Abstract
Our understanding of how CD4+ T cells can regulate CD8+ T cell responses in HIV infection is still incomplete. Recent evidence obtained in mice suggests that CD4+ T cell help is required for efficient CD8+ T cell-mediated immunity in chronic infection: CD8+ T cells primed in the absence of such help release the TNF-related apoptosis-inducing ligand TRAIL and undergo apoptosis. Using a novel ELISPOT assay, in the present study we show that CD8+ T cells are also a source of the antigen-specific TRAIL response in HIV-infected patients with CD4+ T cell counts below 200. In patients with CD4+ T cell counts above 200 TRAIL was not detectable. Accordingly, antigens to which patients have likely been exposed when CD4+ T cell levels were high (e.g., influenza, CMV, and EBV) did not induce TRAIL. Within the HIV-positive donor population with low CD4+ T cell counts a dissociation of the interferon-γ (IFN-γ) and TRAIL response to different HIV peptide epitopes was detectable suggesting impaired immunity to antigens that triggered TRAIL in the absence of IFN-γ. Our findings emphasize that “helpless” CD8+ T cells, i.e., cells that have been primed in the absence of CD4+ T cell help, may play a crucial role in HIV infection. A “helpless” phenotype may impair CD8+ T cell control of HIV and other infections and possibly contribute to the depletion of CD4+ T cells via apoptosis. Immunizations and infections in this “helpless” state might result in ineffective CD8+ T cell responses.
Introduction
The successful control of viral infections such as HIV typically involves synergistic activities of various components of the immune system. In addition to specific antibodies, CD8+ T cells are critical for controlling the infection.1 While the requirement of CD4+ T cell help has been well established for the antibody response of B cells,2–4 recent evidence suggests that CD4+ T cell help is also crucial for efficient CD8+ T cell-mediated immunity.4–6 Data demonstrating the dependence of CD8+ T cell immunity on the presence of CD4+ T cell help were obtained in murine models involving H-2d-restricted recognition of adenovirus-transfected cells,7 the CD8+ T cell responses to cross-presented ovalbumin (OVA)4,8 to the male antigen HY by female mice,9,10 to infection with lymphocytic choriomeningitis virus (LCMV),6 to OVA expressed by Listeria monocytogenes,11 and to vaccinia virus expressing the gp33 determinant of LCMV.13 Similarly, in chronic gamma-herpesvirus infection CD8+ T cell-mediated control of the persistent viremia was also dependent on the presence of CD4+ T cells.12,13 In addition, CD4+ T cell help has been shown to be crucial for effective CD8+ T cell function and memory in the setting of allograft transplantation.14
All of these approaches led to the same conclusions: CD4+ T cell help was not required for engaging naive CD8+ T cells into a primary immune response resulting in similar clonal expansion, interferon (IFN)-γ production, and cytotoxic activity. However, CD8+ T cells primed in the absence of CD4+ T cell help were not capable of undergoing a second round of clonal expansion upon reencountering antigen and secreted low levels of cytokine. Additionally, such helpless8 or “lethargic”9 CD8+ T cells underwent activation-induced cell death upon restimulation, which was mediated by tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL).8
TRAIL, also called APO-2 ligand or TNFSF10,15–19 is a type II transmembrane protein secreted by various cell types, including T lymphocytes,20 natural killer cells,21 dendritic cells,22,23 monocytes, and macrophages.24,25 TRAIL induces rapid apoptosis in virus-infected and tumor cells21,26,27 via the activation of caspases (ICE/Ced-3 proteases).28 In HIV infection, increased plasma levels of TRAIL have been reported affecting monocytes25 and contributing to the characteristic depletion of CD4+ T cells seen in this disease.29–31 At the same time, TRAIL has not been studied yet in the conceptual framework of helpless CD8+ T cells in humans. Because CD4+ T cells are infected and depleted in the course of HIV infection, it seems conceivable that the resulting lack of CD4+ T cell help will impair specific CD8+ T cell immunity to HIV contributing to the inability to control the infection. Our study presented here has addressed this hypothesis.
Materials and Methods
Subjects and sample collection
Peripheral blood was obtained from 29 HIV-positive (2 women, 27 men, ages between 39 and 75 years) from the Special Immunology Unit at University Hospitals (Cleveland, OH). The CD4+ T cell counts ranged from 4 to 1232 cells/mm3. All patients tested were receiving highly active antiretroviral therapy (HAART) at the time point of testing. The patients were categorized according to their CD4+ T cell counts into two groups: CD4+ < 200 (HIV+CD4low; n = 21) and CD4+ > 200 (HIV+CD4high; n = 8). Besides the CD4+ T cell count, the viral load is a major predictor of disease progression in HIV-infected patients. Persistent viremia in the setting of chronic HIV infection may have a major impact on the phenotypic and functional characteristics of virus-specific CD4+ and CD8+ T cells. Therefore, the viral load was determined for each patient using RT-PCR. Following standard guidelines for the diagnosis and treatment of HIV infection levels of viral load can be distributed to the categories low (<10,000 copies/ml), intermediate (10,000–100,000 copies/ml), and high (>100,000 copies/ml). Accordingly, all HIV+CD4high patients displayed low levels of viral load. In the HIV+CD4low group 41.7% of the patients tested showed low levels of viral load, 33.3% intermediate levels, and 25% high levels. The 22 healthy controls tested (HIV−CD4high) (9 women, 13 men, ages between 21 and 64) were health care workers or members of our laboratory and adjoining laboratories. Peripheral blood mononuclear cells (PBMCs) were isolated from 40 to 100 ml of heparinized blood by standard Ficoll density-gradient centrifugation (Isoprep, Robbins Scientific). CD4-, CD8-, or CD3-depleted cells as well as CD8-enriched cell populations were obtained by negative selection using depletion or enrichment cocktails (RosetteSep, StemCell Technologies) and standard Ficoll-density gradient centrifugation. All studies were performed under the approval of the Institutional Review Board for Human Investigation at the University Hospitals of Cleveland.
Antigens
Antigens used were HIV-1 gag 20-mer peptides (10 μg/ml) comprising the HIV-1 subtype B region, with 10-amino acid overlaps between sequential peptides and HIV-1 gag, env, pol, and nef 15-mer peptides with 11-amino acid overlaps between sequential peptides (10 μg/ml). As a control CEF (CMV/EBV/influenza peptide pool) peptides (2 μg/ml) were used. These peptides cover HLA class I restricted determinants of viruses that commonly infect humans, in particular cytomegalovirus (CMV), Epstein–Barr virus (EBV), and influenza (Flu).32 All peptides listed above were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; the CEF control peptide pool was from DAIDS, NIAID. Hepatitis C virus (HCV) antigens (18-mers, 10 μg/ml) were a gift from Donald D. Anthony.
ELISPOT assays
ELISPOT assays were performed as described previously.32 Depending on the cell count ImmunoSpot plates (Cellular Technology Limited) or UNIFILTER small-volume plates (Whatman) were coated with capture antibody diluted in phosphate-buffered saline (PBS) at 4°C overnight. For capturing IFN-γ mAb M700A from Endogen was used at a concentration of 5 μg/ml, for capturing TRAIL polyclonal rabbit antimouse Ab, cross-reactive to human, (abcam) was used at a concentration of 1 μg/ml. Cells were plated in serum-free medium obtained from Cellular Technology Limited. When ImmunoSpot plates were used 2–3 × 105 cells were plated per well; for the small-volume plates 1.4 × 105 cells were plated. All results were normalized to 3 × 105 cells. For IFN-γ biotinylated detection mAb M701 (Endogen; 1 μg/ml) for TRAIL (TNF-related apoptosis-inducing ligand) purified detection mAb 75411.11 (Sigma; 2 μg/ml) served as the secondary antibody. For the TRAIL-ELISPOT antimouse IgGAM (H + L) (Invitrogen) was needed as the tertiary antibody. All plates were developed using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphatase substrate (Kirkegaard & Perry Laboratories). Image analysis was conducted by using an ImmunoSpot Series 3B analyzer (Cellular Technology Limited). For all ELISPOT assays performed the spontaneous medium background was subtracted from the results presented in the figures. In T cell immunology in general, and also for ELISPOT assays, a stimulation index (antigen-induced spot counts/spot counts in the medium control) of >3 is considered a positive response. We used a slightly more stringent cut-off, requiring a positive response to be four times the average of the negative control wells.
Flow cytometric analysis
Flow cytometry was performed as described previously33 using a Becton Dickinson FACScan and staining with FITC-labeled anti-CD8 or anti-CD3 (purified and labeled in our laboratory, clones OKT8 and OKT3) and PE-labeled anti-CD4 antibodies (BD-Pharmingen). A total of 10,000 live cells were analyzed per sample.
Data analysis
Mann–Whitney rank sum test was calculated using SigmaStat (SPSS, Version 7.0) to evaluate the statistical significance of TRAIL secretion among the different subject groups. A probability value of p < 0.05 was considered statistically significant. The cumulative TRAIL secretion of HIV+CD4low patients was compared to that of HFV+CD4high as well as HIV−CD4high subjects. The cumulative IFN-γ secretion of HIV+CD4low, HIV+CD4high, and HIV−CD4high subjects was compared among the different groups. SigmaStat software was also used to assess the strength of correlation between the amount of TRAIL released in individual donors and the levels of viral load. The coefficients of determination (r2 values) were calculated to evaluate the percent of data closest to the line of best fit and thereby to show how well the regression model described the data.
Results
The detection of TRAIL via a novel ELISPOT assay
TRAIL is a cell surface molecule that is shed through cleavage by cystine proteases.19,20,34,35 As with granzyme and perforin molecules that are prestored in activated CD8+ T cells, the detection of antigen-specific TRAIL-producing CD8+ T cells requires the measurement of the antigen-induced secretory activity of the T cell. In many publications, ELISPOT has been shown to be orders of magnitudes more sensitive for such purpose than ELISA because in ELISPOT the analyte is captured at its cellular source before it is diluted into the supernatant, bound to receptors of adjacent cells, or degraded. ELISA in contrast detects the analyte in the supernatant after dilution, absorption, and degradation. Flow cytometry and qRT-PCR do not permit the distinction between expressed and actually secreted molecule. These assays are suited to detect de novo synthesized molecules (such as antigen-induced cytokines),36,37 but are not suited to measure the secretion of preformed molecules such as TRAIL, granzyme, and perforin. TRAIL is constitutively expressed by a high percentage of PBMCs, including CD8+ T cells. None of these techniques permits measurements of the actual secretion by rare individual cells upon stimulation with specific antigen. To this end, we are introducing a novel ELISPOT assay enabling us to detect and visualize peptide-specific TRAIL release by individual cells.
Antigen-specific CD8+ T cells in HIV+CD4low subjects secrete TRAIL
We obtained PBMCs from healthy, non-HIV-infected (n = 22) and HIV-infected individuals (n = 29). The latter were subdivided into two groups: individuals that either displayed less than 200 CD4+ T cells/mm3 (constituting the “HIV+CD4low” group, n = 21) or more than 200 CD4+ T cells/mm3 (“HIV+CD4high”, n = 8). Subsequently, their freshly isolated PBMCs were tested (without additional pre treatment) for HIV peptide-induced secretion of TRAIL or IFN-γ in ELISPOT assays of 24 h duration. The HIV peptide library we used included 49 gag 20-mer as well as 4 env, 4 pol and 6 nef 15-mer peptides. Since all HIV-infected and healthy subjects were seronegative for HCV, a library of 16 HCV peptides (18-mers) served as our negative control. A CEF peptide library containing 23 peptides of CMV/EBV/Flu virus32 was used as our positive control.
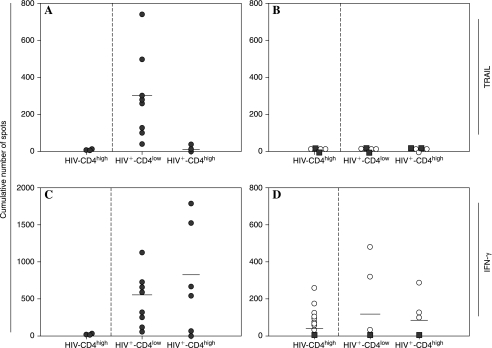
Results are summarized in Fig. 1. The HIV peptides induced TRAIL-producing cells in all HIV+CD4low donors, but in none of the HIV+CD4high subjects (p < 0.001) or in any of the healthy controls (p < 0.001) (Fig. 1A). At the same time, TRAIL secretion proved to be antigen-specific: neither CEF nor HCV peptides triggered TRAIL production in either of the HIV-infected subject groups or in any of the healthy controls (Fig. 1B). Moreover, the HIV peptides did not induce IFN-γ in the healthy controls, but triggered IFN—producing cells in the HIV-infected individuals (p < 0.001). The frequencies of HIV peptide-specific IFN-γ-secreting CD8+ T cells appeared diminished in HIV+CD4low subjects relative to those subjects harboring CD4+ T cells in higher numbers (Fig. 1C), but the difference did not reach statistical significance (p = 0.170). Upon stimulation with CEF peptides IFN-γ secretion occurred in HIV-infected individuals and in healthy controls, whereas HCV peptides did not induce an IFN-γ response in any of the groups (Fig. 1D).
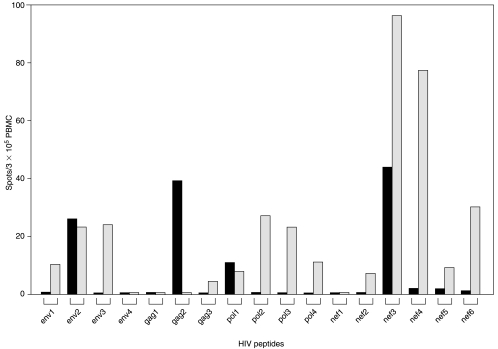
FIG. 1.
HIV peptides induce release of TRAIL in PBMCs of HIV-infected donors with low CD4+ T cell counts. PBMCs of 12 HIV-infected subjects with CD4+ T cell counts below 200 CD4+ T cells/mm3 (HIV+CD4low) and 8 donors with CD4+ T cell counts above 200 CD4 cells/mm3 (HIV+CD4high) as well as 22 non-HIV-infected healthy donors (HIV−CD4high) were tested directly ex vivo in a TRAIL (A, B) and IFN-γ (C, D) ELISPOT assay in the presence of an HIV peptide library (A, C) or of positive (CEF) or negative (HCV) control peptides (B, D) as specified by the symbols. Solid circles show the responses to HIV peptides (A, C) and open circles to CEF peptides (B, D). Solid squares represent the responses to HCV peptides (B, D). Each peptide within the library was tested individually in duplicate wells. The cumulative number of TRAIL or IFN-γ spots induced by all individual peptides within the specific peptide library is represented by the symbols for each donor.
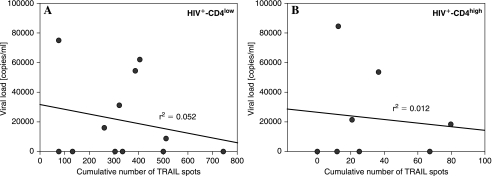
The release of TRAIL in HIV infection might not only depend on the level of the global CD4+ T cell population, i.e., the CD4+ T cell count. A variable antigen exposure/the actual activity of the infection may also contribute to the induction of a helpless CD8+ T cell stage. Therefore, we analyzed to what extent the viral load levels had an influence on TRAIL secretion. Data correlating the viral load levels with the cumulative responses to the 49 gag 20-mer as well as 4 env, 4 pol, and 6 nef 15-mer peptides as tested above are shown in Fig. 2 for HIV+CD4low (Fig. 2A) as well as HIV+CD4high (Fig. 2B) patients. With an r2 value of 0.052 for the former and r2 = 0.012 for the latter no correlation between viral load levels and the release of TRAIL was detectable in either the HIV+CD4low or the HIV+CD4high patient group.
FIG. 2.
Levels of viral load do not correlate with the amount of TRAIL that is released in HIV+CD4low and HIV+CD4high patients. PBMCs of 12 HIV-infected subjects with CD4+ T cell counts below 200 CD4+ T cells/mm3 (HIV+CD4low) (A) and 8 donors with CD4+ T cell counts above 200 CD4 cells/mm3 (HIV+CD4high) (B) were tested directly ex vivo in a TRAIL ELISPOT assay in the presence of an HIV peptide library as described in the legend to Fig. 1. The cumulative number of TRAIL spots induced by all individual peptides within the specific peptide library is represented by the symbols for each donor. The results were correlated with the viral load levels as measured by RT-PCR in each individual donor.
CD8+ T cells are the source of the HIV peptide-induced TRAIL response
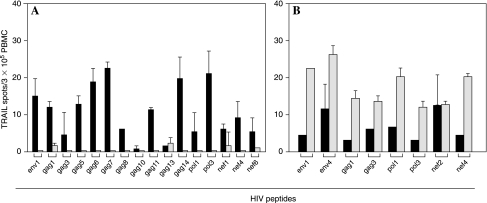
Consecutively, we employed two independent approaches to define the phenotype of the TRAIL-producing cells in response to HIV peptide stimulation in HIV+CD4low subjects. First, we depleted CD8+ T cells from the PBMC population reaching >99% depletion as confirmed by flow cytometry. The peptide-triggered TRAIL responses seen in the PBMCs were no longer detectable in the CD8 T cell-depleted PBMCs (Fig. 3A). On the contrary, CD4 T cell-depleted PBMCs showed increased numbers of HIV peptide-induced TRAIL spots, reflecting the enrichment of CD8+ T cells (data not shown). Second, we enriched CD8+ T cells to <97% purity and tested for reactivity to the individual peptides against which the PBMCs responded. The responses detected in the PBMCs reproduced in the CD8+ T cell population while showing increased spot numbers in the latter reflecting the increased frequencies of CD8+ T cells in the test cell population (Fig. 3B).
FIG. 3.
HIV-peptide-induced TRAIL-secreting cells are CD8 positive. (A) CD8 T cell depletion. Full PBMCs (solid bars) and CD8 cell-depleted PBMCs (open bars, <0.01% residual CD8+ T cells) were tested in a TRAIL ELISPOT assay for reactivity to the specified peptides as described in Fig. 1. Results are shown for an individual donor showing the mean and SD of spot numbers induced by the peptide tested in duplicate wells. The data are representative for 121 individual HIV peptides inducing TRAIL production in 6 donors. (B) CD8+ T cell enrichment. Bulk PBMCs (solid bars) and purified CD8+ T cells as obtained by negative selection (hatched bars, >97% purity) were tested in a TRAIL ELISPOT assay as described above. Results are depicted for an individual donor showing the mean and SD of spot numbers induced by each specified peptide tested in duplicate wells. The data are representative for 72 individual HIV peptides inducing TRAIL production in 4 donors.
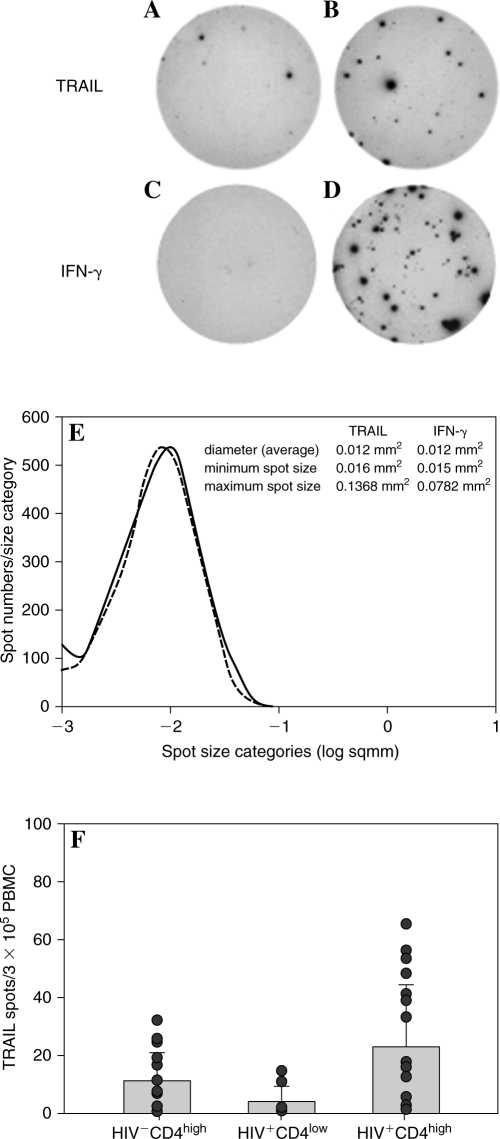
Characteristic TRAIL and IFN-γ spots induced in purified CD8+ T cells by HIV peptides are shown in Fig. 4A and B. The sizes of TRAIL spots were similar for purified CD8+ T cells and PBMCs (data not shown). Size and morphology of the TRAIL spots were also similar to IFN-γ spots (Fig. 4A–D). Spot size histograms are compared in Fig. 4E. Since spot size and morphology in ELISPOT assays reflect the quantity of molecule released by the cell, these data suggest similar per cell TRAIL and IFN-γ productivity. Unlike several ELISPOT assays in which the medium background counts are typically very low [IFN-γ, interleukin (IL)-2, IL-3, IL-4, IL-17], in the TRAIL assay the medium background was somewhat elevated (Fig. 4F) similar to IL-6, IL-10, granzyme, and TNF assays. The elevated TRAIL background is likely to result from spontaneous apoptosis of PBMCs in the cell culture. In spite of the elevated medium background, however, the TRAIL assay provided a very clear HIV antigen-induced signal in HIV-infected subjects over this background (Fig. 1A and B, Fig. 3)
FIG. 4.
Representative ELISPOT wells showing HIV-peptide-induced production of TRAIL or IFN-γ by CD8+ T cells. (A–D) HIV-peptide-induced ELISPOTs are shown on the right with the corresponding medium control wells on the left. (E) Spot size distribution histogram for TRAIL and IFN-γ spots produced by PBMCs in HIV+CD4low donors, as specified. Cumulative data obtained by analysis of 2500 spots in each category are shown. (F) Spontaneous TRAIL release in HIV+CD4low (n = 21), HIV+CD4high (n = 8), and HIV−CD4high (n = 22) control donors. Each data point represents the mean of duplicate wells for a single donor with SD <20%.
Dissociated production of TRAIL and IFN-γ by HIV peptide-specific CD8+ T cells
In murine models, helped CD8+ T cells secrete IFN-γ, but no TRAIL while helpless CD8+ T cells release TRAIL and are impaired in IFN-γ production. We tested in 21 HIV+CD4low subjects whether this dichotomy would become detectable at the determinant specificity level. PBMCs or CD8+ T cells from these donors were challenged with single HIV peptides, performing IFN-γ and TRAIL ELISPOT assays in parallel. Peptides that induced TRAIL-producing cells typically did not induce IFN-γ production, and occasionally IFN-γ production was seen in the absence of TRAIL. Mixed responses also occurred, conceivably indicating a heterogeneous CD8+ T cell population. A characteristic example is provided in Fig. 5 showing TRAIL as well as IFN-γ responses of an individual HIV+CD4low subject upon stimulation with HIV peptides derived from diverse viral regions. This TRAIL release profile is consistent with that of helpless CD8+ T cells. In the CD4+HIVlow subjects cumulatively only 5.4 ± 3.8% (SD) of all HIV peptides tested (11/197) triggered an isolated IFN-γ response (Table 1), suggesting that helplessness affected the majority of the HIV-reactive CD8+ T cells in those subjects.
FIG. 5.
Dissociated production of TRAIL and IFN-γ by HIV peptide-specific CD8+ T cells. (A–D) PBMCs from an individual HIV+CD4low donor were tested for specific HIV-peptide-induced production of TRAIL (open bars) and IFN-γ (solid bars). ELISPOT assays for TRAIL and IFN-γ were performed in parallel. The mean of duplicate wells is represented for each peptide and TRAIL/IFN-γ combination (SD <20%). The data are representative for 34 individual HIV peptides tested for this donor, and for 938 peptides tested in PBMCs of 21 HIV+CD4low donors.
Table 1.
HIV Peptide-Induced Release of IFN-γ and Trail in PBMCs of HIV-Infected Individualsa
| |
% of peptides to which a positive response was detecteda |
|||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | Mean ± SD | |
| IFN-γ | 7 | 10.7 | 4.7 | 7.4 | 0 | 0 | 8.3 | 5.4 ± 3.8 |
| TRAIL | 74.4 | 67.9 | 76.3 | 78.6 | 33.3 | 58.3 | 50 | 62.7 ± 15.3 |
| IFN-γ/TRAIL | 18.6 | 21.4 | 19 | 14.3 | 66.7 | 41.7 | 41.7 | 31.9 ± 17.6 |
PBMCs of seven HIV+CD4low subjects (A–G) were tested in ELISPOT assays for HIV-peptide-induced production of IFN-γ or TRAIL; 78 HIV peptides were tested individually in duplicate wells. In each individual the number of peptides inducing a positive response (out of the 78 peptides tested) was set as 100%. We then calculated the percentage of peptides that induced IFN-γ alone (in the absence of TRAIL), TRAIL alone (in the absence of IFN-γ), or the combination of both. Mean and SD for all subjects have been calculated and are shown in the last column.
IFN-γ single positive CD8+ T cells might reflect determinants of the virus that have undergone mutation: they might have primed CD8+ T cells at a time when the host was still CD4high. After the virus mutated, these determinants ceased to stimulate CD8+ T cells. Functionally, such HIV peptides would behave in a manner similar to CEF peptides. In contrast, viral peptides that are continuously biosynthesized in the infected host will cause ongoing immune stimulation. Because of the ongoing antigen stimulation the CD8+ T cell turnover rate is increased in HIV infection from 1 to 2% in non-HIV-infected individuals to as high as 10% in HIV-infected subjects.38,39
Discussion
Our results not only provide mechanistic insights into the immune pathology of HIV infection, but also suggest clinical relevance. A helpless phenotype is likely to impair the ability of HIV-specific CD8+ T cells to control HIV replication. Increased levels of soluble TRAIL have been measured in the serum of HIV-infected individuals.25,40 While monocytes, DCs, and CD4+ T cells have been implicated in producing TRAIL,22,25,28 our data suggest that HIV-specific CD8+ T cells might also be a primary source. Since TRAIL has proapoptotic effects in virus-infected cells28 its release by HIV-specific CD8+ T cells might be considered an antiviral effector function, possibly complimenting perforin-, granzyme-, Fas-FasL-, and other TNF superfamily member-mediated cytolytic functions. In tissue culture it has been shown that exposure of HIV-infected cells to TRAIL preferentially kills HIV-infected cells and therefore injection of TRAIL has been suggested as a therapeutic strategy.41 However, according to our data, in spite of the high numbers of TRAIL-releasing CD8+ T cells in advanced HIV infection (i.e., in HIV+CD4low subjects) these CD8+ T cells do not appear to be able to control the infection. On the contrary, the release of TRAIL by high numbers of HIV-specific CD8+ T cells might further contribute to the CD4 cell depletion in HIV infection since soluble TRAIL can induce apoptosis in uninfected CD4+ T cells.28 The depletion of CD4+ T cells in HIV infection might underlie a vicious circle: once CD4+ T cell numbers drop below 200 (i.e., our empirically estimated cut-off), HIV-specific CD8+ T cells acquire a helpless phenotype and start secreting large amounts of TRAIL. This, in turn, would further decrease CD4+ T cell counts and thereby enhance helplessness in HIV-specific CD8+ T cells. Supporting this notion it has been reported that CD4+ T cell counts and serum concentrations of TRAIL are inversely correlated.25 Accordingly, in our model helpless CD8+ T cells also produced decreased amounts of IFN-γ, a cytokine that inhibits viral replication.42 If these cells behave identically in murine models and humans, they might have diminished cytolytic activity, in addition to their reduced proliferative potential. Future studies are needed to delineate whether the blockage of the TRAIL pathway, e.g., the neutralization of TRAIL by antibody, might be suited to disrupt this deleterious circle and to restore CTL function.
Overall, in the present study the main hypothesis states that the lack of help occurs at the level of the global CD4+ T cell population and is reflected by the CD4+ T cell count. CD4 cell help of CD8 cell immunity is thought to be mediated by APC activation and/or by local production of growth factors.43–45 In either case specificity is conveyed by specifically and temporarily linked recognition of antigen by CD4+ and CD8+ T cells, as it typically occurs in an antigen-specific T cell response. Thus, we hypothesize that it is the absence of HIV antigen-specific CD4+ T cells that renders HIV-specific CD8+ T cells helpless. The absence of HIV-specific CD4+ T cells occurs due to the global CD4 cell depletion in HIV, and also due to the induction of unresponsiveness induced in these cells by the persisting HIV antigen. However, it also seems possible and needs to be evaluated in future studies whether a defect in HIV-specific CD4+ T cell responses and progressive immune dysfunction can cause HIV-specific CD8+ T cells to secrete TRAIL upon cognate antigen stimulation.
Our observation that most CD8+ T cells in HIV+CD4low subjects display the helpless phenotype suggests that the commitment of CD8+ T cells to the helped phenotype is less stable than suggested by the murine data. In murine models, the studies of helped phenotype stability were confined to one or a few rounds of restimulation.8–11 In humans, the helped CD8+ T cell phenotype may also be maintained for a limited number of CD8+ T cell restimulation cycles. However, the initially helped phenotype of HIV-specific CD8+ T cells acquired in the state of immune competence is apparently lost in the absence of continued CD4+ T cell help and the presence of continuous stimulation by the persisting virus.
Alternatively, the helped HIV-specific CD8+ T cells might become exhausted due to antigen persistence and thus decline in numbers. In this scenario, the HIV-reactive CD8+ T cell pool in HIV+CD4low individuals primarily consists of recent thymic emigrants that have been primed in a CD4+ T cell-deficient environment. On the contrary, infections with CMV, EBV, and influenza in HIV-infected subjects have most likely occurred early in life predating HIV infections. Thus, the CEF peptide reactive CD8+ T cells in these individuals have been primed in an unimpaired CD4+ T cell environment and consequently display the helped phenotype, remaining unaffected by the CD4+ T cell loss. Whichever mechanism may apply, we clearly show that the HIV-specific CD8+ T cells (but not the CEF-reactive CD8+ T cells) acquire the helpless phenotype in HIV+CD4low subjects.
It needs to be established whether de novo infections of HIV+CD4low subjects, e.g., with HCV, are likely to result in a helpless CD8+ T cell response contributing to the inability to control that infection. In addition, primary immunizations of HIV+CD4low subjects may equally not be advisable. Such immunizations might not only fail to induce an effective CD8+ T cell response, but are likely to commit the antigen-specific CD8+ T cells toward the helpless phenotype even after the patient's CD4+ T cells recover due to therapy. Our data suggest that such immunizations should be postponed until CD4+ T cell counts rise above 200.
TRAIL-producing CD8+ T cells were specific for a multitude of peptides in each HIV-infected subject with different peptides being recognized by different donors. Such diverse repertoires have also been observed measuring IFN-γ in the same patients (data not shown) being characteristic of chronic HIV infection.33,46–49 This diversity of peptide recognition results, among other factors, from HLA-polygenism and -polymorphism in the human population. It requires extensive peptide library testing to obtain comprehensive information on the HIV-specific CD8+ T cell pool in each individual. IFN-γ ELISPOT assays have become prevalent in immune monitoring of HIV-specific CD8+ T cells since they are well suited for high throughput screening of peptide libraries as well as because of their unsurpassed sensitivity for detecting the low-frequency HIV peptide-specific CD8+ T cells. While intracellular cytokine staining for IFN-γ can be considered an alternative for IFN-γ ELISPOT assays (measuring the antigen-induced de novo synthesis of the cytokine) TRAIL staining does not suit this purpose because of the constitutive membrane expression of this molecule. TRAIL ELISPOT assays might find their place next to IFN-γ ELISPOT assays for immune monitoring of CD8+ T cells in HIV infection, the former detecting helpless CD8+ T cells, but missing the helped CD8+ T cells, and the latter detecting the helped CD8+ T cells, but missing the helpless CD8+ T cells. It will need to be shown whether the enumeration of both cell types at single cell resolution permits a precise determination of the prevalence of either cell type in the host.
Acknowledgments
We thank all participating HIV patients and healthy donors and D.D. Anthony for providing HCV peptide pool and his valuable discussions. This work was supported by NIH Grant AI-47756 to M.T.L., RES425819 to P.V.L., and in part by the Center for AIDS Research at Case Western Reserve/University Hospitals of Cleveland (AI-36219). S.K. was supported by a fellowship of the Studienstiftung des Deutschen Volkes and by the Koeln Fortune Program of the University of Cologne. Paul V. Lehmann and Magdalena Tary-Lehmann contributed equally to this work.
Disclosure Statement
No competing financial interests exist.
References
- 1.Walker CM. Moody DJ. Stites DP. Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 2.Gray D. Siepmann K. Wohlleben G. CD40 ligation in B cell activation, isotype switching and memory development. Semin Immunol. 1994;6:303–310. doi: 10.1006/smim.1994.1039. [DOI] [PubMed] [Google Scholar]
- 3.Parker DC. T cell-dependent B cell activation. Annu Rev Immunol. 1993;11:331–360. doi: 10.1146/annurev.iy.11.040193.001555. [DOI] [PubMed] [Google Scholar]
- 4.Rocha B. Tanchot C. Towards a cellular definition of CD8+ T-cell memory: The role of CD4+ T-cell help in CD8+ T-cell responses. Curr Opin Immunol. 2004;16:259–263. doi: 10.1016/j.coi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Janssen EM. Lemmens EE. Wolfe T. Christen U. von Herrath MG. Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 6.Shedlock DJ. Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 7.Schoenberger SP. van der Voort EI. Krietemeijer GM. Offringa R. Melief CJ. Toes RE. Cross-priming of CTL responses in vivo does not require antigenic peptides in the endoplasmic reticulum of immunizing cells. J Immunol. 1998;161:3808–3812. [PubMed] [Google Scholar]
- 8.Janssen EM. Droin NM. Lemmens EE. Pinkoski MJ. Bensinger SJ. Ehst BD. Griffith TS. Green DR. Schoenberger SP. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 9.Bourgeois C. Veiga-Fernandes H. Joret AM. Rocha B. Tanchot C. CD8 lethargy in the absence of CD4 help. Eur J Immunol. 2002;32:2199–2207. doi: 10.1002/1521-4141(200208)32:8<2199::AID-IMMU2199>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10.Bourgeois C. Rocha B. Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 11.Sun JC. Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardin RD. Brooks JW. Sarawas SR. Doherty PC. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med. 1996;184:863–871. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson PG. Belz GT. Altman SD. Doherty PC. Virus-specific CD8+ T cell numbers are maintained during gamma-herpesvirus reactivation in CD4-deficient mice. Prof Natl Acad Sci USA. 1998;95:15565–15570. doi: 10.1073/pnas.95.26.15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Y. Wang Y. Wu Z. Kupiec-Weglinsky JW. Defective alloreactive CD8 T cell function and memory response in allograft recipients in the absence of CD4 help. J Immunol. 2007;179:4529–4534. doi: 10.4049/jimmunol.179.7.4529. [DOI] [PubMed] [Google Scholar]
- 15.Jeremias I. Herr I. Boehler T. Debatin KM. TRAIL/Apo-2-ligand-induced apoptosis in human T cells. Eur J Immunol. 1998;28:143–152. doi: 10.1002/(SICI)1521-4141(199801)28:01<143::AID-IMMU143>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Nebbioso A. Clarke N. Voltz E. Germain E. Ambrosino C. Bontempo P. Alvarez R. Schiavone EM. Ferrara F. Bresciani F. Weisz A. de Lera AR. Gronemeyer H. Altucci L. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 17.Pitti RM. Marsters SA. Ruppert S. Donahue CJ. Moore A. Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 18.Ryan LA. Peng H. Erichsen DA. Huang Y. Persidsky Y. Zhou Y. Gendelman HE. Zheng J. TNF-related apoptosis-inducing ligand mediates human neuronal apoptosis: Links to HIV-1-associated dementia. J Neuroimmunol. 2004;148:127–139. doi: 10.1016/j.jneuroim.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Wiley SR. Schooley K. Smolak PJ. Din WS. Huang CP. Nicholl JK. Sutherland GR. Smith TD. Rauch C. Smith CA. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 20.Kayagaki N. Yamaguchi N. Nakayama M. Kawasaki A. Akiba H. Okumura K. Yagita H. Involvement of TNF-related apoptosis-inducing ligand in human CD4+ T cell-mediated cytotoxicity. J Immunol. 1999;162:2639–2647. [PubMed] [Google Scholar]
- 21.Smyth MJ. Cretney E. Takeda K. Wiltroutn RH. Sedger LM. Kayagaki N. Yagita H. Okumura K. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon gamma-dependent natural killer cell protection from tumor metastasis. J Exp Med. 2001;193:661–670. doi: 10.1084/jem.193.6.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbeuval JP. Hardy AW. Boasso A. Anderson SA. Dolan MJ. Dy M. Shearer GM. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: Role of type I IFN-producing plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2005;102:13974–13979. doi: 10.1073/pnas.0505251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidalain PO. Azocar O. Lamouille B. Astier A. Rabourdin-Combe C. Servet-Delprat C. Measles virus induces functional TRAIL production by human dendritic cells. J Virol. 2000;74:556–559. doi: 10.1128/jvi.74.1.556-559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbeuval JP. Lambert C. Sabido O. Cottier M. Fournel P. Dy M. Genin C. Macrophages from cancer patients: Analysis of TRAIL, TRAIL receptors, and colon tumor cell apoptosis. J Natl Cancer Inst. 2003;95:611–621. doi: 10.1093/jnci/95.8.611. [DOI] [PubMed] [Google Scholar]
- 25.Herbeuval JP. Boasso A. Grivel JC. Hardy AW. Anderson SA. Dolan MJ. Chougnet C. Lifson JD. Shearer GM. TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected patients and its in vitro production by antigen-presenting cells. Blood. 2005;105:2458–2464. doi: 10.1182/blood-2004-08-3058. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa E. Nakazawa M. Yoshinari M. Minami M. Role of tumor necrosis factor-related apoptosis-inducing ligand in immune response to influenza virus infection in mice. J Virol. 2005;79:7658–7663. doi: 10.1128/JVI.79.12.7658-7663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JY. Huerta-Yepez S. Vega M. Baritaki S. Spandidos DA. Bonavida B. The NO TRAIL to YES TRAIL in cancer therapy. Int J Oncol. 2007;31:685–691. [PubMed] [Google Scholar]
- 28.Jeremias I. Herr I. Boehler T. Debatin KM. TRAIL/Apo-2-ligand-induced apoptosis in human T cells. Eur J Immunol. 1998;28:143–152. doi: 10.1002/(SICI)1521-4141(199801)28:01<143::AID-IMMU143>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Herbeuval JP. Grivel JC. Boasso A. Hardy AW. Chougnet Dolan MJ. Yagita H. Lifson JD. Shearer GM. CD4+ T cell death induced by infectious and noninfectious HIV-1: Role of type I interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood. 2005;106:3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy AW. Graham DR. Shearer GM. Herbeuval JP. HIV turns plasmacytoid dendritic cells (pDC) into TRAIL-expressing killer pDC and down-regulates HIV coreceptors by Toll-like receptor 7-induced IFN-alpha. Proc Natl Acad Sci USA. 2007;104:17453–17458. doi: 10.1073/pnas.0707244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim N. Dabrowska A. Jenner RG. Aldovini A. Human and simian immunodeficiency virus-mediated upregulation of the apoptotic factor TRAIL occurs in antigen-presenting cells from AIDS-susceptible but not from AIDS-resistant species. J Virol. 2007;81:7584–7597. doi: 10.1128/JVI.02616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynn KJ. Belz GT. Altman JD. Ahmed R. Woodland DL. Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 33.Kleen TO. Asaad R. Landry SJ. Boehm BO. Tary-Lehmann M. Tc1 effector diversity shows dissociated expression of granzyme B and interferon-gamma in HIV infection. AIDS. 2004;18:383–392. doi: 10.1097/00002030-200402200-00003. [DOI] [PubMed] [Google Scholar]
- 34.Mariani SM. Krammer PH. Surface expression of TRAIL/Apo-2 ligand in activated mouse T and B cells. Eur J Immunol. 1998;28:1492–1498. doi: 10.1002/(SICI)1521-4141(199805)28:05<1492::AID-IMMU1492>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 35.Mariani SM. Krammer PH. Differential regulation of TRAIL and CD95 ligand in transformed cells of the T and B lymphocyte lineage. Eur J Immunol. 1998;28:973–982. doi: 10.1002/(SICI)1521-4141(199803)28:03<973::AID-IMMU973>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 36.Jung T. Schauer U. Heusser C. Neumann C. Rieger C. Detection of intracellular cytokines by flow cytometry. J Immunol Methods. 1993;159:197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- 37.Labalette-Houache M. Torpier G. Capron A. Dessaint JP. Improved permeabilization procedure for flow cytometric detection of internal antigens. Analysis of interleukin-2 production. J Immunol Methods. 1991;138:143–153. doi: 10.1016/0022-1759(91)90162-9. [DOI] [PubMed] [Google Scholar]
- 38.Hellerstein M. Hanley MB. Cesar D. Siler S. Papageorgopoulos C. Wieder E. Schmidt D. Hoh R. Neese R. Macallan D. Deeks S. McCune JM. Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat Med. 1999;5:83–89. doi: 10.1038/4772. [DOI] [PubMed] [Google Scholar]
- 39.Sachsenberg N. Perelson AS. Yerly S. Schockmel GA. Leduc D. Hirschel B. Perrin L. Turnover of CD4+ and CD8+ T lymphocytes in HIV-1 infection as measured by Ki-67 antigen. J Exp Med. 1998;187:1295–1303. doi: 10.1084/jem.187.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liabakk NB. Sundan A. Torp S. Aukrust P. Froland SS. Espevik T. Development, characterization and use of monoclonal antibodies against sTRAIL: Measurement of sTRAIL by ELISA. J Immunol Methods. 2002;259:119–128. doi: 10.1016/s0022-1759(01)00501-4. [DOI] [PubMed] [Google Scholar]
- 41.Creery D. Weiss W. Lim WT. Aziz Z. Angel JB. Kumar A. Down-regulation of CXCR-4 and CCR-5 expression by interferon-gamma is associated with inhibition of chemotaxis and human immunodeficiency virus (HIV) replication but not HIV entry into human monocytes. Clin Exp Immunol. 2004;137:156–165. doi: 10.1111/j.1365-2249.2004.02495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarol LC. Imai K. Asamitsu K. Tetsuka T. Barzaga NG. Okamoto T. Inhibitory effects of IFN-gamma on HIV-1 replication in latently infected cells. Biochem Biophys Res Commun. 2002;291:890–896. doi: 10.1006/bbrc.2002.6532. [DOI] [PubMed] [Google Scholar]
- 43.Gray D. Matzinger P. T cell memory is short-lived in the absence of antigen. J Exp Med. 1991;174:969–974. doi: 10.1084/jem.174.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shedlock DJ. Whitmire JK. Tan J. MacDonald AS. Ahmed R. Shen H. Role of CD4 T cell help and costimulation in CD8 T cell responses during Listeria monocytogenes infection. J Immunol. 2003;170:2053–2063. doi: 10.4049/jimmunol.170.4.2053. [DOI] [PubMed] [Google Scholar]
- 45.Epstein MM. Di Rosa F. Jankovic D. Sher A. Matzinger P. Successful T cell priming in B cell-deficient mice. J Exp Med. 1995;182:915–922. doi: 10.1084/jem.182.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen TM. Watkins DI. New insights into evaluating effective T-cell responses to HIV. AIDS. 2001;15(Suppl. 5):S117–S126. doi: 10.1097/00002030-200100005-00015. [DOI] [PubMed] [Google Scholar]
- 47.Cao J. McNevin J. Holte S. Fink L. Corey L. McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77:6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalod M. Dupuis M. Deschemin JC. Sicard D. Salmon D. Delfraissy JF. Venet A. Sinet M. Guillet JG. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8(+) responses in HIV type 1-infected patients: Comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Draenert R. Altfeld M. Brander C. Basgoz N. Corcoran C. Wurcel AG. Stone DR. Kalams SA. Trocha A. Addo MM. Goulder PJ. Walker BD. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J Immunol Methods. 2003;275:19–29. doi: 10.1016/s0022-1759(02)00541-0. [DOI] [PubMed] [Google Scholar]