antibodies against the N terminus of LC3 (Nanotools), the C terminus of LC3 (Abgent), GAPDH (Abcam), lamin A/C (Cell Signaling Technology), or actin (Sigma-Aldrich). Phospho-specific polyclonal antibodies were produced by injecting keyhole limpet hemacyanin–conjugated phosphopeptide into New Zealand rabbits (Abgent). Peptide competition was performed to verify specificity of the phospho-specific antisera (Fig. S2 c). For 2D immunoblots, isoelectric focusing was performed using Immobiline DryStrips (GE Healthcare), pH 3–10, followed by SDS-PAGE.
Recombinant LC3 purification, in vitro kinase assay, and LC3 metabolic labeling
Recombinant GST-LC3 (pGEX-6P-1-LC3; provided by M. Shibata and Y. Uchiyama, Osaka University) was induced using IPTG in DH5α bacteria and purified on an immobilized glutathione column (Thermo Fisher Scientific) according to the manufacturer’s specifications. LC3 was cleaved from the GST using PreScission Protease (GE Healthcare) and incubated with recombinant PKA catalytic subunit (New England Biolabs, Inc.) for 2 h at 30°C in 50 mM Tris-HCl and 10 mM MgCl2 with 200 µM ATP. Phosphorylation was detected by autoradiography.
For detection of LC3 phosphorylation in cells, 293T cells were transfected with HA-LC3 followed by treatment with DMSO, forskolin, or forskolin and H89 in the presence of 1 mCi/ml 32P as H332PO4. All treatments were in the presence of rolipram, and for the final hour of incubation, okadaic acid and FK506. Cells were lysed in PBS with 1% Triton X-100 (PBST) and incubated with anti-HA antibody and agarose beads overnight for immunoprecipitation. After washing with PBST, proteins were eluted in sample buffer and separated by SDS-PAGE. Gels were silver stained, and HA-LC3 bands were excised. 32P incorporation was measured as counts/minute (LS3801; Beckman Coulter) and normalized to band intensity.
Identification of LC3 phosphorylation site by MS
Gel bands were excised and destained, disulfides were reduced with TCEP (Tris(2-carboxyethyl)phosphine hydrochloride)), and sulfhydryls were alkylated with iodoacetamide as previously described (Shevchenko et al., 2006). Gel plugs were digested overnight with 8 ng/µl chymotrypsin (Roche) at room temperature. Peptides were extracted with 1:2 (vol/vol) 5% formic acid/acetonitrile. Samples were desalted using a ZipTip (C18; Millipore) and spotted for mass spectrometric analysis on a MALDI-TOF/TOF analyzer (4800; Applied Biosystems). Peptide phosphorylation was detected as a gain of 80 D by MALDI-TOF-MS. Amino acid sequences were identified by subjecting peptides to collision-induced dissociation during MALDI-TOF-MS/MS.
Statistical analysis
All graphical data have been compiled from multiple independent experiments and are presented as mean ± SEM. Student’s t test was used to compare means between two groups. One-way analysis of variance was used for multiple groups followed by posthoc t test with Bonferroni corrections for multiple comparisons.
Online supplemental material
Fig. S1 shows that autophagy mediates neurite injury induced by LRRK2 G2019S or MPP+, PKA inhibition increases basal autophagy, and cAMP opposes rapamycin-induced early AVs. Fig. S2 shows phospho-LC3 site identification and phosphoantibody characterization and bafilomycin effects in rapamycin and MPP+-treated cells. Fig. S3 shows PKA consensus site comparison among LC3 homologues. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002108/DC1.
Acknowledgments
We thank the scientists listed in Materials and methods for providing reagents.
This work was supported in part by the National Institutes of Health (grants AG026389, DC009120, NS064728, NS065789, and RR024153). C.T. Chu is the recipient of an American Federation for Aging Research/Ellison Medical Foundation Julie Martin Mid-Career Award in Aging Research. J. Mountzouris is a former employee of Abgent, Inc.
Footnotes
Abbreviations used in this paper:
- AV
- autophagic vacuole
- db-cAMP
- dibutyryl cAMP
- MALDI
- matrix-assisted laser desorption/ionization
- MS
- mass spectrometry
- PD
- Parkinson disease
- TOF
- time of flight
- WT
- wild type
References
- Bok J., Zha X.M., Cho Y.S., Green S.H. 2003. An extranuclear locus of cAMP-dependent protein kinase action is necessary and sufficient for promotion of spiral ganglion neuronal survival by cAMP. J. Neurosci. 23:777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B., Nixon R.A. 2006. Neuronal macroautophagy: from development to degeneration. Mol. Aspects Med. 27:503–519 10.1016/j.mam.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Budovskaya Y.V., Stephan J.S., Reggiori F., Klionsky D.J., Herman P.K. 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279:20663–20671 10.1074/jbc.M400272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y.V., Stephan J.S., Deminoff S.J., Herman P.K. 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 102:13933–13938 10.1073/pnas.0501046102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalovich E.M., Zhu J.H., Caltagarone J., Bowser R., Chu C.T. 2006. Functional repression of cAMP response element in 6-hydroxydopamine-treated neuronal cells. J. Biol. Chem. 281:17870–17881 10.1074/jbc.M602632200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.T., Plowey E.D., Dagda R.K., Hickey R.W., Cherra S.J., III, Clark R.S. 2009. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 453:217–249 10.1016/S0076-6879(08)04011-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M. 2004. Autophagy: in sickness and in health. Trends Cell Biol. 14:70–77 10.1016/j.tcb.2003.12.002 [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D.J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen I., Gordon P.B., Strømhaug P.E., Seglen P.O. 1996. Role of cAMP in the regulation of hepatocytic autophagy. Eur. J. Biochem. 236:163–170 10.1111/j.1432-1033.1996.00163.x [DOI] [PubMed] [Google Scholar]
- Jaeger P.A., Wyss-Coray T. 2009. All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Mol. Neurodegener. 4:16 10.1186/1750-1326-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouno T., Mizuguchi M., Tanida I., Ueno T., Kanematsu T., Mori Y., Shinoda H., Hirata M., Kominami E., Kawano K. 2005. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 280:24610–24617 10.1074/jbc.M413565200 [DOI] [PubMed] [Google Scholar]
- MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. 2006. The familial parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 52:587–593 10.1016/j.neuron.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Plowey E.D., Cherra S.J., III, Liu Y.J., Chu C.T. 2008. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 105:1048–1056 10.1111/j.1471-4159.2008.05217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. 2008. Role of non-canonical beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15:1318–1329 10.1038/cdd.2008.51 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Stephan J.S., Yeh Y.Y., Ramachandran V., Deminoff S.J., Herman P.K. 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA. 106:17049–17054 10.1073/pnas.0903316106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. 2004. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 9:611–618 10.1111/j.1356-9597.2004.00750.x [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., Deretic V. 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 28:2244–2258 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.J., Ding Y., Kohtz D.S., Kohtz S., Mizushima N., Cristea I.M., Rout M.P., Chait B.T., Zhong Y., Heintz N., Yue Z. 2006. Induction of autophagy in axonal dystrophy and degeneration. J. Neurosci. 26:8057–8068 10.1523/JNEUROSCI.2261-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Suzuki N.N., Hanada T., Ichimura Y., Kumeta H., Fujioka Y., Ohsumi Y., Inagaki F. 2007. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282:8036–8043 10.1074/jbc.M611473200 [DOI] [PubMed] [Google Scholar]
- Yang Y., Fukui K., Koike T., Zheng X. 2007. Induction of autophagy in neurite degeneration of mouse superior cervical ganglion neurons. Eur. J. Neurosci. 26:2979–2988 10.1111/j.1460-9568.2007.05914.x [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Zaman S., Broach J.R., Klionsky D.J. 2007. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:4180–4189 10.1091/mbc.E07-05-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Friedman L., Komatsu M., Tanaka K. 2009. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta. 1793:1496–1507 10.1016/j.bbamcr.2009.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A.D., Xie X., Ma D., Yuan J. 2007. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. USA. 104:19023–19028 10.1073/pnas.0709695104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C.T. 2007. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 170:75–86 10.2353/ajpath.2007.060524 [DOI] [PMC free article] [PubMed] [Google Scholar]