Cyclin A outcompetes inhibitory spindle assembly checkpoint proteins for binding to the APC/C ubiquitin ligase coactivator Cdc20 to promote its self-destruction even when the checkpoint is active (see also a paper from van Zon et al., in this issue).
Abstract
The anaphase-promoting complex/cyclosome (APC/C) is the ubiquitin ligase essential to mitosis, which ensures that specific proteins are degraded at specific times to control the order of mitotic events. The APC/C coactivator, Cdc20, is targeted by the spindle assembly checkpoint (SAC) to restrict APC/C activity until metaphase, yet early substrates, such as cyclin A, are degraded in the presence of the active checkpoint. Cdc20 and the cyclin-dependent kinase cofactor, Cks, are required for cyclin A destruction, but how they enable checkpoint-resistant destruction has not been elucidated. In this study, we answer this problem: we show that the N terminus of cyclin A binds directly to Cdc20 and with sufficient affinity that it can outcompete the SAC proteins. Subsequently, the Cks protein is necessary and sufficient to promote cyclin A degradation in the presence of an active checkpoint by binding cyclin A–Cdc20 to the APC/C.
Introduction
Ubiquitin-mediated proteolysis is fundamental to the proper control of mitosis. The anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase ensures the correct ordering of events by targeting specific proteins at specific times (Pines, 2006). The APC/C responds to the spindle assembly checkpoint (SAC) such that it targets securin and cyclin B1 for destruction only when all of the chromosomes have attached to the mitotic spindle, thereby ensuring that sister chromatids segregate to opposite poles and that cells cannot exit mitosis until sister chromatids have separated. The SAC prevents the APC/C from ubiquitylating securin and cyclin B1 by targeting its coactivator, Cdc20 (Musacchio and Salmon, 2007). However, Cdc20 is not completely inactivated because it can still work with the APC/C to ubiquitylate substrates such as cyclin A, Nek2A, and HOXC10 (den Elzen and Pines, 2001; Geley et al., 2001; Hames et al., 2001; Gabellini et al., 2003; Wolthuis et al., 2008). How APC/CCdc20 is able to recognize some substrates independently of the SAC but not others is crucial to our understanding of how the APC/C and SAC cooperate to ensure genomic stability.
One important insight is that the N terminus of Cdc20 is able to activate the APC/C in Xenopus laevis egg extracts (Kimata et al., 2008) to recognize the Nek2A protein that binds directly to the APC/C (Hayes et al., 2006). An analogous mechanism may underlie the degradation of cyclin A, which requires the Cks protein to be degraded (Swan et al., 2005; Wolthuis et al., 2008). Cks proteins contain an anion-binding pocket implicated in binding to phosphoproteins (Bourne et al., 1996), including the APC/C (Sudakin et al., 1997), whose phosphorylation by cyclin B1–Cdk at mitosis is crucial to its activity (Kraft et al., 2003). Because some cyclin A bound Cdc20 before mitosis, we proposed that its associated Cks subunit might direct the Cdc20–cyclin A complex to the newly phosphorylated APC/C, thereby triggering cyclin A destruction at mitotic entry (Wolthuis et al., 2008), but at that time, we did not have any experimental proof for this model.
Our model also raised two important questions. First, if the Cks subunit does target Cdc20–cyclin A–Cdk to the APC/C, is this sufficient for destruction, or does the Cdk contribute? Cyclin A cannot be degraded in prometaphase if it can’t bind its Cdk or if the Cdk can’t bind Cks (Stewart et al., 1994; den Elzen and Pines, 2001; Geley et al., 2001; Wolthuis et al., 2008), but whether the requirement to bind a Cdk is solely to recruit Cks is not known. The second question is that because only a fraction of cyclin A is bound to Cdc20 when cells enter mitosis, how is the rest of cyclin A degraded while the SAC is active?
The simplest hypothesis is that cyclin A can compete with the SAC proteins to bind Cdc20 and then be recruited to the APC/C via Cks. In this study, we provide strong support for this hypothesis by showing that cyclin A does not have to bind to Cdc20 before mitosis to be degraded because the N terminus of cyclin A binds directly to Cdc20 and can outcompete the SAC complex for binding to Cdc20. Furthermore, we show that only Cks, and not the Cdk subunit, is required for proteolysis because fusing the N terminus of cyclin A to a Cks protein is sufficient to target it to the APC/C and confer destruction in an SAC-resistant manner.
Results and discussion
Cdc20 binds directly to the N terminus of cyclin A
Previously, we showed that cyclin A bound to Cdc20 in G2 phase and that both Cdc20 and Cks were required for its degradation (Wolthuis et al., 2008). However, we did not determine the mechanism by which cyclin A could be degraded in an SAC-resistant manner. To address this question, we first sought to identify the regions of cyclin A required to bind to Cdc20.
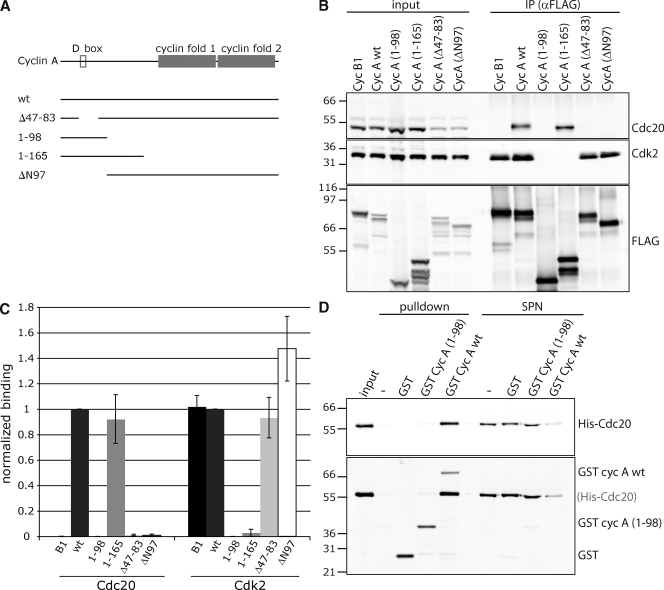
An extended sequence in the N terminus of cyclin A is required to target cyclin A for degradation in prometaphase (den Elzen and Pines, 2001; Geley et al., 2001; Jacobs et al., 2001). We generated several cyclin A deletion mutants (Fig. 1 A) tagged at the C terminus with the Flag epitope and a fluorescent protein (Venus; Nagai et al., 2002). As a control, we used cyclin B1, which is degraded in an SAC-sensitive manner (Clute and Pines, 1999). To ensure that the proteins were expressed at close to endogenous levels, we generated stable inducible HeLa cell lines in which a single copy of the plasmid was integrated into the genome at a flippase recognition target (FRT) site. The proteins were immunoprecipitated from prometaphase cells using an anti-Flag antibody and probed for Cdc20 and Cdk2. As expected, the N-terminal fragments lacking the cyclin box could not bind Cdk2 (Fig. 1, B and C).
Figure 1.
The N terminus of cyclin A binds Cdc20. (A) Schematic representation of cyclin A mutants. (B) Stable inducible HeLa FRT cell lines expressing cyclin B1, and the cyclin A mutants were synchronized in mitosis by a single thymidine block and released in the presence of nocodazole. To prevent cyclin A degradation, MG132 was added 2 h before collecting mitotic cells by shake off. Cells were lysed, and anti-Flag immunoprecipitates (IP) were probed for Cdc20, Cdk2, and Flag. (C) Cdc20 and Cdk2 levels in the anti-Flag immunoprecipitates (B) were quantified using an Odyssey scanner, corrected for the level of Flag-tagged protein, and normalized to the amount bound to wt cyclin A. Error bars indicate mean ± SEM from three experiments. (D) GST–cyclin A was purified from bacteria and incubated for 2 h at 4°C with His6-Cdc20 purified from insect cells. Cyclin A was isolated on glutathione beads, and the beads and supernatants (SPN) analyzed by immunoblots were probed for Cdc20 and GST. Values are representative of seven experiments. Molecular mass markers are shown on the left (kilodaltons).
Consistent with our previous findings (Wolthuis et al., 2008), cyclin A, but not cyclin B1, strongly bound to Cdc20, but neither the N-terminal fragment (1–98) containing the extended motif required for cyclin A degradation nor the complementary mutant ΔN97 were able to bind Cdc20 (Fig. 1, B and C), indicating that a more complex region was required. A longer N-terminal fragment, 1–165, did bind Cdc20 to a similar extent as full-length cyclin A (Fig. 1, B and C), which is in agreement with results from a yeast two-hybrid analysis (Ohtoshi et al., 2000). Moreover, cyclin A and Cdc20 bound directly to each other (Fig. 1 D).
Complementing these data, an internal deletion that removed the extended D-box motif (Δ47–83) abrogated Cdc20 binding (Fig. 1, B and C) when expressed at similar levels to endogenous cyclin A. We conclude that the extended D-box motif contributes to Cdc20 binding, but is not itself sufficient, and that another region between residues 98 and 165 is required.
Cyclin A competes with BubR1 for Cdc20 binding
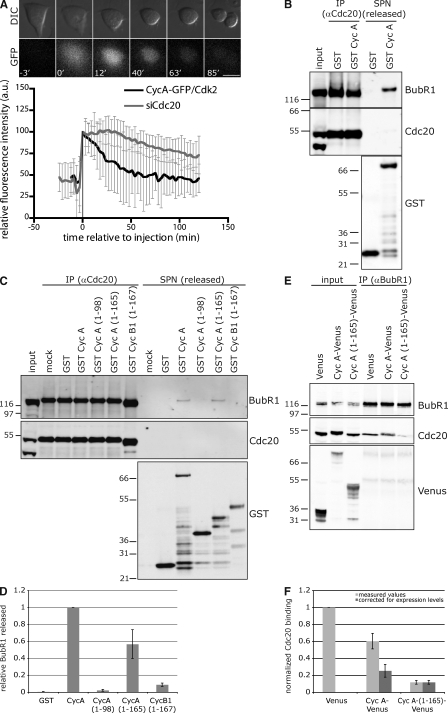
We previously showed that some, but by no means all, cyclin A binds to Cdc20 in G2 phase (Wolthuis et al., 2008), raising the question of what happened to cyclin A that was not bound to Cdc20. To answer this, we injected purified cyclin A–GFP–Cdk2 lacking Cdc20 into prometaphase cells and found that this was degraded with normal kinetics in a Cdc20-dependent manner (Fig. 2 A). Thus, cyclin A does not have to bind to Cdc20 before mitosis to be targeted for degradation in an SAC-resistant manner. This supports our hypothesis that cyclin A binds to Cdc20 even while the checkpoint is active. The first step in the SAC pathway is that Mad2 binds to Cdc20 (Musacchio and Salmon, 2007); therefore, the simplest mechanism would be that cyclin A and Mad2 bind to overlapping regions of Cdc20. But we find that cyclin A binds to the WD40 repeat at the C terminus of Cdc20 (unpublished data), whereas the Mad2-binding region maps to the N terminus (aa 124–137; Yu, 2007).
Figure 2.
Cyclin A competes with BubR1 for binding to Cdc20. (A) The cyclin A–GFP–Cdk2 complex was purified from baculovirus-infected insect cells and injected into prometaphase HeLa cells identified by DIC (top). Time is relative to time of injection (min). Bar, 10 µm. GFP fluorescence through mitosis was measured (bottom) for normal (black curve) and Cdc20-depleted (gray curve) cells and set to 100 at the time of injection. Error bars indicate mean ± SD of 22 cells from two experiments (control) and 26 cells from two experiments (siCdc20). (B) Cdc20 bound in the SAC complex was immunoprecipitated (IP) with anti-Cdc20 antibodies from nocodazole-arrested cells and incubated with GST alone or GST–cyclin A for 2 h at 4°C. The supernatant (SPN) was removed and analyzed by immunoblotting for BubR1, Cdc20, and GST. Black line indicates that intervening lanes have been spliced out. (C) Cdc20 immunoprecipitates (obtained as in B) were incubated with buffer (mock), GST, the indicated GST–cyclin A mutants, or GST–cyclin B1 (1–167) and analyzed as in B. (D) The amount of BubR1 released from the immunocomplex (C) was quantified using an Odyssey scanner, corrected for the amount of Cdc20 immunoprecipitated and normalized to that released by wt cyclin A. Error bars indicate mean ± SEM from at least five experiments. (E) Stable cell lines expressing either Venus alone, cyclin A–Venus, or cyclin A (1–165)–Venus were treated with nocodazole, and MG132 was added 2 h before lysis. Anti-BubR1 immunoprecipitates were probed for BubR1, Cdc20, and Venus. (F) The amount of Cdc20 associated with immunoprecipitated BubR1 (E) was quantified as in D and corrected for the amount of immunoprecipitated BubR1 (light gray). To compare the wt and 1–165 samples, the values were corrected for expression levels, setting the 1–165-Venus level to 1 (dark gray). Error bars indicate mean ± SEM from four experiments. (B, C, and E) Input represents 1/10 of the immunoprecipitation. Molecular mass markers are shown on the left (kilodaltons).
Because cyclin A and Mad2 did not compete for the same binding site on Cdc20, we tested the possibility that cyclin A could compete with the SAC complex for Cdc20 (note that by competition, we include mechanisms by which cyclin A could preferentially bind Cdc20 by inducing an allosteric change). Because we find only a minor fraction of Mad2 bound to Cdc20 (Nilsson et al., 2008), we tested whether cyclin A could displace BubR1. We immunoprecipitated Cdc20 from nocodazole-treated cells, added purified GST fusion proteins, and assayed their effect on the integrity of the Cdc20–SAC complex. Consistent with our competition hypothesis, excess cyclin A displaced BubR1 from Cdc20 (Fig. 2 B), and this depended on its ability to interact with Cdc20; full-length cyclin A and the 1–165 fragment were both able to displace BubR1, whereas neither the 1–98 fragment of cyclin A nor the N terminus of cyclin B1 containing its D box (1–167) could do so (Fig. 2, C and D). We added a large excess (10–50-fold over Cdc20-coimmunoprecipitated BubR1) of the proteins in these in vitro reactions, but it is reasonable to suppose that the in vitro conditions do not recapitulate those in vivo and that competition in vivo may be more efficient.
To test whether cyclin A could compete with BubR1 for Cdc20 in vivo, we overexpressed cyclin A or cyclin A 1–165 ∼4–12-fold over endogenous and found that both greatly reduced the level of Cdc20 in BubR1 immunoprecipitates (Fig. 2, E and F). Thus, we conclude that both in vitro and in vivo, cyclin A is able to compete with the SAC proteins for binding to Cdc20, even when Cdc20 is already bound to the SAC complex.
Binding to Cdc20 is necessary but not sufficient for the proper timing of cyclin A degradation
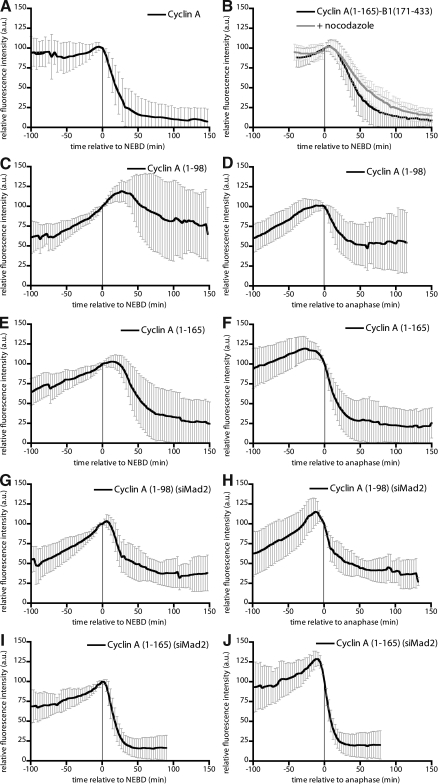
Having identified a region of cyclin A sufficient to bind to Cdc20, regardless of whether Cdc20 was free or bound in an SAC complex, we asked whether this was sufficient to target a protein for destruction in prometaphase. We used Venus-tagged proteins to assay the precise timing of destruction in mitosis and compared full-length cyclin A, which is degraded as soon as cells begin prometaphase (Fig. 3 A; den Elzen and Pines, 2001; Geley et al., 2001; Wolthuis et al., 2008), with both the N-terminal fragments and a chimeric protein replacing the N terminus of cyclin B1 with that of cyclin A.
Figure 3.
Binding to Cdc20 only confers SAC-dependent destruction. (A) HeLa cells were injected in G2 phase with cyclin A–Venus-Flag, and the fluorescence was measured through mitosis. Fluorescence at NEBD is set to 100. Error bars indicate mean ± SD of 36 cells. Time is relative to NEBD. (B) Mean degradation curve of a chimera between the N terminus of cyclin A (1–165) and C terminus of cyclin B1 (171–433) obtained as in A in the absence (black curve) or presence (gray curve) of nocodazole. Error bars indicate mean ± SD of 49 cells from three experiments (untreated) and 27 cells from two experiments (nocodazole). (C–F) Mean degradation curve of cyclin A (1–98)–Venus-Flag (C and D) and cyclin A (1–165)–Venus-Flag (E and F) obtained as in A. Time is relative to NEBD (C and E) or anaphase (D and F), and fluorescence at NEBD or anaphase is set to 100. Error bars indicate mean ± SD of 12 cells from three experiments (C and D) and 36 cells from three experiments (E and F). (G–J) Mean degradation curve of cyclin A (1–98)–Venus-Flag (G and H) and cyclin A (1–165)–Venus-Flag (I and J) in Mad2-depleted cells were obtained as in A except that the Mad2 siRNA oligonucleotides were transfected during synchronization. Because inactivating the SAC accelerates progression from NEBD to anaphase (not depicted; Meraldi et al., 2004), we only considered those cells in which NEBD to anaphase was shortened from the 40 min in control cells to <20 min. Time is relative to NEBD (G and I) or anaphase (H and J). Error bars indicate mean ± SD of 10 cells (G and H) and 13 cells from three experiments (I and J).
In support of our hypothesis, the N terminus of cyclin A conferred the ability to be degraded in an SAC-resistant manner on cyclin B1 (Fig. 3 B), whereas the cyclin A 1–98 fragment that could not bind Cdc20 was not degraded until anaphase (Fig. 3, C and D; note that data are plotted twice, once normalized to nuclear envelope breakdown [NEBD] and once to anaphase). In contrast, the 1–165 fragment that bound Cdc20 was degraded earlier, but notably, this was in metaphase not prometaphase (Fig. 3, E and F). Because metaphase equates to when the SAC has been satisfied, we tested whether this destruction was under the control of the SAC. We depleted Mad2 to inactivate the SAC and found this advanced cyclin A (1–165) degradation to begin just after NEBD (Fig. 3, I and J), which is consistent with regulation by the SAC. In contrast, degradation of the 1–98 fragment was still delayed until anaphase (Fig. 3, G and H).
We conclude that although the 1–165 fragment can compete with BubR1 for Cdc20, it can only be degraded when the SAC is satisfied. Therefore, binding directly to Cdc20 is not in itself sufficient to confer SAC-resistant proteolysis. Thus, additional factors must contribute to the SAC-resistant degradation of cyclin A.
Cks1 is essential to mediate checkpoint-resistant degradation
To identify the missing component that confers SAC-resistant destruction, we focused first on the Cks protein because full-length cyclin A must bind to its Cdk partner and consequently to Cks to be degraded (Wolthuis et al., 2008). Moreover, the N terminus of cyclin A was sufficient to confer SAC-resistant degradation on cyclin B1 (Fig. 3 B) that binds Cks via its Cdk1 partner. However, it was unclear whether the Cdk subunit itself contributed to the destruction of cyclin A.
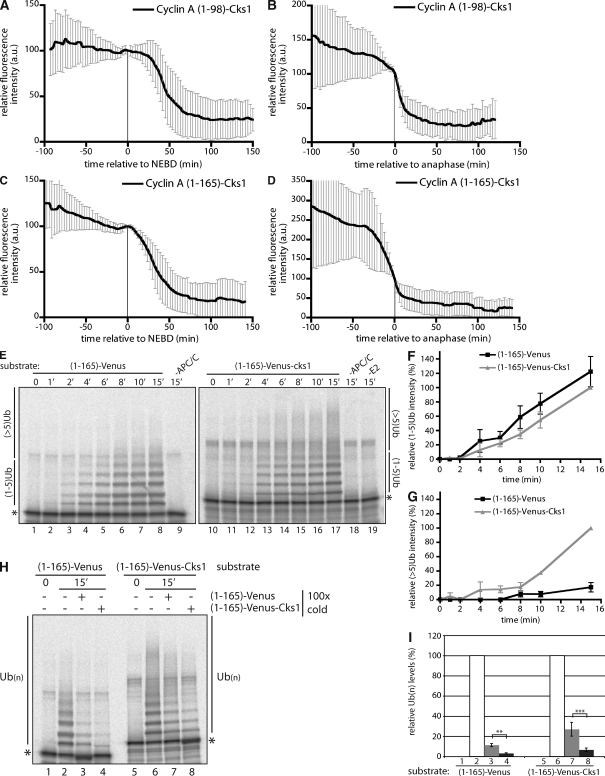
Because we hypothesized that the role of Cks was to bind the APC/C, we predicted that we could bypass the Cdk and target the cyclin A 1–165 fragment for degradation even in the presence of an active SAC by fusing it to a Cks protein. In confirmation of this, a cyclin A (1–165)–Venus-Cks1 fusion protein was degraded just after NEBD (Fig. 4, C and D) with the same timing as wild-type (wt) cyclin A. Furthermore, fusing Cks1 to cyclin A (1–98) that could not bind Cdc20 had no effect; it was still degraded in anaphase (Fig. 4, A and B). Similarly, fusing Cks1 to the N terminus of cyclin B1 that cannot bind Cdc20 when the SAC is active did not change the timing of its destruction to prometaphase, although it made it a more efficient substrate (unpublished data; see van Zon et al. in this issue).
Figure 4.
Cyclin A–associated Cks1 increases the efficiency of cyclin A ubiquitylation. (A–D) Cells were injected in G2 phase with cyclin A (1–98)–Venus-Cks1 (A and B) or cyclin A (1–165)–Venus-Cks1 (C and D), and the fluorescence was measured through mitosis. Time is relative to NEBD (A and C) or anaphase (B and D). Error bars indicate mean ± SD of 19 cells from two experiments (A and B) or 24 cells from two experiments (C and D). (E) In vitro ubiquitylation assay of cyclin A (1–165)–Venus (lanes 1–9) or cyclin A (1–165)–Venus-Cks1 (lanes 10–19). Reactions were performed for the indicated time (shown in minutes) before analysis by SDS-PAGE and phosphoimaging. Control reactions are without APC/C (lanes 9 and 18) or E2 (lane 19). (F and G) Quantification of ubiquitin conjugates in E with one to five ubiquitin (F) and more than five ubiquitin (G) molecules were normalized to the total amount of ubiquitylated substrate. (F and G) Error bars indicate mean ± SEM from four experiments. (H) Ubiquitylation reactions for cyclin A (1–165)–Venus (lanes 1–4) and cyclin A (1–165)–Venus-Cks1 (lanes 5–8). 100 times excess of unlabeled cyclin A (1–165)–Venus (lanes 3 and 7) or cyclin A (1–165)–Venus-Cks1 (lanes 4 and 8) was added as a competitor at the beginning of the reaction. (I) Quantification of reactions in H. Numbers correspond to lanes in H. *, unmodified substrate; **, P < 0.01; ***, P < 0.05 (calculated using Student’s t test).
These results demonstrate that Cks1 is sufficient to confer SAC-resistant degradation on a cyclin A–Cdc20 complex, bypassing any requirement for Cdk binding or activity (Fig. S1, A and B). Thus, the primary reason why cyclin A has to bind to its Cdk to be degraded is to be targeted to the APC/C via the Cks protein.
Fusion to Cks1 promotes cyclin A ubiquitylation
If Cks does target cyclin A to the APC/C, one might predict that it should enhance cyclin A ubiquitylation. Therefore, we compared the in vitro ubiquitylation of cyclin A (1–165)–Venus (Fig. 4 E, lanes 1–9) and cyclin A (1–165)–Venus-Cks1 proteins (Fig. 4 E, lanes 10–19). Cks increased the processivity of cyclin A ubiquitylation because although the amount of mono- and oligoubiquitin species (one to five ubiquitins) was similar for the two substrates (Fig. 4 F), the proportion of polyubiquitin chains was higher for the Cks1 fusion (Fig. 4 G). Moreover, cyclin A (1–165)–Cks1 was clearly a better competitor in in vitro APC/C-dependent ubiquitylation reactions than cyclin A (1–165; Fig. 4, H and I).
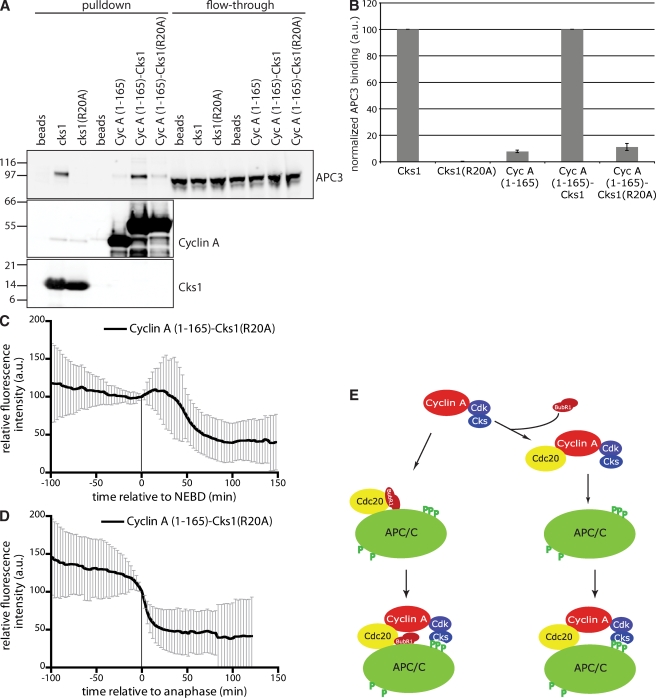
To test whether the Cks protein had to bind to the APC/C to promote cyclin A degradation in vivo, we mutated the anion-binding site of Cks1 (R20A; Watson et al., 1996). This mutant was severely disabled in its ability to bind to the APC/C, whereas binding to a Cdk was not affected (Fig. 5, A and B; and not depicted). When fused to cyclin A (1–165), Cks1 (R20A) was unable to confer SAC-resistant degradation; like the 1–165 fragment alone, the R20A fusion protein could only be degraded in metaphase once the SAC was satisfied (Fig. 5, C and D). Thus, the anion-binding site of Cks1 was essential to confer SAC-resistant degradation on a cyclin A–Cdc20 complex. This result strongly indicates that Cks1 mediates the recruitment of cyclin A to a phosphorylated APC/C subunit. Several APC/C subunits are phosphorylated during mitosis (Kraft et al., 2003; Steen et al., 2008), of which APC3 is most likely to bind Cks.
Figure 5.
Cks directly recruits cyclin A to the APC/C. (A) Purified His6-Cks1 and an anion-binding site mutant (R20A) and fusion proteins between cyclin A (1–165)–Venus and wt or (R20A) mutant Cks1 were immobilized on beads and incubated with extract from nocodazole-treated HeLa cells. Beads were analyzed by SDS-PAGE and immunoblotted for APC3, cyclin A, and Cks1. Molecular mass markers are shown on the left (kilodaltons). (B) APC3 binding to Cks1 (A) was quantified on an Odyssey scanner and corrected for the amount of Cks1. Values were normalized to wt Cks1. Error bars indicate mean ± SEM from three experiments. (C and D) Degradation of cyclin A (1–165)–Venus-Cks1 (R20A) measured as in Fig. 4 (A and B). Time is relative to NEBD (C) or anaphase (D). Error bars indicate mean ± SD of 28 cells from two experiments. (E) Working model. The cyclin A–Cdk–Cks complex binds to soluble or APC/C-associated Cdc20 by out competing the SAC complex. Recruitment to the APC/C is mediated by Cks interaction with phosphorylated APC/C.
We conclude that two main factors contribute to the SAC-resistant degradation of cyclin A. First, cyclin A can bind directly to Cdc20 with sufficient affinity to displace SAC proteins. Second, the Cks protein targets a cyclin A–Cdk complex to the APC/C in a manner that allows Cdc20 bound to cyclin A to activate the APC/C.
Currently, we do not know exactly how cyclin A competes with the SAC proteins for Cdc20, in part because there is some disagreement over how the SAC proteins themselves bind Cdc20. We and others find that the majority of Cdc20 binds to an SAC complex with BubR1 and Bub3 but only a small fraction of Mad2, whereas others find that Mad2 forms a stoichiometric part of the complex (the reason for this discrepancy is unclear but may be attributable to the methods used to lyse the cells and isolate the SAC complexes or the methods used to calculate the amounts of protein in the complex; Nilsson et al., 2008; Herzog et al., 2009; Kulukian et al., 2009). However, there is general agreement that Mad2 is required for Cdc20 to bind to BubR1 in vivo; therefore, if cyclin A blocks access to the Mad2-binding site on Cdc20, either sterically or by inducing a conformational change, this would prevent BubR1 from binding. Alternatively, as suggested by our in vitro assays, cyclin A and BubR1 themselves might compete for Cdc20.
Because both cyclin A and cyclin B1 require Cdc20 for their degradation, is the difference between them simply that cyclin A has a greater affinity for Cdc20, or might it bind to Cdc20 in a different manner that allows SAC-independent destruction? We think the latter idea is unlikely because the SAC inhibits destruction of cyclin A (1–165) that binds Cdc20. Thus, our interpretation is that cyclin A can be degraded by either or both of two pathways (Fig. 5 E). Cyclin A can either compete for Cdc20 before being targeted to the APC/C by Cks, or a cyclin A–Cdk–Cks complex could bind to an APC/C to which Cdc20 is already bound as part of an SAC complex, and, once bound, the N terminus of cyclin A could displace the SAC proteins from Cdc20, thereby activating the APC/C.
Materials and methods
Cell culture and synchronization
HeLa cells were cultured in advanced DME (Invitrogen) supplemented with 2% fetal bovine serum. Cells were synchronized by a thymidine/aphidicolin protocol as previously described (Di Fiore and Pines, 2007). Cells were blocked with 2.5 mM thymidine (Sigma-Aldrich) for 24 h, released for 12 h, and blocked again with 5 µg/ml aphidicolin (Sigma-Aldrich) for 24 h. Cells were released into fresh medium. For prometaphase arrest, 0.1 µg/ml nocodazole (Sigma-Aldrich) or 100 nM taxol (Sigma-Aldrich) was added during release from a single thymidine block, and 12 h later, cells were either harvested by mitotic shake off or 10 µM MG132 (EMD) was added, and cells collected by mitotic shake off 2 h later. The HeLa-FRT cell lines (provided by S. Taylor, University of Manchester, Manchester, England, UK) were transfected using the FLIP-in system (Invitrogen) to generate stable inducible cell lines. Cells were induced with 1 µg/ml tetracycline (EMD) 12 h before harvesting.
Plasmids
Cyclin A and its mutants were cloned into a modified version of pCDNA5/FRT/TO (Invitrogen) containing Venus-Flag tag using the Gateway system. Fusions between cyclin A fragments and Cks1 were constructed in modified versions of the pEYFP-N1 plasmid, resulting in the YFP/Venus sequence expressed between cyclin A and Cks1. For the cyclin A-B1 fusion protein, cyclin A N terminus (aa 1–165) was fused to the C terminus of cyclin B1 (aa 171–433) in a modified version of pEYFP-N1 plasmid. For protein purification, pGEX (GE Healthcare), a modified version of pET30a (EMD) containing a strep tag, and pRSET (Invitrogen) vectors were used.
Microinjection and time-lapse imaging and analysis
For microinjection and time-lapse microscopy, the culture medium was replaced with Leibovitz’s L-15 medium (Invitrogen). Plasmids or recombinant protein were microinjected into G2 or prometaphase cells, respectively, using a semiautomatic microinjector (Eppendorf) on a microscope (DMIRBE or DMIRE2; Leica). Differential interference contrast (DIC) and fluorescence images were captured every 3 min with a charge-coupled device camera (QuantEM 512B; Photometrics) using Slidebook software (Intelligent Imaging Innovations). A modified version of ImageJ software (National Institutes of Health) was used to quantify the fluorescence after background subtraction. DIC microscopy was used to monitor mitotic phases.
RNA interference
siRNA duplex against Mad2 (5′-GGAAGAGUCGGGACCACAGUU-3′; Thermo Fisher Scientific), Cdc20 (5′-CGGAAGACCTGCCGTTACA-3′; Thermo Fisher Scientific), and control siRNA duplex against GAPDH (Applied Biosystems) were transfected at a final concentration of 100 nM with Oligofectamine (Invitrogen) according to the manufacturer’s protocol. Transfection was performed during synchronization 5 h after release from the thymidine block. For Cdc20, an extra transfection was performed before starting the synchronization protocol.
Protein expression and pull-downs
Recombinant proteins were induced with 0.5 mM IPTG in BL21 (DE3) at 25°C for 5 h and purified on glutathione Sepharose 4B beads (GE Healthcare), Strep-Tactin matrix (IBA), or nickel beads (QIAGEN) according to the manufacturer’s protocols. His-tagged Cdc20 was purified from baculovirus-infected Sf9 cells as previously described (Nilsson et al., 2008). For GST pull-downs, GST proteins were immobilized on glutathione beads and incubated with purified Cdc20 in buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, and 1 mM DTT) with 10 µg BSA for 2 h at 4°C. For His6-Cks1 and cyclin A–Cks-strep–binding assays, purified proteins were coupled to nickel or Strep-Tactin beads and incubated in buffer (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, and 0.2% NP-40) for 2 h at 4°C with extracts from nocodazole-treated HeLa cells. For protein microinjection, cyclin A–GFP–His–Cdk2 complex was purified from baculovirus-infected insect cells using nickel beads, and the folding of the purified complex was assessed on a gel filtration column (Superdex 200; GE Healthcare).
Immunoprecipitation and competition experiments
Cells were lysed in 50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 0.5% NP-40 plus protease inhibitor cocktail (Roche), and 10 nM microcystin (LGC-Promotech) for 20 min on ice and clarified by a 12,000 g spin for 15 min at 4°C. Complexes were immunoprecipitated for 2 h at 4°C with anti-Flag (M2; Sigma-Aldrich) or anti-BubR1 (BD) antibody covalently coupled to protein G (Dynabeads; Invitrogen). After five washes in lysis buffer, proteins were eluted from beads by incubating for 5 min at 65°C in sample buffer. For the competition experiments, Cdc20 complexes were isolated using anti-Cdc20 (H-7; Santa Cruz Biotechnology, Inc.) antibody. After the washes, beads with associated complexes were divided into several tubes, and 1–2 µM purified GST fusion protein was added to the beads and incubated in buffer A for 2 h at 4°C. The supernatant containing the unbound GST proteins and proteins released from the immunoprecipitated complexes were analyzed by immunoblotting. Beads were washed three times with buffer A, and the complexes still associated eluted in sample buffer for 5 min at 65°C.
Immunoblotting
After electrophoresis on 4–12% gradient Bis-Tris acrylamide gels, proteins were transferred to a PVDF membrane. Membrane saturation and all of the following incubation steps were performed in 5% low fat milk in 0.1% PBS-Tween. Anti-Flag (M2; Sigma-Aldrich), anti-GST (B-14; Santa Cruz Biotechnology, Inc.), anti–cyclin A (Cancer Research UK), and anti-APC3 (BD) were used at 1:1,000. Anti-Cdc20 (Bethyl Laboratories), anti-BubR1 (Bethyl Laboratories and BD), and anti-Cdk2 (Koop, 2007) were used at 1:500. Anti-Cks1 (Invitrogen) was used at 1:150. Alexa Fluor 680 (Invitrogen)– and IRDye (800CW; LI-COR Biosources)-conjugated secondary antibodies were used at 1:5,000. The antibody signal was detected using the infrared imaging system (Odyssey; LI-COR) for quantitative immunoblotting.
APC/C purification and ubiquitylation assays
Taxol-treated HeLa cells were resuspended in 20 mM Hepes, pH 7.8, 175 mM NaCl, 2.5 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail (Roche), 2 µM ocadaic acid, 10 nM microcystin LR, and phosphatase inhibitor cocktail II (EMD) and ruptured using nitrogen cavitation. APC/C was immunoprecipitated from 10 mg of extract using anti-APC4 antibodies immobilized on protein G (Dynabeads; Invitrogen). Substrates were purified on Strep-Tactin beads (IBA) and radiolabeled with 33P by phosphorylation of a muscle kinase tag (HMK) using protein kinase A (Sigma-Aldrich). Ubiquitylation reactions were performed at 37°C in 15 µl of QPIP buffer (50 mM Pipes, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 10% glycerol, 1 mM DTT, and 1 mM EGTA) containing 5 µl APC4 beads, 300 nM E1, 2.6 µM UbcH10, 150 µM ubiquitin, 1 µM BSA, 100 nM CDC20, 100 nM radiolabeled substrate, 2 mM ATP, 2.3 µM creatine kinase, and 10 mM creatine phosphate as previously described (Garnett et al., 2009). For the competition with cold (unlabeled) substrate, a 100 times excess over the labeled substrate was added to each reaction at time 0. Reactions were stopped with SDS sample buffer, processed for SDS-PAGE, and analyzed on a phosphoimager (FLA-5000; Fujifilm).
Online supplemental material
Fig. S1 shows the strong reduction in Cdk2 binding of the cyclin A (1–165)–Venus-Cks1 protein compared with full-length cyclin A. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001083/DC1.
Acknowledgments
We thank Steven Taylor for the gift of the parental HeLa-FRT cell line. We are also grateful to all members of our laboratory for stimulating discussions and to Claire Acquaviva, who contributed to the early stage of the project.
This work was supported by a program grant from Cancer Research UK (to J. Pines) and by a core grant (to the Gurdon Institute) from Cancer Research UK and The Wellcome Trust.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DIC
- differential interference contrast
- FRT
- flippase recognition target
- NEBD
- nuclear envelope breakdown
- SAC
- spindle assembly checkpoint
- wt
- wild type
References
- Bourne Y., Watson M.H., Hickey M.J., Holmes W., Rocque W., Reed S.I., Tainer J.A. 1996. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 84:863–874 10.1016/S0092-8674(00)81065-X [DOI] [PubMed] [Google Scholar]
- Clute P., Pines J. 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1:82–87 10.1038/10049 [DOI] [PubMed] [Google Scholar]
- den Elzen N., Pines J. 2001. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 153:121–136 10.1083/jcb.153.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B., Pines J. 2007. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 177:425–437 10.1083/jcb.200611166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D., Colaluca I.N., Vodermaier H.C., Biamonti G., Giacca M., Falaschi A., Riva S., Peverali F.A. 2003. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 22:3715–3724 10.1093/emboj/cdg340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11:1363–1369 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S., Kramer E., Gieffers C., Gannon J., Peters J.-M., Hunt T. 2001. Anaphase-promoting complex/cyclosome–dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153:137–148 10.1083/jcb.153.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R.S., Wattam S.L., Yamano H., Bacchieri R., Fry A.M. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20:7117–7127 10.1093/emboj/20.24.7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M.J., Kimata Y., Wattam S.L., Lindon C., Mao G., Yamano H., Fry A.M. 2006. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat. Cell Biol. 8:607–614 10.1038/ncb1410 [DOI] [PubMed] [Google Scholar]
- Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J.-M. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 323:1477–1481 10.1126/science.1163300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H.W., Keidel E., Lehner C.F. 2001. A complex degradation signal in cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 20:2376–2386 10.1093/emboj/20.10.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Baxter J.E., Fry A.M., Yamano H. 2008. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell. 32:576–583 10.1016/j.molcel.2008.09.023 [DOI] [PubMed] [Google Scholar]
- Koop L. 2007. The role of Cyclin A in the entry to mitosis. PhD thesis. University of Cambridge, Cambridge, England, UK: 176 pp [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.M. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609 10.1093/emboj/cdg627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtoshi A., Maeda T., Higashi H., Ashizawa S., Hatakeyama M. 2000. Human p55(CDC)/Cdc20 associates with cyclin A and is phosphorylated by the cyclin A-Cdk2 complex. Biochem. Biophys. Res. Commun. 268:530–534 10.1006/bbrc.2000.2167 [DOI] [PubMed] [Google Scholar]
- Pines J. 2006. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 16:55–63 10.1016/j.tcb.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Steen J.A., Steen H., Georgi A., Parker K., Springer M., Kirchner M., Hamprecht F., Kirschner M.W. 2008. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proc. Natl. Acad. Sci. USA. 105:6069–6074 10.1073/pnas.0709807104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E., Kobayashi H., Harrison D., Hunt T. 1994. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34cdc2. EMBO J. 13:584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Shteinberg M., Ganoth D., Hershko J., Hershko A. 1997. Binding of activated cyclosome to p13(suc1). Use for affinity purification. J. Biol. Chem. 272:18051–18059 10.1074/jbc.272.29.18051 [DOI] [PubMed] [Google Scholar]
- Swan A., Barcelo G., Schüpbach T. 2005. Drosophila Cks30A interacts with Cdk1 to target cyclin A for destruction in the female germline. Development. 132:3669–3678 10.1242/dev.01940 [DOI] [PubMed] [Google Scholar]
- van Zon W., Ogink J., ter Riet B., Medema R.H., te Riele H., Wolthuis R.M.F. 2010. The APC/C recruits cyclin B1–Cdk1–Cks in prometaphase before D box recognition to control mitotic exit. J. Cell Biol. 190:587–602 10.1083/jcb.200912084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.H., Bourne Y., Arvai A.S., Hickey M.J., Santiago A., Bernstein S.L., Tainer J.A., Reed S.I. 1996. A mutation in the human cyclin-dependent kinase interacting protein, CksHs2, interferes with cyclin-dependent kinase binding and biological function, but preserves protein structure and assembly. J. Mol. Biol. 261:646–657 10.1006/jmbi.1996.0490 [DOI] [PubMed] [Google Scholar]
- Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. 2008. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell. 30:290–302 10.1016/j.molcel.2008.02.027 [DOI] [PubMed] [Google Scholar]
- Yu H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 27:3–16 10.1016/j.molcel.2007.06.009 [DOI] [PubMed] [Google Scholar]