Abstract
In this issue, a study by Groehler and Lannigan (2010. J. Cell Biol. doi:10.1083/jcb.201002124) sheds light on the regulation of proliferating cell nuclear antigen (PCNA) turnover and how it is counteracted by the small chromatin-bound kinase ERK8 (extracellular signal-regulated kinase 8). Importantly, inactivation of ERK8 results in genome instability and is associated with cell transformation.
Almost 30 yr ago, proliferating cell nuclear antigen (PCNA) was first identified in dividing cells using sera derived from patients suffering from systemic lupus erythematosus (Takasaki et al., 1981). A few years later, the “mother” of all cancer markers had been associated with DNA synthesis (Madsen and Celis, 1985), but it wasn’t until 1988 that Bauer and Burgers (1988) and Prelich and Stillman (1988) discovered that the homotrimeric clamp served as a processivity factor for DNA polymerases. In 1992, Shivji et al. (1992) showed that PCNA was required for DNA repair, and 10 yr later, it was identified as a target of ubiquitin and SUMO (small ubiquitin-like modifier) conjugation after exposure to ultraviolet light (Hoege et al., 2002). For a protein that has been in the spotlight of modern biochemistry, it is quite remarkable that almost nothing is known about its normal cellular turnover.
Insight into this process comes now from the study of an unlikely regulator. In this issue, Groehler and Lannigan (2010) demonstrate that the relatively poorly characterized ERK8 (extracellular signal-regulated kinase 8) takes center stage in the regulation of PCNA stability in primary mammary epithelial cells. The ERK family of kinases belongs to the mitogen-activated protein kinase superfamily and carries a Thr-Glu-Tyr (T-E-Y) activation motif that needs to be phosphorylated to enable kinase activity (Abe et al., 2002). Interestingly, ERK8 also needs to bind to chromatin to become active. The authors identified a highly conserved PXXXP motif in the C-terminal half of ERK8 that appeared to confer autoinhibition, an activity which is relieved upon chromatin binding. Relatively close by, in the middle of ERK8, resides a PCNA-interacting peptide (PIP) box required for the interaction with PCNA (Warbrick, 1998). Curiously, only the chromatin-bound fraction of ERK8 bound to the chromatin-bound fraction of PCNA. However, a functional PIP box was not required for ERK8 to associate with nuclear DNA in the cell. These results argue that ERK8 is not anchored to chromatin by PCNA but associates with it independently. Moreover, they strongly suggest that ERK8’s PIP box binds to PCNA only when the kinase is associated with chromatin. Importantly, overexpression of an ERK8 PIP box mutant resulted in destabilization of PCNA. The effect on PCNA stability seemed to be highly specific, as depletion of ERK8 caused codepletion of PCNA but did not lead to a decrease in steady-state levels of a variety of other cell cycle regulators.
Why is the interaction with PCNA confined to chromatin? The reason is likely due to the fact that ERK8’s PIP box is buried in the middle of the protein. Most PCNA-interacting proteins carry their PIP box either at the N or C terminus (Vivona and Kelman, 2003). One other well-studied example for a protein with an internal PIP box is the essential replication factor MCM10 (minichromosome maintenance protein 10). MCM10 undergoes cell cycle–regulated modification, which probably induces a conformational switch that is necessary for the PIP box–mediated interaction with PCNA (Das-Bradoo et al., 2006). In the same vein, it is conceivable that chromatin association and the accompanying relief of autoinhibition of ERK8 cause the middle portion of the kinase to change its configuration, thereby assuming a functional PIP box domain that can be recognized by PCNA. In situations in which the rapid unloading of PCNA is required, regulation of ERK8 may be the most effective way to dispose of chromatin-bound PCNA, which is known to have an exceedingly low exchange rate (Sporbert et al., 2002). Despite the fact that interaction with ERK8 is necessary to stabilize chromatin-bound PCNA, it remains unclear whether PCNA is a direct target of ERK8-mediated phosphorylation.
The next goal of Groehler and Lannigan (2010) was to dissect the mechanism underlying the ERK8-regulated degradation of PCNA. Based on the consideration that physical contact between the kinase and PCNA was an integral part of the protection, they hypothesized that ERK8 might compete with an E3 ubiquitin ligase that may target PCNA via its own PIP box. This turned out to be a smart guess because the only candidate to test was the E3 ligase HDM2, the human homologue of murine double minute 2 (Momand et al., 1992). In a set of well-controlled experiments, the authors not only demonstrate that HDM2 interacts directly with and degrades PCNA when ERK8 is absent, but they also exclude indirect effects by p53 and retinoblastoma (Rb) on this process. p53 is a direct target of HDM2 and is stabilized when their interaction is inhibited (Tao and Levine, 1999). Elevated levels of p53 trigger cell cycle arrest concomitant with hypophosphorylation of Rb, but none of these changes affect the stability of PCNA. It is not hard to imagine that the loss of chromatin-bound PCNA has severe consequences for the functionality of DNA replication and repair, resulting in chromosome breakage. The authors argued that a similar level of genome instability should be visible in ERK8-depleted cells. This was indeed the case as visualized by the accumulation of γ-H2AX foci and broken DNA (Rogakou et al., 1998). Importantly, Groehler and Lannigan (2010) observed similar effects in the ERK8 PIP box mutant, further lending credence to their model. It is worthwhile pointing out that the turnover of PCNA expands the spectrum of replication factors whose degradation is tightly linked to chromatin. CDT1, a member of the prereplication complex (Cook, 2009), is rapidly degraded in the face of DNA damage. Its degradation occurs exclusively on the chromatin-associated fraction of the protein pool and is dependent on CDT1 binding to PCNA (Arias and Walter, 2005; Hu and Xiong, 2006; Senga et al., 2006).
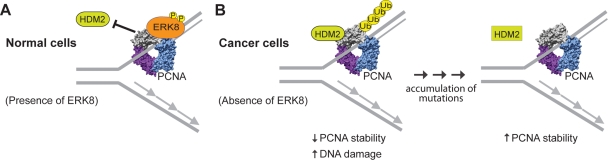
An important question that this study raises is of course to what extent, if at all, is PCNA turnover deregulated in cancer cells? The commonly high levels of PCNA in transformed cells would be most compatible with a deregulation of ERK8 and/or HDM2 to provide a significant growth advantage. Indeed, the authors show in the last part of their study that in at least two transformed cell lines, PCNA is rendered inert to the presence of ERK8. They speculate that the underlying reason is a defect in HDM2, and although this is the most likely explanation, it still needs to be validated. It will be interesting to see how common the misregulation of PCNA turnover is in cancer tissues. At this point, it is intriguing to envision a dynamic scenario in which a two-step mechanism facilitates cell transformation (Fig. 1). Initially, deregulation of ERK8 may cause PCNA levels to decrease. This would contribute to genome instability and the accumulation of new mutations, including those affecting proper function of HDM2. In step two, deregulation of HDM2 may turn things around and result in an increase of PCNA, supporting rapid proliferation.
Figure 1.
Role of ERK8 in maintaining genome stability. (A) In normal cells, chromatin-bound ERK8 interacts with the chromatin fraction of PCNA, which resides at the replication fork (here just shown at the leading strand for simplicity). ERK8 binding inhibits the E3 ubiquitin ligase HDM2 from interacting with PCNA. (B) In cancer cells, inactivation of ERK8 enables HDM2 to interact with and ubiquitinate PCNA, targeting it for degradation. A decrease in PCNA levels causes an increase in DNA damage, resulting in the accumulation of new mutations. These new mutations may render HDM2 nonfunctional (rectangular form), which ultimately results in an increase of PCNA stability and facilitates cell proliferation. The homotrimeric PCNA structure (Protein Data Bank ID 2OD8) was generated using the Chimera software program (Pettersen et al., 2004).
Acknowledgments
The authors acknowledge support by National Institutes of Health grant GM074917 and the Leukemia and Lymphoma Society to A.K. Bielinsky.
References
- Abe M.K., Saelzler M.P., Espinosa R., III, Kahle K.T., Hershenson M.B., Le Beau M.M., Rosner M.R. 2002. ERK8, a new member of the mitogen-activated protein kinase family. J. Biol. Chem. 277:16733–16743 10.1074/jbc.M112483200 [DOI] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19:114–126 10.1101/gad.1255805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G.A., Burgers P.M. 1988. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. USA. 85:7506–7510 10.1073/pnas.85.20.7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.G. 2009. Replication licensing and the DNA damage checkpoint. Front. Biosci. 14:5013–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S., Ricke R.M., Bielinsky A.K. 2006. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 26:4806–4817 10.1128/MCB.02062-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groehler A.L., Lannigan D.A. 2010. A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J. Cell Biol. 190:575–586 10.1083/jcb.201002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Hu J., Xiong Y. 2006. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281:3753–3756 10.1074/jbc.C500464200 [DOI] [PubMed] [Google Scholar]
- Madsen P., Celis J.E. 1985. S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication? FEBS Lett. 193:5–11 10.1016/0014-5793(85)80068-5 [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti G.P., Olson D.C., George D., Levine A.J. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 69:1237–1245 10.1016/0092-8674(92)90644-R [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Prelich G., Stillman B. 1988. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 53:117–126 10.1016/0092-8674(88)90493-X [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Senga T., Sivaprasad U., Zhu W., Park J.H., Arias E.E., Walter J.C., Dutta A. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281:6246–6252 10.1074/jbc.M512705200 [DOI] [PubMed] [Google Scholar]
- Shivji K.K., Kenny M.K., Wood R.D. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 69:367–374 10.1016/0092-8674(92)90416-A [DOI] [PubMed] [Google Scholar]
- Sporbert A., Gahl A., Ankerhold R., Leonhardt H., Cardoso M.C. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365 10.1016/S1097-2765(02)00729-3 [DOI] [PubMed] [Google Scholar]
- Takasaki Y., Deng J.S., Tan E.M. 1981. A nuclear antigen associated with cell proliferation and blast transformation. J. Exp. Med. 154:1899–1909 10.1084/jem.154.6.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Levine A.J. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA. 96:3077–3080 10.1073/pnas.96.6.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivona J.B., Kelman Z. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167–172 10.1016/S0014-5793(03)00622-7 [DOI] [PubMed] [Google Scholar]
- Warbrick E. 1998. PCNA binding through a conserved motif. Bioessays. 20:195–199 [DOI] [PubMed] [Google Scholar]