Abstract
C-terminal binding proteins (CtBPs) are cellular corepressors that are targeted by adenovirus E1A. A conserved motif of E1A (PLDLS) interacts with an N-terminal hydrophobic cleft of CtBPs. Many cellular cofactors also interact with CtBPs through PLDLS-like motifs. E1A interaction with CtBP2 changed the composition of the CtBP2 protein complex and enhanced CtBP2 acetylation. We have identified a mutant of CtBP2 (M48A) that fails to interact with cellular cofactors while interacting normally with E1A. Other cleft mutations in CtBP2 affected interaction of both cellular cofactors and E1A. The M48A mutant did not repress the cellular E-cadherin promoter but inhibited transactivation mediated by the E1A N-terminal region through interaction with the E1A PLDLS motif. In vitro, E1A enhanced CtBP2 acetylation by p300 via a mechanism involving dissociation of acetylated CtBP2 from p300. E1A enhanced nuclear localization of CtBP1 as well as a cytoplasmically localized acetylation-deficient mutant of CtBP2 (3KR-CtBP2) through PLDLS-dependent interaction. Chromatin immunoprecipitation assays revealed presence of CtBP2 on E-cadherin and c-fos promoters. While E1A did not significantly alter targeting of CtBP2 to the E-cadherin and c-fos promoters, it dramatically enhanced promoter targeting of 3KR-CtBP2. Our results raise a possibility that E1A may gain access to cellular promoters through PLDLS-dependent interaction with CtBPs.
Keywords: E1A, CtBP, p300, acetylation, E-cadherin, c-fos
Introduction
The adenovirus E1A proteins are expressed during the early phase of the viral life cycle and induce cell proliferation. This E1A activity is mediated by interaction with various cellular growth regulatory proteins. The N-terminal region of E1A along with the conserved region 1 (CR1) interacts with histone acetyl transferases (HATs) to modulate chromatin modification and transcription, whereas CR2 interacts with the tumor suppressor protein pRb and related proteins, p130 and p107. Through these interactions, E1A exon 1 drives cell cycle into the S-phase and cooperates with other oncogenes to transform primary cells. The C-terminal CR4 domain of E1A (exon 2) contains the conserved PLDLS motif (Schaeper et al., 1995) that interacts with C-terminal binding proteins (CtBPs). Mutations of E1A that encompass the PLDLS-motif result in enhanced Ras cooperative transformation (Subramanian et al., 1989; Boyd et al., 1993; Schaeper et al., 1995). The mechanism by which CR4 modulates oncogenic transformation in conjunction with CtBP remains an active area of investigation.
In the vertebrates, there are two highly related CtBP homologs, CtBP1 and CtBP2. They primarily differ at a short N-terminal region. They are functionally redundant for a number of developmental processes and are unique with regard to certain others (Hildebrand and Soriano, 2002). CtBPs function predominantly as transcriptional corepressors (Chinnadurai, 2002). They are structurally related to D2-hydroxy acid dehydrogenases and bind to nicotinamide adenine dinucleotides (NAD(H)) that promote their dimerization. CtBPs link DNA-binding repressors and histone-modifying enzymes to mediate transcriptional repression. A nuclear protein complex assembled by CtBP1 contained DNA-binding proteins such as ZEB and various enzymatic constituents such as histone deacetylases (HDAC1/2), histone methyl transferases (HMTases) that mediate deacetylation and methylation of histone H3-Lys9 (H3-K9), and lysine-specific demethylase 1 (Shi et al., 2003).
Structural analysis of CtBP1 has revealed that the N-terminal region forms a hydrophobic cleft, which in conjunction with the C-terminal region constitutes the major cofactor recruitment site (designated here as PLDLS-binding cleft) (Kumar et al., 2002; Nardini et al., 2003; Lundblad, 2006). More recently, a second protein interaction surface groove has been identified which interacts with an RRT motif (Quinlan et al., 2006). While DNA-binding proteins such as ZEB1/2 interact with CtBP primarily through PLDLS-like motifs (Postigo and Dean, 1999), factors such as Znf217 interact through both PLDLS-like and RRT motifs (Quinlan et al., 2006). The precise mode of interaction of different DNA-binding factors and factors that mediate repression by CtBP remains to be elucidated. Since viral CtBP-interacting proteins such as E1A would be expected to have an evolutionary advantage over the cellular PLDLS-motif proteins, E1A is an excellent tool to probe the assembly of CtBP cofactors.
Among the various CtBP target promoters, the E-cadherin (E-cad) promoter has been better studied (Grooteclaes and Frisch, 2000; Shi et al., 2003). CtBP-dependent recruitment of E-box repressors such as ZEB1/2 and enzymes that mediate H3-K9 deacetylation (HDAC1/2) and methylation (HMTases, G9a-GLP) has been linked to repression of the E-cad promoter (Shi et al., 2003). Similarly, the c-fos promoter has been shown to be regulated by CtBP (Criqui-Filipe et al., 1999). Here, we have used these two promoters for analysis of promoter binding by CtBP and potential regulation by E1A.
Results
Differential transcriptional repression activities of CtBP2 mutants
We mutagenized the PLDLS-binding cleft of CtBP2 to identify residues that may dictate differential interaction with E1A and prototypical cellular CtBP-binding proteins. The PLDLS-binding cleft of CtBP2 is lined with several hydrophobic residues (Nardini et al., 2003). CtBP2 mutants (L35A, M48A, A58E and V72A) were generated and examined for their ability to inhibit transactivation by the E1A CR1 through interaction with the E1A PLDLS motif (Zhao et al., 2006a). CtBP-null mouse cells MEF90 (Hildebrand and Soriano, 2002) were transfected with the reporter (pG5-MLP-Luc), Gal4-E1AΔ91-199 (Sollerbrant et al., 1996) and various CtBP2 constructs. Luciferase assay showed that mutant A58E did not repress while M48A repressed to the same extent as CtBP2 wild type (Figure 1a). Mutant L35A had a lower level of repression. Mutant V72A behaved like CtBP2 wild type in this and in other related assays (data not shown) and hence was not pursued further. To examine the effects on the E-cad promoter, the reporter pE-cad-Luc (Shi et al., 2003) was co-transfected with various CtBP2 constructs (Figure 1b). All three mutants, A58E, L35A and M48A were deficient for repression. Thus, mutant M48A of CtBP2 is unique in that it retains the ability to suppress the E1A CR1-mediated transactivation but was defective in repressing the E-cad promoter. In contrast, the A58E mutation abolished repression of both the E-cad promoter and the E1A CR1-mediated transactivation.
Figure 1.
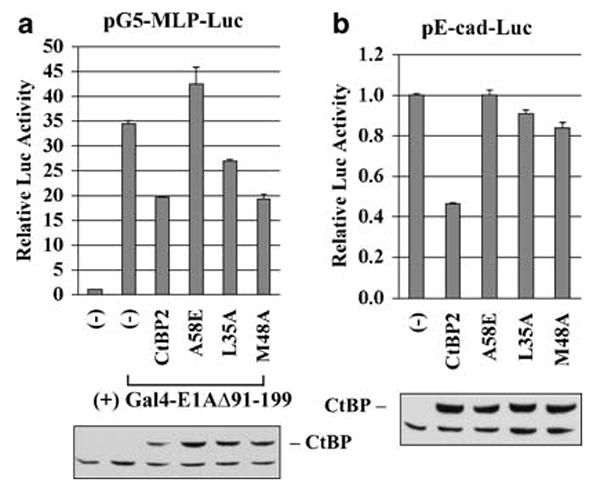
Differential transcriptional activities of CtBP2 mutants. (a) Effect on E1A CR1-mediated transactivation. MEF90 cells were transfected with pG5-MLP-Luc, phRL-0 internal control, pGal4-E1AΔ91-199 and different CtBP constructs as indicated. Dual-luciferase assays were performed. Lower panel: western blot with the HA antibody for Flag-HA-tagged CtBP proteins. (b) Effect on E-cadherin promoter activity. The reporter assays were performed using pE-cad-Luc.
Interaction of CtBP2 mutants with E1A and cellular factors
We examined the ability of the above-described and additional mutants to interact with E1A and cellular factors by transfection of HeLa cells followed by coimmunoprecipitation (with the Flag antibody) and western blot analysis (Figure 2a). The efficiency of interaction of ZEB1 (hereafter referred as ZEB), CoREST and HDAC2 was similar for CtBP2 (lane 2), 3KR-CtBP2 (lane 3), ΔN-CtBP2 (lane 4) and CtBP1 (lane 6). In contrast, the cleft mutants A58E (lane 5), L35A (lane 7) and M48A (lane 8) were defective in interaction with ZEB and HDAC2 (Figure 2a). These mutations also abolished interaction of G9a (not shown). The interaction between mutant M48A and CoREST was greatly diminished. Thus, mutations in Leu35, Met48 and Ala58 of the CtBP2 cleft strongly prevented association of cellular factors such as ZEB, CoREST, HDAC2 and G9a.
Figure 2.
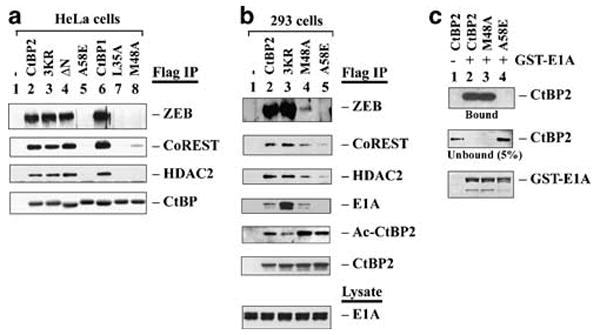
Interaction of CtBP with cellular factors and E1A. (a) Interaction with cellular factors. HeLa cells were transfected with different CtBP constructs for 24 h, and cell lysates were immunoprecipitated with the Flag antibody and examined by western blots using indicated antibodies. (b) Interaction with E1A and cellular factors. A total of 293 cells were transfected with indicated CtBP plasmids, and CtBP complexes were analysed as in (a). Acetylated CtBP2 (Ac-CtBP2) was detected by the pan-Ac-Lys antibody. (c) GST pull-down assay. GST-E1A was bound to glutathione-agarose beads, and incubated with H6-tagged CtBP2, M48A or A58E mutants. CtBP2 proteins in the bound and unbound fractions were analysed by western blot using the CtBP2 antibody. GST-E1A was detected by Coomassie blue staining.
Since M48A-CtBP2 repressed E1A CR1-mediated transcriptional activation (Figure 1a), we examined its interaction with E1A endogenously expressed in 293 cells. As shown (Figure 2b), the pattern of ZEB binding by CtBP2 and mutants was comparable as that in HeLa cells (Figure 2a). Interestingly, in 293 cells, A58E (lane 5) retained a low level of binding to both CoREST and HDAC2, and M48A (lane 4) bound to CoREST and HDAC2 more weakly than wild type CtBP2 (lane 2). Importantly, CtBP2 (lane 2) and M48A (lane 4) bound to endogenous E1A at comparable levels, while A58E (lane 5) binding to E1A was undetectable. In contrast, 3KR-CtBP2 (lane 3) bound to E1A more efficiently than CtBP2 or M48A. These results suggest that M48A mutant retained the ability to interact with E1A while A58E mutant did not. Similar pattern of interaction of M48A and A58E with transiently expressed E1A was obtained in HeLa cells (data not shown).
To ascertain the effects of M48A and A58E mutations on interaction with E1A, a glutathione S-transferase (GST) pull-down assay was performed using GST-E1A and H6-tagged CtBP2 (Figure 2c). As shown, GST-E1A bound the wild type CtBP2 (lane 2) and M48A (lane 3) efficiently while no binding to A58E was detected (lane 4). Thus, M48A interacts with E1A both in vitro and in vivo, while A58E fails to interact under both conditions.
E1A-enhanced CtBP2 acetylation and dissociation of Ac-CtBP2 from p300
We previously reported that E1A facilitates CtBP2 interaction with p300 and enhances CtBP2 acetylation (Zhao et al., 2006b). To examine the mechanism of E1A-mediated enhancement of CtBP2 acetylation, we performed an in vitro acetylation/p300-binding assay using partially purified Flag-HA-tagged p300, GST-E1A and H6-CtBP2 (Figure 3a). p300 was first bound to the Flag antibody beads, and then used to acetylate H6-CtBP2 in the presence or absence of GST-E1A. Ac-CtBP2 that remained bound to the immobilized p300 as well as in the supernatant was examined by western blot with the pan-Ac-Lys antibody (Figure 3b, second panel). As shown, a strong Ac-CtBP2 signal was associated with p300 only when both CtBP2 and Ac-CoA were present (lane 5). When both GST-E1A and CtBP2 were present, a stronger Ac-CtBP2 was detected (lane 7). GST-E1A together with the 3KR-CtBP2 mutant resulted in a lower level of Ac-3KR-CtBP2 (lane 8). GST-E1A(K239A) also enhanced CtBP2 acetylation (lane 9).
Figure 3.
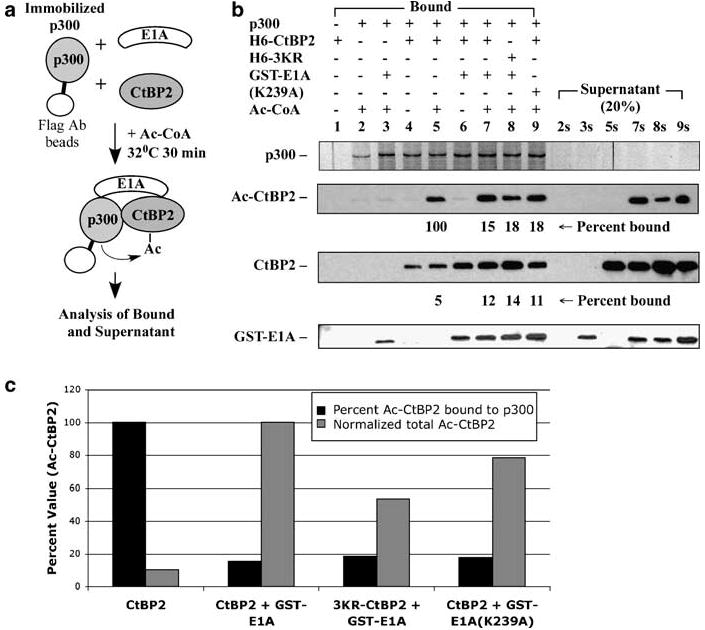
Effect of E1A-induced CtBP2 acetylation on CtBP2 binding to p300. (a) Scheme of the procedure. Flag-HA-p300 was bound to Flag antibody beads, and incubated with H6-tagged CtBP2 and 3KR-CtBP2 in the presence or absence of GST-E1A and Ac-CoA. (b) Binding of CtBP2 with p300. The fraction bound to the p300-containing beads and the supernatant fraction (20% loaded compared to the bound fraction) were examined by western blots as indicated. CtBP2 and GST-E1A were detected by western blots. Acetylated CtBP2 (Ac-CtBP2) was detected with the pan-Ac-Lys antibody. p300 was detected by silver staining. (c) Quantification of CtBP2 binding with p300. Total amount of Ac-CtBP2 was normalized to the level of Ac-CtBP2 in the presence of GST-E1A (lanes 4 and 4s).
Examination of the reaction supernatants revealed a striking difference between CtBP2 acetylation in the presence and absence of GST-E1A (Figure 3b, lanes 2s–9s). In the absence of GST-E1A, little, if any, Ac-CtBP2 was detected in the supernatant (lane 5s) despite the presence of Ac-CtBP2 in the bound fraction (lane 5). In contrast, when GST-E1A was present, abundant Ac-CtBP2 was detected in the supernatant (lane 7s). Similarly, significant amounts of Ac-3KR-CtBP2 in the presence of GST-E1A (lane 8s) and Ac-CtBP2 in the presence of GST-E1A(K239A) (lane 9s) were detected in the supernatant fractions. Quantification of the Ac-CtBP2 signals suggested that in the absence of GST-E1A, most of the Ac-CtBP2 remained bound to p300, whereas in the presence of GST-E1A or GST-E1A(K239A), only less than 20% of Ac-CtBP2 remained associated with p300 (Figures 3b and c). Comparison of the total amounts of Ac-CtBP2 suggested that GST-E1A enhanced CtBP2 acetylation by approximately tenfold (Figure 3c). These results support a model that interaction between E1A and CtBP2 promotes acetylation of CtBP2 and subsequent dissociation of Ac-CtBP2 from p300. This might be a dynamic ‘loading and unloading’ mechanism for E1A-enhanced CtBP2 acetylation.
While the E1A N-terminal region mediate interaction with p300, the C-terminal PLDLS motif mediates interaction with CtBP. In vitro binding assays (Figure 4a) suggested that while GST-E1AΔ2-74 interacted well with CtBP2 (lane 2), GST-E1A-C* (with a PLDLS→PLASS mutation) failed to interact (lane 3). During in vitro acetylation and p300-binding assays (Figure 4b), GST-E1A (lane 5) enhanced CtBP2 acetylation and a larger fraction of the acetylated CtBP2 was found in the reaction supernatant. In contrast, neither GST-E1AΔ2-74 (lane 6) nor GST-E1A-C* (lane 7) enhanced CtBP2 acetylation. These results support the conclusion that E1A-mediated enhancement of CtBP2 acetylation requires simultaneous interaction of E1A with p300 and CtBP2.
Figure 4.
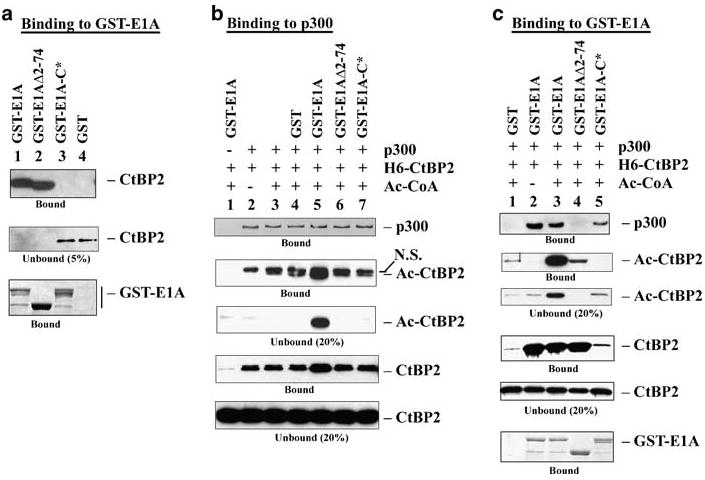
Effect of E1A mutations on CtBP2 acetylation. (a) In vitro binding of CtBP2 and E1A mutants. In vitro binding was performed by the GST pull-down assay as described in Figure 2c. CtBP2 was detected by western blot and GST-E1A detected by Coomassie blue staining. (b) Effects of E1A on CtBP2 acetylation and association with p300. The assay was performed under conditions described in Figure 3, and various proteins were detected by western blots with the antibodies indicated in Figure 3. p300 was detected with the Flag antibody. (c) Interaction of GST-E1A with acetylated CtBP2. The binding studies were carried out under conditions described in Figure 4b, except GST-E1A proteins were first bound to glutathione beads. p300 was detected with the p300 monoclonal antibody and GST-E1A was detected by Coomassie blue staining.
To examine the effects of CtBP2 acetylation on interaction with E1A, an in vitro assay was performed (Figure 4c). GST-E1A associated with acetylated CtBP2 efficiently (lane 3), although acetylated CtBP2 was also detected in the unbound fraction. GST-E1AΔ2-74 (lane 4) bound to CtBP2 normally but the bound CtBP2 was only acetylated at a low level. Correspondingly, GST-E1AΔ2-74 (lane 4) did not bind p300. The GST-E1A-C* mutant (lane 5) did not bind CtBP2 but interacted well with p300. Thus, acetylated CtBP2 appears to be capable of interacting with E1A.
Effect of E1A on CtBP subcellular localization and promoter targeting
Transcriptional repression by CtBP has been thought to require targeting of CtBP to promoters indirectly through DNA-binding repressors. It is also presumed that E1A may relieve CtBP-mediated repression either by sequestration of free CtBP or by interfering with the activity of promoter bound CtBP. We examined CtBP2 interaction with the promoters for E-cad and c-fos by chromatin immunoprecipitation (ChIP) assays (Figure 5a). A HeLa cell line stably expressing Flag-tagged CtBP2 was used together with control HeLa cells in ChIP with the Flag antibody. As shown in Figure 5a, CtBP2 bound specifically to both E-cad and c-fos promoters.
Figure 5.
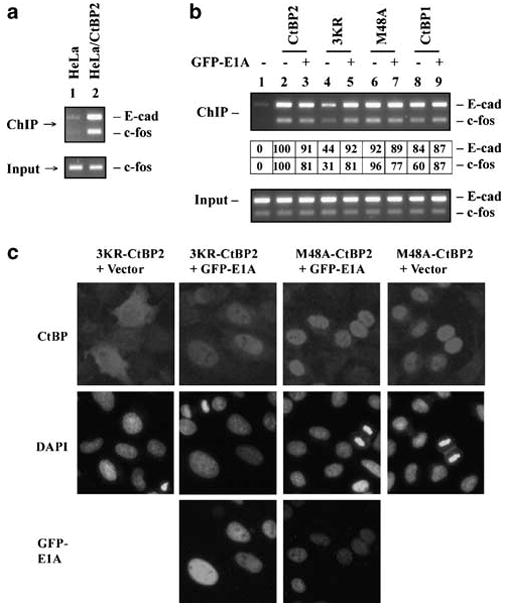
Localization of CtBP at the cellular promoters. (a) ChIP analysis of CtBP2 binding to E-cad and c-fos promoters. Lane 1, normal HeLa cells. Lane 2, HeLa cells stably expressing Flag-HA-tagged CtBP2. ChIP was performed with the Flag antibody. PCR reactions for E-cad and c-fos promoters (370 and 190 bp, respectively) were carried out in the same reactions. (b) Localization of CtBP at E-cad and c-fos promoters. ChIP analysis was performed with HeLa cells that were transiently transfected with various CtBP constructs in the presence or absence of GFP-E1A. For the ChIP data table (middle of panel), signal in lane 1 was subtracted from other lanes and the signal in lane 2 was normalized to 100 for both E-cad and c-fos. (c) Effect of E1A on subcellular localization of CtBP2 mutants. Immunofluorescence analysis was carried out using HeLa cells transfected with the indicated plasmids. The cells were stained with Flag-Cy3 antibody to localize CtBP proteins 24 h after transfection.
To examine promoter binding by various CtBP proteins, HeLa cells were transfected with CtBP constructs in the absence or presence of GFP-E1A (Figure 5b). ChIP assays revealed that all CtBP proteins, except 3KR-CtBP2 (lane 4) were bound to the E-cad and c-fos promoters efficiently. Strikingly, E1A only slightly inhibited promoter binding by CtBP2 (lane 3) and M48A-CtBP2 (lane 7). In contrast, E1A significantly enhanced promoter targeting by 3KR-CtBP2 (compare lanes 4 and 5). The effect of E1A on 3KR-CtBP2 may reflect enhanced nuclear localization of 3KR-CtBP2 by E1A (Figure 5c). E1A also did not reduce promoter localization of CtBP1 (lane 9). Instead there was a modest increase in localization of CtBP1 with the c-fos promoter. The efficient localization of M48A-CtBP2 to the promoters of both E-cad and c-fos was in sharp contrast to the inability of M48A-CtBP2 to bind the prototypical DNA-binding repressor ZEB (Figure 2) suggesting alternate mechanisms/factors for localization of M48A to these promoters (see ‘Discussion’).
Interaction with nuclear cofactors has been suggested as one of the mechanisms for nuclear localization of CtBP1 (Bergman et al., 2006; Verger et al., 2006). As reported earlier (Zhao et al., 2006b), CtBP1 was localized predominantly in the nucleus with partial cytoplasmic localization (Figure 6a). Coexpression of GFP-E1A with CtBP1 resulted in enhanced nuclear localization of CtBP1. In contrast, coexpression of CtBP1 with GFP-E1A-C* had a smaller effect on the nuclear localization of CtBP1. Surprisingly, coexpression of CtBP1 with the GFP-E1A(K239A) mutant (which does not have a functional nuclear localization signal, (Madison et al., 2002) resulted in cytoplasmic retention of CtBP1. In contrast, coexpression of GFP-E1A(K239A) with CtBP2 enhanced nuclear localization of GFP-E1A(K239A). The ability of GFP-E1A and its mutants to influence the subcellular localization of CtBP1 was correlated with their interaction with CtBP1 through the PLDLS-motif (Figure 6b). In 293 cells, which express endogenous E1A, both CtBP1 and CtBP2 were localized predominantly in the nucleus. We previously reported that in E1A-negative cell lines (HeLa and A549 cells), CtBP2 is nuclear while CtBP1 displays a low level of cytoplasmic localization (Zhao et al., 2006b). Thus, it seems that the endogenous E1A in 293 cells may facilitate nuclear localization of CtBP1.
Figure 6.
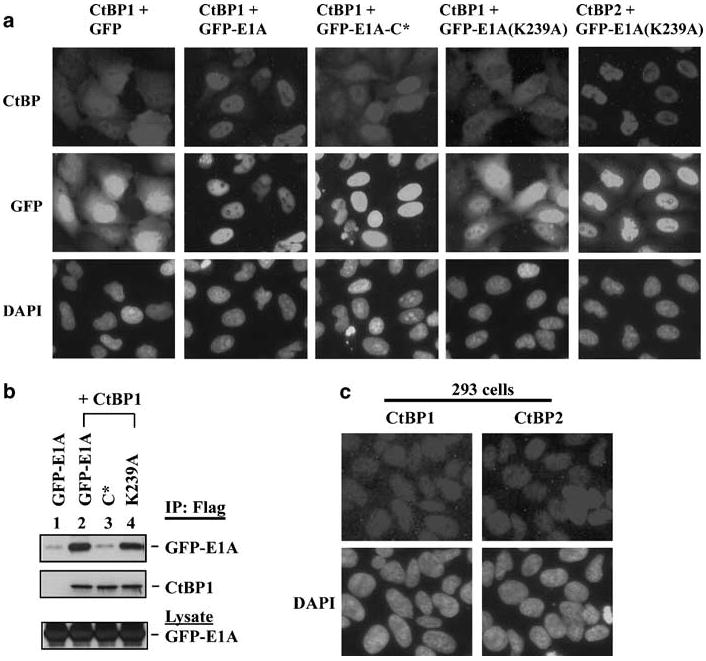
Effect of E1A on subcellular localization of CtBP1. (a) Immunofluorescence analysis. HeLa cells were transfected with different GFP-E1A plasmids along with CtBP1 or CtBP2. Transfected cells were stained with Flag-Cy3 antibody for the localization of CtBP proteins as in Figure 5. (b) Coimmunoprecipitation of CtBP1 and GFP-E1A. HeLa cells were co-transfected with CtBP1 and different GFP-E1A mutants as indicated. Cell lysates were immunoprecipitated with the Flag antibody and analysed by western blots with the Flag antibody to detect CtBP1 or GFP antibody to detect GFP-E1A. (c) Subcellular localization of CtBP1 and CtBP2 in 293 cells. All the cells were examined by indirect immunofluorescence staining with mAbs against CtBP1 or CtBP2.
Discussion
The PLDLS-like motif is critical for interaction of E1A and several cellular factors with CtBP. It is possible that E1A might have a selective advantage over cellular proteins in targeting CtBP to meet acute needs during viral replication. The amino acid residues in CtBP2, Leu35, Met48 and Ala58 appear to be critical for recruitment of cellular factors such as ZEB, G9a, CoREST and HDAC (Figure 2). In contrast, Met48 is not critical for interaction of E1A. These results are consistent with the observation that M48A-CtBP2 was defective in repressing the E-cad promoter while preserving the ability to interact with E1A to inhibit transactivation mediated by the E1A N-terminal domain (Figure 1). Thus, M48A mutation serves as a useful tool to distinguish between CtBP2 interaction with E1A and with cellular cofactors.
In addition to the critical roles of the PLDLS-motifs, other factors that are simultaneously recruited into the complex might also influence the relative affinity for CtBP. It was reported that interaction of E1A with CtBP1 was reduced by p300-mediated acetylation of Lys239 of E1A (Zhang et al., 2000). However, the cytoplasmic localization of K239A-E1A may complicate the interpretation (Madison et al., 2002). A recent study using K239-acetylated E1A peptides suggested that Lys239 acetylation may modestly reduce the affinity for CtBP in vitro by inducing structural changes (Molloy et al., 2006).
Our present results (Figure 3) are consistent with the interpretation that E1A-facilitated, p300-mediated CtBP2 acetylation may lead to dissociation of CtBP2 from p300. Dissociation of Ac-CtBP2 from p300 in the presence of E1A may be a prerequisite for efficient CtBP2 acetylation. E1A-enhanced CtBP2 acetylation may help to inhibit normal CtBP2 functions. Alternatively, Ac-CtBP2 may acquire novel functions through altered interactions with cofactors. Identification of a specific promoter regulated by CtBP2 may facilitate elucidation of the role of acetylation in regulating CtBP2 functions.
The weaker interaction of the CtBP2 and M48A-CtBP2 with E1A in 293 cells as compared to the 3KR-CtBP2 mutant (Figure 2b) might indicate that CtBP2 acetylation destabilizes CtBP2 interaction with E1A. However, in vitro acetylation/E1A-binding assay (Figure 4c) suggested that E1A is capable of binding to acetylated CtBP2. However, due to the limitations of the in vitro assay, the effect of CtBP2 acetylation on E1A interaction remains inconclusive. Since the level of CtBP2 acetylation in vivo reflects a dynamic balance between acetylation and deacetylation, the effects of CtBP2 acetylation on in vivo CtBP2 interaction with cofactors may also be transient.
Despite efficient CtBP interaction with E1A, the consequences of such interaction remain to be fully explored. Our results that E1A recruits p300 into the CtBP2 complex and enhances CtBP2 acetylation (Zhao et al., 2006a; current study) suggest potential effects of E1A on chromatin remodeling and gene expression through interaction with CtBP. Specific CtBP2 binding to cellular E-cad and c-fos promoters was demonstrated by ChIP assays (Figure 5). Coexpression of CtBP2 with E1A did not significantly affect CtBP2 localization at these promoters. Coexpression of E1A enhanced the nuclear localization of 3KR-CtBP2 as well as its binding to promoters (Figure 5), suggesting that E1A may be capable of associating with the promoters by targeting CtBP. This notion is consistent with a previous observation that E1A (12S) and associated HATs were localized at cdc6 and cyclin A promoters (Ghosh and Harter, 2003). This hypothesis contrasts with the belief that E1A displaces CtBP from the associated repressors and promoters. The observation that M48A-CtBP2 was still localized to the promoters was surprising since M48A-CtBP2 was deficient in interaction with cellular repressors such as ZEB. It may be possible that M48A-CtBP2 localizes at the promoters through alternative mechanisms including interaction with RRT-motif-containing repressors such as Znf217 (Quinlan et al., 2006).
We observed that E1A/CtBP1 coexpression results in enhanced CtBP1 nuclear localization (Figure 6). The observation that E1A(K239A) mutant blocks nuclear localization of CtBP1 also suggests that CtBP1 nuclear localization is at least partially determined by the cellular cofactors that bind to CtBP1 through the PLDLS-like motif. The nuclear localization of endogenous CtBP1 in 293 cells suggested that E1A may target CtBP1 to the nucleus in these cells. Since E1A is transiently produced at a relatively high level during adenovirus replication, it is likely that this E1A might also facilitate nuclear localization of the endogenous CtBP1 as well as a splice variant of CtBP2 (CtBP2-S) that lacks the N-terminal nuclear retention signal (Verger et al., 2006).
Materials and methods
Protein expression
CtBP, H6-CtBP, GFP-E1A and GST-E1A expression constructs were prepared as described previously (Zhao et al., 2006a, b).
In vitro CtBP2 acetylation and p300-binding assays
Baculovirus-expressed p300 was bound to Flag antibody beads, and used for in vitro acetylation assay as described earlier (Zhao et al., 2006a). The p300-associated proteins were eluted with the HAT buffer containing 1μg/μl of the Flag peptide.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described previously using the Flag antibody (Zhao et al., 2006a). The E-cad primer set (Shi et al., 2003) and the c-fos primer set (N202 GCACTG-CACCCTCGGTGTTGGCTG and N203 GCAGTTCCTGTCTCAGAGGTCTCG) were used simultaneously.
Acknowledgments
We thank M Kuppuswamy for critical reading of the manuscript and R Subramanian for technical assistance. This study was supported by research grants CA-84941 and CA-33616.
References
- Bergman LM, Morris L, Darley M, Mirnezami AH, Gunatilake SC, Blaydes JP. Role of the unique N-terminal domain of CtBP2 in determining the subcellular localisation of CtBP family proteins. BMC Cell Biol. 2006;7:35. doi: 10.1186/1471-2121-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh MK, Harter ML. A viral mechanism for remodeling chromatin structure in G0 cells. Mol Cell. 2003;12:255–260. doi: 10.1016/s1097-2765(03)00225-9. [DOI] [PubMed] [Google Scholar]
- Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, et al. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Lundblad JR. Structural determinants of CtBP function. In: Chinnadurai G, editor. CtBP Family Proteins. Landes Bioscience and Springer Science-Business Media; 2006. pp. 83–92. [Google Scholar]
- Madison DL, Yaciuk P, Kwok RP, Lundblad JR. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J Biol Chem. 2002;277:38755–38763. doi: 10.1074/jbc.M207512200. [DOI] [PubMed] [Google Scholar]
- Molloy D, Mapp KL, Webster R, Gallimore PH, Grand RJ. Acetylation at a lysine residue adjacent to the CtBP binding motif within adenovirus 12 E1A causes structural disruption and limited reduction of CtBP binding. Virology. 2006;355:115–126. doi: 10.1016/j.virol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Nardini M, Spano S, Cericola C, Pesce A, Massaro A, Millo E, et al. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, et al. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–8172. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Sollerbrant K, Chinnadurai G, Svensson C. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 1996;24:2578–2584. doi: 10.1093/nar/24.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian T, La Regina M, Chinnadurai G. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene. 1989;4:415–420. [PubMed] [Google Scholar]
- Verger A, Quinlan KG, Crofts LA, Spano S, Corda D, Kable EP, et al. Mechanisms directing the nuclear localization of the CtBP family proteins. Mol Cell Biol. 2006;26:4882–4894. doi: 10.1128/MCB.02402-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Subramanian T, Chinnadurai G. Changes in C-terminal binding protein 2 (CtBP2) corepressor complex induced by E1A and modulation of E1A transcriptional activity by CtBP2. J Biol Chem. 2006a;281:36613–36623. doi: 10.1074/jbc.M603550200. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J Biol Chem. 2006b;281:4183–4189. doi: 10.1074/jbc.M509051200. [DOI] [PubMed] [Google Scholar]


