Abstract
Mesenchymal stem cells (MSCs) are a promising cell source for cartilage tissue engineering given their chondrogenic potential. This potential has yet to be fully realized, as the mechanical properties of MSC-based constructs are lower than those of chondrocyte-based constructs cultured identically. The aim of this study was to better understand the transcriptional underpinnings of this functional limitation. Matched chondrocytes and MSCs from three donors were cultured in agarose in a defined medium containing transforming growth factor β3 (TGF-β3). We evaluated the compressive mechanical properties and matrix deposition of maturing constructs over 56 days. Transcriptional differences between the two cell types were assessed on day 0 and 28 via microarray analysis and real-time polymerase chain reaction; differential deposition of matrix molecules was assessed by immunohistochemistry. Although the mechanical and biochemical properties of cell-seeded constructs improved with culture duration, MSC values plateaued at day 28, and remained lower than chondrocyte values. Using microarray analysis, 324 genes were identified as mis-expressed during chondrogenesis. Differential expression of 18 genes was validated, and differential deposition of proteoglycan 4 and TGF-beta-induced 68 kDa protein (TGFBI) was confirmed. Temporal expression profiles of these 18 genes showed that some genes were never expressed (chondromodulin), some were expressed at lower levels (proteoglycan 4), and some were expressed only at later time points (TGFBI) in MSCs compared to chondrocytes. These findings further define the complex transcriptional topography of MSC chondrogenesis, and provide new benchmarks for optimizing the growth of MSC-based engineered cartilage.
Introduction
Articular cartilage lines the surfaces of diarthrodial joints, dissipating and transferring loads engendered with locomotion. The prevalence of degenerative diseases and the lack of endogenous repair1 has focused regenerative strategies on the production of engineered cartilage. Recent efforts have generated neo-cartilage constructs with near-native compressive properties using juvenile chondrocytes from animal sources.2,3 Despite this promise, the use of autologous chondrocytes is clinically limited given the scarcity of healthy cells and their reduced capacity for tissue formation with aging and disease.4 Adult bone-marrow-derived mesenchymal stem cells (MSCs) are an attractive alternative, as they can undergo chondrogenesis and take on a chondrocyte-like phenotype.5–7
If MSC-based solutions for cartilage degeneration are to be successful, certain critical considerations must be evaluated. Because cartilage exists in a harsh load-bearing environment, engineered MSC-based neo-cartilage must replicate key functional (i.e., mechanical) features of the native tissue. Toward that end, MSCs have been cultured in a number of three-dimensional (3D) hydrogels.8 These materials support chondrogenesis and the deposition of an increasingly stiff matrix when cultured in the presence of transforming growth factor β (TGF-β) superfamily members.9–14 While this chondrogenic potential in 3D culture is encouraging, comparisons to differentiated chondrocytes cultured identically consistently show that MSC-based constructs do not achieve the same functional properties.15,16 Standard means of enhancing functional growth in chondrocyte-based constructs, such as increasing cell-seeding density or the application of long-term dynamic compressive loading protocols, have thus far not improved the compressive properties of MSC-based constructs.16,17
These findings underscore the inherent differences between chondrocytes and chondrogenically induced MSCs, and suggest the need for a more complete evaluation of chondrogenesis in relation to differentiated cells, and how this phenotypic transition relates to mechanical function. To date, most work in the field has focused on whether or not chondrogenesis has occurred (defined by the expression of a few key cartilaginous genes and the deposition of proteoglycans and collagen type II), and on identifying genes associated with this differentiation process. For example, several studies have characterized the temporal profiles of known chondrogenic markers during MSC chondrogenesis, at both the molecular and tissue levels.18–20 Other recent microarray studies identified new candidate markers by comparing undifferentiated MSCs and cartilage,21 while still others have employed microarray analysis to evaluate differentiation and subsequent dedifferentiation of MSCs to identify differentiation and stemness genes through multiple lineage progression.22
Collectively, these studies provide important information on the molecular events underlying MSC chondrogenesis, and have identified key factors in this process. However, most have taken a “yes/no” approach in defining chondrogenesis and have not assessed the differences in functional capacity between chondrogenically differentiated MSCs and fully differentiated chondrocytes in a 3D tissue engineering context. The current standard of reference for chondrogenic induction of MSCs is often an undifferentiated MSC or a healthy or osteoarthritic chondrocyte. To our knowledge, there exists no study comparing the molecular fingerprints of donor-matched healthy MSCs and chondrocytes maintained under identical culture conditions in vitro in 3D culture. As it is under these conditions that we observe robust growth of chondrocyte-based constructs2,3 and functional limitations in MSC chondrogenesis,15 it is under these conditions that we must identify the underlying molecular differences that define these functional disparities.
To address this issue, the current study was undertaken to identify markers of functional MSC chondrogenesis. A genome-wide screen using bovine microarrays was carried out using healthy donor-matched cells (chondrocytes and MSCs) seeded in 3D constructs. Three donors were evaluated after long-term culture under identical pro-chondrogenic conditions, and the transcriptional profiles of undifferentiated MSCs, chondrogenically differentiated MSCs, and chondrocytes were evaluated. Through this process, we defined a novel set of factors that were differentially regulated between these groups and showed that these differences in expression existed in both the absolute and temporal sense. This study provides critical new insight into the complexity of chondrogenesis on a transcriptional level, and provides new targets for enhancing their functional maturation and clinical potential.
Materials and Methods
Cell isolation
Bone-marrow-derived MSCs were isolated from the carpal bones of three separate 3–6-month-old calves (Research 87, Boylston, MA).16 MSCs from each donor were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 1% penicillin/streptomycin/fungizone and 10% fetal bovine serum and expanded up to passage 3. Articular cartilage was harvested from the carpometacarpal joints of the same animals and matched to MSC populations. Cartilage was digested with pronase and collagenase.23 Chondrocytes were encapsulated immediately upon isolation, with a separate gel cast for each donor.
Construct fabrication and long-term 3D culture
Sterile type VII agarose (49°C, 4% w/v; Sigma, St. Louis, MO) in phosphate-buffered saline was combined 1:1 with primary chondrocytes or MSCs in chemically defined medium (CM), and cell-seeded constructs (20 million cells/mL, Ø4 × 2.25 mm) were formed as in.15,16 CM consisted of Dulbecco's modified Eagle's medium supplemented with 1 × penicillin/streptomycin/fungizone, 0.1 μM dexamethasone, 50 μg/mL ascorbate 2-phosphate, 40 μg/mL L-proline, 100 μg/mL sodium pyruvate, and 1 × insulin-transferrin selenous acid+ (ITS+) (6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL selenous acid, 1.25 mg/mL bovine serum albumin, and 5.35 μg/mL linoleic acid). Contructs were cultured for 56 days in 1 mL/disk of CM supplemented with 10 ng/mL TGF-β3 (CM+; R&D Systems, Minneapolis, MN). At biweekly intervals, constructs were evaluated for mechanical properties and biochemical content. Gene expression of day 0 and 28 MSC-seeded constructs as well as day 28 chondrocyte-seeded constructs were evaluated using microarrays and real-time polymerase chain reaction (PCR). In subsequent studies assessing gene expression over long-term culture, MSC- and chondrocyte-seeded constructs were fabricated as above with cells from three donors combined. Cell-seeded constructs were cultured in CM+ for 56 days. Gene expression was evaluated biweekly, and mechanical properties were assessed at day 56.
Mechanical testing
Using a custom apparatus,15 constructs were tested in unconfined compression between two impermeable platens. Following equilibration in creep (0.02 N for 5 min), samples were subjected to 10% strain applied at 0.05%/s, followed by relaxation for 1000 s. Dynamic testing was performed by superimposing a 1% sinusoidal deformation at a frequency of 1.0 Hz. Equilibrium and dynamic modulus were determined as in Ref.15 After mechanical testing, constructs were frozen at −20°C for biochemical evaluation.
Biochemical analysis
For biochemical analysis, constructs were papain digested,16 and sulfated glycosaminoglycan (GAG) and collagen contents were evaluated using the 1,9-dimethylmethylene blue dye-binding24 and orthohydroxyproline25 assays, respectively. For collagen, a 1:7.14 orthohydroxyproline:collagen ratio was used.26 DNA content was determined using the PicoGreen dsDNA assay (Molecular Probes, Eugene, OR).
Real-time PCR
Total RNA was extracted by two sequential isolations in TRIZOL-chloroform and quantified (ND-1000; Nanodrop Technologies, Wilmington, DE). Reverse transcription and cDNA amplification by real-time PCR was performed as in Ref.16 For initial analyses, expression of four known chondrogenic genes (aggrecan and collagen types II, IX, and XI) were determined and normalized to glyceraldehyde 3-phophate dehydrogenase. For subsequent analyses (after microarray), additional intron-spanning primer sets were generated from bovine sequences (www.ensembl.org) using Primer Express software (version 3.0; Applied Biosystems). All primers are provided in Supplemental Table S1 (available online at www.liebertonline.com/ten).
Microarray: Target preparation and hybridization
Total RNA was extracted and quantified as described above. Microarray services were provided by the Penn Microarray Facility, including quality control tests of total RNA by Agilent Bioanalyzer and Nanodrop spectrophotometry. All protocols were conducted as described in the NuGEN Ovation Manual and the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, 100 ng of total RNA was converted to first-strand cDNA using reverse transcriptase primed by a poly(T) oligomer that incorporated a synthetic RNA sequence. Second-strand cDNA synthesis was followed by ribo-SPIA (Single Primer Isothermal Amplification; NuGEN Technologies, Inc., San Carlo, CA) for linear amplification of each transcript, and the resulting cDNA was fragmented, assessed by Bioanalyzer, and biotinylated. cDNA yields ranged from 5 to 10 μg, and 5 μg was added to Affymetrix hybridization cocktails, heated at 99°C for 2 min, and hybridized for 16 h at 45°C to nine Bovine GeneChips (Affymetrix, Inc., Santa Clara, CA). Microarrays were washed at low (6 × standard saline phosphate EDTA [SSPE]) and high (100 mM 2-(N-morpholino ethane sulfonic acid [MES], 0.1 M NaCl) stringency and stained with streptavidin–phycoerythrin. Fluorescence was amplified by adding biotinylated antistreptavidin and an additional aliquot of streptavidin–phycoerythrin stain. Fluorescent signal was collected with excitation at 570 nm using an Affymetrix Gene Chip Scanner 3000 (Affymetrix, Inc.).
Microarray data analysis
Output (.cel) files from scanning were processed (Partek, St. Louis, MO) and gene expression was assessed after normalization with the Robust Multi-Chip Average (RMA) algorithm. Principal component analysis was carried out to determine global variation. To determine statistical significance, a two-way mixed model ANOVA was applied with donor and cell type as the independent variables. Three pairwise contrasts were evaluated to assess fold changes in gene expression levels. Changes in expression were assessed between donor-matched undifferentiated MSCs (day 0) and differentiated MSCs (day 28), undifferentiated MSCs (day 0) and chondrocytes (day 28), as well as differentiated MSCs (day 28) and chondrocytes (day 28). p-Values were generated for donor, cell type, and each pairwise contrast. To determine false discovery rates (FDRs), the Benjamini Hochberg method was applied to the generated p-values and step-up p-values were calculated. Genes that were considered under-expressed during chondrogenesis were defined as those that were at least twofold greater in chondrocytes compared to both day 0 and 28 MSCs and no more than twofold less or threefold greater in day 28 MSCs compared to day 0 MSCs. Genes that were considered over-expressed were defined as genes that were at least twofold greater in both day 0 and day 28 MSCs compared to chondrocytes, and no more than twofold greater in day 28 MSCs compared to day 0 MSCs. Genes of interest were identified using an FDR of 10% (Spotfire software; Tibco, Somerville, MA). Expression was verified by real-time PCR for the genes listed in Figure 5C. All microarray data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE18394 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18394).
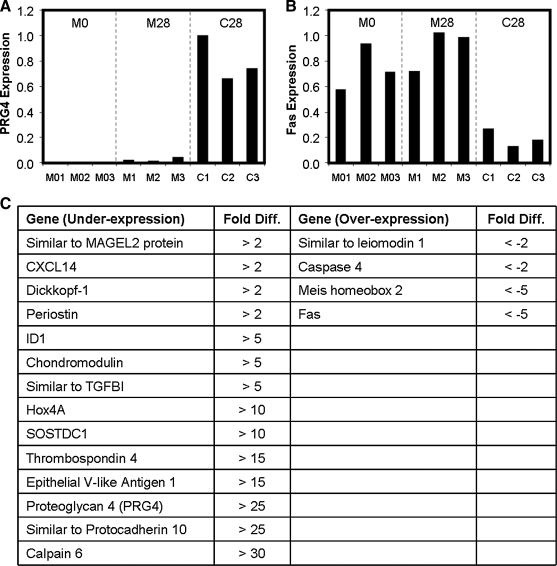
FIG. 5.
Gene expression of (A) proteoglycan 4 (PRG4) and (B) Fas, illustrating genes that were under- or over-expressed in MSC-seeded constructs relative to chondrocyte-seeded constructs. (C) Genes confirmed by real-time polymerase chain reaction to be differentially expressed with fold difference indicated for C28 compared to M0 and M28.
Histology and immunohistochemistry
Paraffin sections (8 μm) were deparaffinized, rehydrated, and stained with hematoxylin and eosin (Sigma), Alcian Blue (pH 1.0), or Picrosirius Red for cell distribution, sulfated proteoglycans, and collagens, respectively. For immunohistochemical analysis, antigen retrieval was performed by incubating sections in citrate buffer (10 mM citric acid with 0.05% Tween 20 at pH 6.0) heated to 99°C for 25 min, cooling for 20 min (25°C), and incubating in hyaluronidase.27 Collagen type I (MAB3391; Millipore, Billerica, MA), collagen type II (11-116B3; Developmental Studies Hybridoma Bank, Iowa City, IA), proteoglycan 4 (PRG4; ab28484; Abcam, Cambridge, MA), and TGF-beta induced 68 kDa protein (TGFBI; ab66957; Abcam) primary antibodies were used to identify matrix components. Primary and secondary antibody hybridization and subsequent color development with 3,3′-diaminobenzidine (DAB) chromogen was carried out as in Ref.27 Color images were captured at 10× magnification as in Ref.16
Statistical analysis
Statistical analysis was performed on mechanical and biochemical data using three-way ANOVA with time, cell type, and donor as independent variables. Where significance was indicated, Tukey's posthoc tests were carried out. For all measures, significance was determined at p < 0.05. All values are reported as mean ± standard deviation.
Results
Mechanical properties
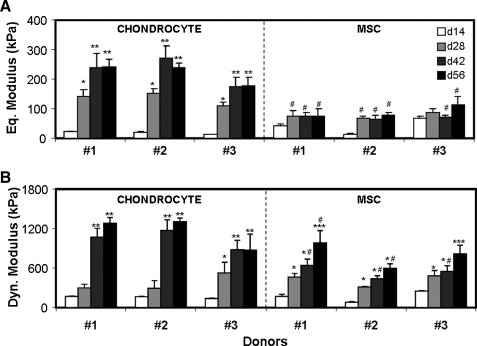
Three donor-matched sets of chondrocytes and MSCs were encapsulated in agarose and cultured in a defined medium containing TGF-β3 for 56 days. Consistent with previous studies, the compressive properties of cell-seeded constructs improved with time in culture. For all donors, the equilibrium modulus of chondrocyte-seeded constructs increased through day 42, while MSC-seeded constructs did not improve after day 28 (p = 1.0, Fig. 1A). Chondrocyte-seeded constructs attained equilibrium moduli of 180–240 kPa, while MSC-seeded constructs reached moduli of 75–114 kPa. In all constructs, the dynamic modulus continued to develop beyond day 28, regardless of cell type (Fig. 1B). Both equilibrium and dynamic properties were significantly lower for all MSC groups than for chondrocytes at the final time point, with the exception of donor 3 dynamic properties.
FIG. 1.
Time-dependent compressive (A) equilibrium and (B) dynamic modulus (kPa) of chondrocyte-seeded and mesenchymal stem cell (MSC)-seeded constructs with culture in a chemically defined medium supplemented with 10 ng/mL transforming growth factor β3 (TGF-β3). d14–d56 indicate days 14–54. *Greater than d14 within donors and cell type (p < 0.001); **greater than d28 within donors and cell type (p < 0.02); ***greater than d28 within donors and cell type (p < 0.05); #lower compared to donor-matched chondrocytes at the same time point (p < 0.01). Data represent the mean and standard deviation of four samples per group per time point.
Biochemical content and chondrogenic gene expression
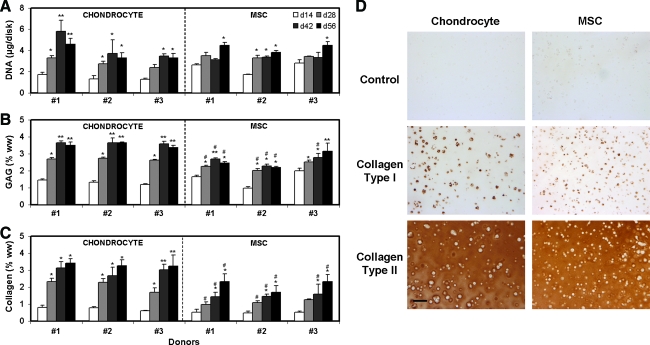
Consistent with mechanical data, the biochemical content for chondrocyte- and MSC-seeded constructs increased with time, with chondrocyte-seeded constructs increasing more rapidly and achieving significantly higher levels by day 42. DNA content was comparable between groups (Fig. 2A). While GAG and collagen content improved for both chondrocyte- and MSC-seeded constructs, MSC-seeded groups failed to attain the levels achieved by their donor-matched chondrocyte constructs (Fig. 2B, C). Histological analysis showed uniform GAG and collagen distribution throughout each construct (Supplemental Fig. S1, available online at www.liebertonline.com). At the final time point, all constructs stained strongly for collagen type II and weakly for collagen type I, regardless of cell type or donor (Fig. 2D). Expression of cartilage-specific extracellular matrix (ECM) structural components (aggrecan and type II, IX and XI collagen) at day 28 showed higher expression in differentiated MSC-seeded constructs than in chondrocyte-seeded constructs for each donor (Fig. 3). These genes were expressed at negligible levels in undifferentiated MSCs (data not shown).
FIG. 2.
Biochemical composition of constructs with variation in time in culture, donor, and cell type. (A) DNA content (μg/disk), (B) glycosaminoglycan (GAG) content (% ww), and (C) collagen content (% ww) of chondrocyte- and MSC-seeded constructs. (D) Immunohistochemical analysis of collagen types I and II distribution in cell-seeded constructs for a single donor. *Greater than d14 within donors and cell type (p < 0.015); **greater than d28 within donors and cell type (p < 0.025); #lower compared to donor-matched chondrocytes at the same time point (p < 0.04). Data represent the mean and standard deviation of four samples per group per time point. Scale bar: 100 μm. Color images available online at www.liebertonline.com/ten.
FIG. 3.
Expression of cartilage extracellular matrix (ECM) genes in MSC-seeded constructs normalized to chondrocytes-seeded constructs after 28 days of culture in TGF-β3 containing medium. For all three donors, aggrecan and type II, IX, and XI collagen expression levels were higher in MSC-seeded constructs.
Microarray screening
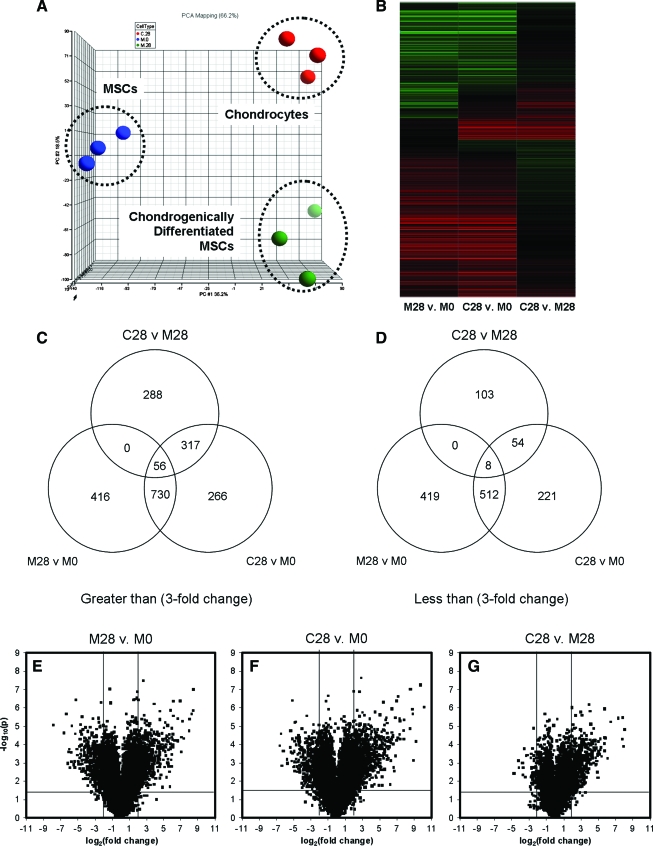
Microarray screening was then carried out to identify additional genes that are mis-expressed during MSC chondrogenesis. In this screen, day 0 MSCs (M0, undifferentiated MSCs), day 28 MSCs (M28, chondrogenically differentiated MSCs), and day 28 chondrocytes (C28) in 3D culture were processed for three donors. Samples were chosen for analysis at day 28 as this marks the point where the mechanical properties of chondrocyte- and MSC-based constructs began to diverge significantly (Fig. 1), with MSC-based construct properties plateauing after this point. Principal component analysis of the microarray data indicated that while chondrogenically differentiated MSCs and chondrocytes were more similar to one another compared to undifferentiated MSCs (principal component 1 [PC1]), significant differences persisted between these two groups (principal component 2 [PC2], Fig. 4A). Heat map observation further confirmed this observation, with higher and lower levels of expression for individual genes depicted in red and green, respectively (Fig. 4B).
FIG. 4.
(A) Principal component analysis and (B) heat map generated from microarray data showing differential gene expression (green indicates greater; red indicates lower) between undifferentiated MSCs at day 0 (M0), chondrogenically differentiated MSC-seeded constructs at day 28 (M28), and chondrocyte-seeded constructs at day 28 (C28). (C, D) Venn diagrams and (E–G) volcano plots for M0, M28, and C28 indicates number of genes and dispersion of genes that were differentially regulated with chondrogenic induction in three-dimensional culture. Color images available online at www.liebertonline.com/ten.
With chondrogenic induction, 1202 genes were upregulated in M28 compared to M0 (Fig. 4C). Of these genes, 730 genes were comparably expressed by M28 relative to C28, while 56 genes failed to attain C28 expression levels. Of the 730 genes induced, several were known cartilage markers, including COMP and SERPINA1 (>300-fold relative to M0), chondroadherin (>200-fold relative to M0), collagen type XI (>100-fold relative to M0), collagen types II and IX, and aggrecan 1 (>40-fold relative to M0). Microarray analysis also revealed 317 genes that were not changed during MSC chondrogenesis at day 28. Although these 317 genes were expressed by chondrocytes, they remained not expressed or poorly expressed (<3-fold change) by MSCs compared to chondrocytes, regardless of MSC differentiation status. A similar analysis of genes suppressed during chondrogenesis identified 939 genes that were lower in M28 compared to M0 (Fig. 4D). Within this group, 512 genes were similar between M28 and C28, while only 8 genes were highly expressed in M0, moderately expressed in M28, and poorly expressed in C28. There were 54 genes that were over-expressed in both M0 and M28, and the expression of these genes at day 28 was not affected by chondrogenesis.
These observations were visually confirmed by volcano plots summarizing fold-changes in gene expression and statistical criteria (Fig. 4E–G). Most notably, the C28 to M28 comparison indicated that the most significant differences were those of higher expression in the C28 group. Using specific criteria (>2-fold change) and an FDR of 10%, 252 genes and 72 genes were identified as potentially under-expressed or over-expressed during MSC chondrogenesis, respectively (Supplemental Tables S2 and S3, available online at www.liebertonline.com). A subset of these genes was selected for further analysis and real-time PCR was used to validate the results for 18 of these genes. Of these genes, 14 were poorly expressed in day 0 and 28 MSC-seeded constructs relative to chondrocyte-seeded constructs, while 4 genes remained highly expressed in the MSC populations even after chondrogenesis but were absent or very low in chondrocytes (Fig. 5C). Representative results for PRG4 and Fas demonstrate the patterns of this transcriptional mis-regulation during MSC chondrogenesis (Fig. 5A, B).
Gene expression profiles
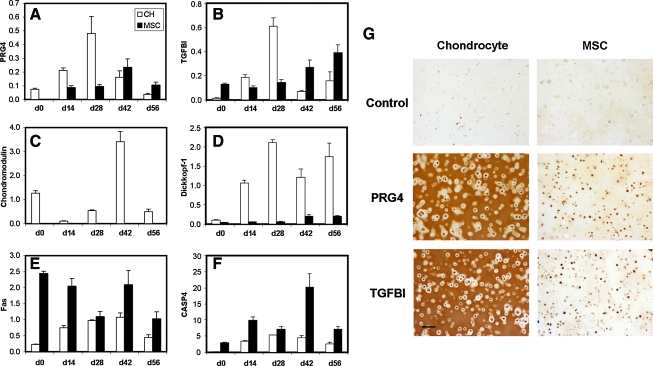
One limitation in this previous analysis was that only a single time point was analyzed (day 28). To assess the temporal profiles of identified genes, chondrocyte- and MSC-seeded constructs were maintained in the chondrogenic medium for 56 days and the expression levels of the previously identified genes (Fig. 5) were evaluated biweekly. Distinct patterns of expression emerged from this analysis, as demonstrated by representative graphs of PRG4, TGFBI, chondromodulin, and dickkopf-1. While chondromodulin and dickkopf-1 expression remained markedly under-expressed in MSC-seeded constructs compared to chondrocyte-seeded constructs at every time point assayed (Fig. 6C, D), delayed expression of PRG4 and TGFBI was observed in MSC-seeded constructs, with increasing levels of expression at later time points (Fig. 6A, B). PRG4 expression in MSC-seeded constructs was highest at day 42 (compared to day 28 for chondrocyte-seeded constructs), although the peak expression level in MSC-seeded constructs remained lower than the peak level in chondrocyte-seeded constructs for the time points evaluated. Conversely, TGFBI expression increased continually in MSC-seeded constructs at day 56. It is unknown whether the expression level of this gene would continue to increase past day 56 and reach eventual parity with peak chondrocyte-seeded construct levels (day 28). Temporal expression profiles of Fas and CASP4 showed that expression of these genes in MSC-seeded constructs remained consistently higher than chondrocyte-seeded constructs throughout the culture duration (Fig. 6E, F). Temporal profiles for all the genes identified in Fig. 5 can be found in Supplementary Figure S3.
FIG. 6.
Expression profiles of (A) PRG4, (B) TGF-beta induced 68 kDa protein (TGFBI), (C) chondromodulin, (D) Dickkopf-1, (E) Fas, and (F) CASP4 show different temporal patterns for chondrogenically induced MSC-seeded constructs compared to chondrocyte-seeded constructs cultured identically for 56 days. (G) Immunohistochemical detection of PRG4 and TGFBI for day 56 cell-seeded constructs shows robust staining in chondrocyte-seeded constructs and weak staining in MSC-seeded constructs. Scale bar: 100 μm. Color images available online at www.liebertonline.com/ten.
To evaluate whether these differences in gene expression were translated to matrix formation, two molecules, PRG4 and TGFBI, were selected for further analysis. For all donors, chondrocyte-seeded constructs on day 56 stained intensely for both PRG4 and TGFBI, although deposition was localized to the center of the constructs. In contrast, extracellular deposition of PRG4 and TGFBI was not observed for any of the MSC-seeded constructs (Fig. 6G). Consistent with previous reports,28 PRG4 immunostaining was discernible in superficial regions of articular cartilage section taken from juvenile bovine carpal joints, with little to no staining in the middle or deep zones (not shown).
Discussion
The ability of bone-marrow-derived MSCs to undergo chondrogenesis and accumulate functional matrix has provided impetus for their use in cartilage tissue engineering. While promising, the extent of MSC conversion toward the chondrogenic phenotype remains in question; these concerns are sparked in part by the limited functional capacity of MSCs in 3D hydrogel culture.15,16 To better understand the basis of this limitation and develop new benchmarks for chondrogenesis, we characterized the molecular profiles of chondrogenically induced MSCs. In particular, we focused on the molecular differences distinguishing differentiated MSCs from donor-matched articular chondrocytes, as identification of these deficits may lead to potential targets for therapeutic intervention.
Consistent with previous findings, after long-term culture in pro-chondrogenic conditions, the compressive properties and biochemical content of MSC-laden constructs fell short of those attained by chondrocytes for all three donors. Standard cartilage markers, such as aggrecan and collagen types II, IX, and XI, were consistently upregulated during MSC chondrogenesis and expression was maintained at high levels at day 28. Surprisingly, MSCs expressed higher levels of these genes than did donor-matched chondrocytes at this timepoint, suggesting that the observed discrepancy in functional properties was not due to expression deficits of these standard matrix molecules. Collagen type II immunostaining further confirmed this observation, as chondrocyte- and MSC-laden constructs stained with equal intensity for this important ECM protein. These findings suggest that while standard markers are expressed, they are insufficient predictors of mechanical functionality of chondrogenically induced MSC populations in 3D culture. On the basis of these results, we hypothesized that other elements important in matrix assembly or turnover may underlie the functional discrepancy between chondrocytes and MSCs.
Therefore, to better elucidate the topography of chondro-induction, we carried out a genome-wide screen via microarray analysis and identified 324 genes that were transcriptionally misregulated over the course of chondrogenesis. These genes were either twofold higher or lower in undifferentiated and chondrogenically differentiated MSC-seeded constructs relative to chondrocyte-seeded constructs at day 28. Of these genes, a subset of 18 genes was selected based on their relevance to mechanical function (matrix elements) or MSC phenotype as established in the literature. These genes were confirmed by real-time PCR and their temporal expression profiles assessed over 56 days, as construct properties typically equilibrate by this time.15 While some genes such as chondromodulin and Dikkopf-1 were never expressed by MSCs undergoing chondrogenesis, others including PRG4 and TGFBI showed delayed patterns of expression. Immunostaining at the terminal timepoint showed robust deposition of PRG4 and TGFBI in chondrocyte-seeded constructs and no staining in MSC-seeded constructs for all three donors. This is consistent with the findings of Gleghorn et al., who demonstrated that PRG4 was retained in chondrocyte-seeded, not MSC-seeded, alginate.29 Interestingly, that study also found greater PRG4 secretion into the medium by MSCs compared to chondrocytes. The lack of synthesis or retention of these minor molecules may play a critical role in functional outcomes, particularly if their role is to nucleate ECM formation or regulate ECM organization.
In addition, these molecules may also be indicative of the state of phenotypic conversion. For example, TGFBI, a type II collagen binding protein,30 inhibits mineralization and maintains the chondrocytic phenotype in hypertrophic chondrocytes.31 Further, expression of this gene is highest in the prehypertrophic stage of developing cartilage.32 Poor expression of TGFBI in MSCs may thus reflect incomplete or incorrect induction toward the chondrogenic phenotype. Indeed, recent data suggest that the phenotype achieved by differentiated MSCs may be more akin to that of transient rather than permanent chondrocytes. During development, transient chondrocytes undergo hypertrophy and eventual ossification, while permanent chondrocytes maintain a fixed chondrogenic phenotype.33 Microarray analysis of mouse articular cartilage and growth plate cartilage showed considerable differences in gene expression between these chondrocyte populations, though standard cartilage markers, including aggrecan, were expressed by both.34 Notably, collagens types II, IX, and XI were expressed at higher levels in the transient chondrocytes than in permanent articular chondrocytes, paralleling the findings of the current study with differentiated MSCs and articular chondrocytes. Other data also suggest that the chondrogenic commitment of induced MSCs may be flexible in vivo.35,36 Implantation of chondrogenically differentiated MSCs resulted in extensive mineralization of the cartilage matrix, mirroring transient chondrocyte phenotypic transitions. In contrast, articular chondrocytes showed no signs of phenotypic instability in vivo, further delineating the differences between fully committed articular chondrocytes and chondrogenically differentiated MSCs.
In this study, we identified a subset of genes that is mis-expressed during MSC chondrogenesis. While important, this subset may not represent the entirety of the transcriptional set that distinguishes MSC chondrogenesis from normal chondrocyte function, as all of the genes described here were identified from analysis of a single time point, day 28. From temporal expression profiles, it is apparent that even within this subset of genes, patterns of expression vary dramatically. Microarray analyses of early and later stage chondrogenesis will be necessary to capture the complete topography of transcriptional dysfunction and may well identify additional targets for consideration. In addition, it is as yet unclear what role these genes play with respect to the mechanical maturation of MSC-based constructs; future knockdown studies will be required to establish functional correlations between gene expression and construct mechanical properties. Once correlation is established, the manipulation of the expression of these genes may enhance the functional capacity of MSCs for cartilage repair.
Taken together, our studies establish a better understanding of the complex molecular topography of MSC chondrogenesis and, in particular, our limitation in the creation of mechanically functional constructs based on this cell source. Having now established new molecular benchmarks of chondrogenesis, these features can be applied to gauge the efficacy of a given culture environment or medium supplement. For example, we have recently probed small molecule libraries for novel biochemical mediators of chondrogenesis using high-throughput screening37; these new markers may prove useful in such optimizations. Alternatively, these same markers may be helpful in the tuning of interactions between MSCs and their biomaterial microenvironment14,38 as well as the timing and magnitude of mechanical perturbation.8,17,39,40 For example, as a subset of the identified genes are sensitive to mechanical stimulation (Supplemental Fig. S4, available online at www.liebertonline.com), the modulation of their expression in response to loading can be used to optimize loading parameters for chondrogenesis. Benchmarking all such efforts against molecular profiles that generate functional neo-cartilage constructs will improve MSC-based cartilage tissue engineering and lead to a mechanically competent, phenotypically stable cartilage replacement.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 EB008722 and R03 AR053668), the Penn Center for Musculoskeletal Disorders, and a Graduate Research Fellowship from the National Science Foundation (A.H.H.). The type II collagen antibody developed by Thomas F. Linsenmayer was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by the Department of Biology, University of Iowa, Iowa City, IA. The authors also gratefully acknowledge the Penn Microarray Core Facility and Dr. Don Baldwin and Dr. John Tobias for their help with microarray processing and data analysis.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hunziker E.B. Articular cartilage repair: are the intrinsic biological constraints undermining this process insuperable? Osteoarthritis Cartilage. 1999;7:15. doi: 10.1053/joca.1998.0159. [DOI] [PubMed] [Google Scholar]
- 2.Byers B.A. Mauck R.L. Chiang I.E. Tuan R.S. Transient exposure to transforming growth factor beta 3 under serum-free conditions enhances the biomechanical and biochemical maturation of tissue-engineered cartilage. Tissue Eng Part A. 2008;14:1821. doi: 10.1089/ten.tea.2007.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lima E.G. Bian L. Ng K.W. Mauck R.L. Byers B.A. Tuan R.S. Ateshian G.A. Hung C.T. The beneficial effect of delayed compressive loading on tissue-engineered cartilage constructs cultured with TGF-beta3. Osteoarthritis Cartilage. 2007;15:1025. doi: 10.1016/j.joca.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran-Khanh N. Hoemann C.D. McKee M.D. Henderson J.E. Buschmann M.D. Aged bovine chondrocytes display a diminished capacity to produce a collagen-rich, mechanically functional cartilage extracellular matrix. J Orthop Res. 2005;23:1354. doi: 10.1016/j.orthres.2005.05.009.1100230617. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone B. Hering T.M. Caplan A.I. Goldberg V.M. Yoo J.U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D. Moorman M.A. Simonetti D.W. Craig S. Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Prockop D.J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 8.Huang A.H. Farrell M.J. Mauck R.L. Mechanics and mechanobiology of mesenchymal stem cell-based engineered cartilage. J Biomech. 2010;43:128. doi: 10.1016/j.jbiomech.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung C. Burdick J.A. Influence of three-dimensional hyaluronic Acid microenvironments on mesenchymal stem cell chondrogenesis. Tissue Eng Part A. 2009;15:243. doi: 10.1089/ten.tea.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buxton A.N. Zhu J. Marchant R. West J.L. Yoo J.U. Johnstone B. Design and characterization of poly(ethylene glycol) photopolymerizable semi-interpenetrating networks for chondrogenesis of human mesenchymal stem cells. Tissue Eng. 2007;13:2549. doi: 10.1089/ten.2007.0075. [DOI] [PubMed] [Google Scholar]
- 11.Kavalkovich K.W. Boynton R.E. Murphy J.M. Barry F. Chondrogenic differentiation of human mesenchymal stem cells within an alginate layer culture system. In Vitro Cell Dev Biol Anim. 2002;38:457. doi: 10.1290/1071-2690(2002)038<0457:cdohms>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Kisiday J.D. Kopesky P.W. Evans C.H. Grodzinsky A.J. McIlwraith C.W. Frisbie D.D. Evaluation of adult equine bone marrow- and adipose-derived progenitor cell chondrogenesis in hydrogel cultures. J Orthop Res. 2008;26:322. doi: 10.1002/jor.20508. [DOI] [PubMed] [Google Scholar]
- 13.Erickson I.E. Huang A.H. Chung C. Li R.T. Burdick J.A. Mauck R.L. Differential maturation and structure-function relationships in mesenchymal stem cell- and chondrocyte-seeded hydrogels. Tissue Eng Part A. 2009;15:1041. doi: 10.1089/ten.tea.2008.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson I.E. Huang A.H. Sengupta S. Kestle S. Burdick J.A. Mauck R.L. Macromer density influences mesenchymal stem cell chondrogenesis and maturation in photocrosslinked hyaluronic acid hydrogels. Osteoarthritis Cartilage. 2009;17:1639. doi: 10.1016/j.joca.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauck R.L. Yuan X. Tuan R.S. Chondrogenic differentiation and functional maturation of bovine mesenchymal stem cells in long-term agarose culture. Osteoarthritis Cartilage. 2006;14:179. doi: 10.1016/j.joca.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Huang A.H. Stein A. Tuan R.S. Mauck R.L. Transient exposure to TGF-beta3 improves the mechanical properties of MSC-laden cartilage constructs in a density dependent manner. Tissue Eng Part A. 2009;15:3461. doi: 10.1089/ten.tea.2009.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorpe S.D. Buckley C.T. Vinardell T. O'Brien F.J. Campbell V.A. Kelly D.J. Dynamic compression can inhibit chondrogenesis of mesenchymal stem cells. Biochem Biophys Res Commun. 2008;377:458. doi: 10.1016/j.bbrc.2008.09.154. [DOI] [PubMed] [Google Scholar]
- 18.Xu J. Wang W. Ludeman M. Cheng K. Hayami T. Lotz J.C. Kapila S. Chondrogenic differentiation of human mesenchymal stem cells in three-dimensional alginate gels. Tissue Eng Part A. 2008;14:667. doi: 10.1089/tea.2007.0272. [DOI] [PubMed] [Google Scholar]
- 19.Sekiya I. Vuoristo J.T. Larson B.L. Prockop D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci USA. 2002;99:4397. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry F. Boynton R.E. Liu B. Murphy J.M. Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp Cell Res. 2001;268:189. doi: 10.1006/excr.2001.5278. [DOI] [PubMed] [Google Scholar]
- 21.Boeuf S. Steck E. Pelttari K. Hennig T. Buness A. Benz K. Witte D. Sültmann H. Poustka A. Richter W. Subtractive gene expression profiling of articular cartilage and mesenchymal stem cells: serpins as cartilage-relevant differentiation markers. Osteoarthritis Cartilage. 2007;16:48. doi: 10.1016/j.joca.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Song L. Webb N.E. Song Y. Tuan R.S. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells. 2006;24:1707. doi: 10.1634/stemcells.2005-0604. [DOI] [PubMed] [Google Scholar]
- 23.Mauck R.L. Wang C.C. Oswald E.S. Ateshian G.A. Hung C.T. The role of cell seeding density and nutrient supply for articular cartilage tissue engineering with deformational loading. Osteoarthritis Cartilage. 2003;11:879. doi: 10.1016/j.joca.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Farndale R.W. Buttle D.J. Barrett A.J. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 25.Stegemann H. Stalder K. Determination of hydroxyproline. Clin Chim Acta. 1967;18:267. doi: 10.1016/0009-8981(67)90167-2. [DOI] [PubMed] [Google Scholar]
- 26.Neuman R.E. Logan M.A. The determination of hydroxyproline. J Biol Chem. 1950;184:299. [PubMed] [Google Scholar]
- 27.Huang A.H. Yeger-McKeever M. Stein A. Mauck R.L. Tensile properties of engineered cartilage formed from chondrocyte- and MSC-laden hydrogels. Osteoarthritis Cartilage. 2008;16:1074. doi: 10.1016/j.joca.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumacher B.L. Hughes C.E. Kuettner K.E. Caterson B. Aydelotte M.B. Immunodetection and partial cDNA sequence of the proteoglycan, superficial zone protein, synthesized by cells lining synovial joints. J Orthop Res. 1999;17:110. doi: 10.1002/jor.1100170117. [DOI] [PubMed] [Google Scholar]
- 29.Gleghorn J.P. Jones A.R. Flannery C.R. Bonassar L.J. Boundary mode frictional properties of engineered cartilaginous tissues. Eur Cell Mater. 2007;14:20. doi: 10.22203/ecm.v014a02. discussion 8. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto K. Noshiro M. Ohno S. Kawamoto T. Satakeda H. Akagawa Y. Nakashima K. Okimura A. Ishida H. Okamoto T. Pan M. Shen W. Yan W. Kato Y. Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochim Biophys Acta. 1997;1355:303. doi: 10.1016/s0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- 31.Ohno S. Doi T. Fujimoto K. Ijuin C. Tanaka N. Tanimoto K. Honda K. Nakahara M. Kato Y. Tanne K. RGD-CAP (betaig-h3) exerts a negative regulatory function on mineralization in the human periodontal ligament. J Dent Res. 2002;81:822. doi: 10.1177/154405910208101205. [DOI] [PubMed] [Google Scholar]
- 32.Ohno S. Doi T. Tsutsumi S. Okada Y. Yoneno K. Kato Y. Tanne K. RGD-CAP ((beta)ig-h3) is expressed in precartilage condensation and in prehypertrophic chondrocytes during cartilage development. Biochim Biophys Acta. 2002;1572:114. doi: 10.1016/s0304-4165(02)00286-6. [DOI] [PubMed] [Google Scholar]
- 33.Pacifici M. Koyama E. Iwamoto M. Gentili C. Development of articular cartilage: what do we know about it and how may it occur? Connect Tissue Res. 2000;41:175. doi: 10.3109/03008200009005288. [DOI] [PubMed] [Google Scholar]
- 34.Yamane S. Cheng E. You Z. Reddi A.H. Gene expression profiling of mouse articular and growth plate cartilage. Tissue Eng. 2007;13:2163. doi: 10.1089/ten.2006.0431. [DOI] [PubMed] [Google Scholar]
- 35.Pelttari K. Winter A. Steck E. Goetzke K. Hennig T. Ochs B.G. Aigner T. Richter W. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54:3254. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 36.Jukes J.M. Both S.K. Leusink A. Sterk L.M. van Blitterswijk C.A. de Boer J. Endochondral bone tissue engineering using embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:6840. doi: 10.1073/pnas.0711662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang A.H. Motlekar N.A. Stein A. Diamond S.L. Shore E.M. Mauck R.L. High-throughput screening for modulators of mesenchymal stem cell chondrogenesis. Ann Biomed Eng. 2008;36:1909. doi: 10.1007/s10439-008-9562-4. [DOI] [PubMed] [Google Scholar]
- 38.Connelly J.T. Garcia A.J. Levenston M.E. Interactions between integrin ligand density and cytoskeletal integrity regulate BMSC chondrogenesis. J Cell Physiol. 2008;217:145. doi: 10.1002/jcp.21484. [DOI] [PubMed] [Google Scholar]
- 39.Mouw J.K. Connelly J.T. Wilson C.G. Michael K.E. Levenston M.E. Dynamic compression regulates the expression and synthesis of chondrocyte-specific matrix molecules in bone marrow stromal cells. Stem Cells. 2007;25:655. doi: 10.1634/stemcells.2006-0435. [DOI] [PubMed] [Google Scholar]
- 40.Huang A.H. Farrel M.J. Kim M. Mauck R.L. Long-term dynamic loading improves the mechanical properties of chondrogenic mesenchymal stem cell-laden hydrogel. Eur Cell Mat. 2010;19:72. doi: 10.22203/ecm.v019a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.