Abstract
Several genome-wide association studies identified the chr15q25.1 region, which includes three nicotinic cholinergic receptor genes (CHRNA5-B4) and the cell proliferation gene (PSMA4), for its association with lung cancer risk in Caucasians. A haplotype and its tagging single nucleotide polymorphisms (SNPs) encompassing six genes from IREB2 to CHRNB4 were most strongly associated with lung cancer risk (OR = 1.3; P < 10−20). In order to narrow the region of association and identify potential causal variations, we performed a fine-mapping study using 77 SNPs in a 194 kb segment of the 15q25.1 region in a sample of 448 African-American lung cancer cases and 611 controls. Four regions, two SNPs and two distinct haplotypes from sliding window analyses, were associated with lung cancer. CHRNA5 rs17486278 G had OR = 1.28, 95% CI 1.07–1.54 and P = 0.008, whereas CHRNB4 rs7178270 G had OR = 0.78, 95% CI 0.66–0.94 and P = 0.008 for lung cancer risk. Lung cancer associations remained significant after pack-year adjustment. Rs7178270 decreased lung cancer risk in women but not in men; gender interaction P = 0.009. For two SNPs (rs7168796 A/G and rs7164594 A/G) upstream of PSMA4, lung cancer risks for people with haplotypes GG and AA were reduced compared with those with AG (OR = 0.56, 95% CI 0.38–0.82; P = 0.003 and OR = 0.73, 95% CI 0.59–0.90, P = 0.004, respectively). A four-SNP haplotype spanning CHRNA5 (rs11637635 C, rs17408276 T, rs16969968 G) and CHRNA3 (rs578776 G) was associated with increased lung cancer risk (P = 0.002). The identified regions contain SNPs predicted to affect gene regulation. There are multiple lung cancer risk loci in the 15q25.1 region in African-Americans.
INTRODUCTION
Lung carcinoma is the leading cause of cancer death in the USA with African-Americans having the highest incidence (1–4). Genetic susceptibility is one possible factor influencing this disparity (5). Recently, three genome-wide association studies in Caucasian populations have implicated the chr15q25.1 region in lung cancer risk (6–8). How genomic variations in this region might affect lung cancer risk in the African-American population is poorly understood because single nucleotide polymorphisms (SNPs) implicated in Caucasians have very different allele frequencies and linkage patterns in African-Americans (9).
The candidate chr15q25.1 region includes six genes. Three of these genes, CHRNA5, CHRNA3 and CHNB4, are nicotinic receptor subunit genes, whereas PSMA4, a proteosome subunit encoding gene, has recently been associated with in vitro lung cancer cell proliferation and apoptosis (10). The region also contains IREB2, an iron responsive element-binding protein, and LOC123688 (an aminoglicoside phototransferase domain). The effects of variants in this region on lung cancer risk are multifaceted (9–19). The very high linkage disequilibrium (LD) among these SNPs in Caucasians has complicated identifying the causal variants. Among African-Americans, SNPs within chr15q25.1 have low linkage compared with Caucasians (9), which means that studies of African-Americans could provide the opportunity to refine the location of lung cancer risk alleles in this region. Consequently, we investigated the association of 77 SNPs in the chr15q25.1 region with lung cancer in African-American cases and controls in the San Francisco Bay Area. We included appropriate adjustment for potential ancestry differences among cases and controls with an extensive panel of ancestry informative markers.
RESULTS
Demographics
Cases were more likely than controls to have smoked, smoked more pack-years (Table 1), have a lower household income and were less likely to have achieved a degree beyond high school. There was no notable or significant difference between cases and controls in distributions of age, gender or % African ancestry.
Table 1.
Demographic and smoking characteristics of the study participants, Northern California Lung Cancer Study 1998–2008
| Cases (n = 448) | Controls (n = 611) | P-valuea | |
|---|---|---|---|
| Percentage (%) | |||
| Male | 45.8 | 45.7 | 0.975 |
| Ever smoked | 92.9 | 63.7 | <0.001 |
| Household income ≥ $20 000 | 55.0 | 65.0 | 0.002 |
| Higher degree than high school | 39.4 | 55.8 | <0.001 |
| Mean ± SE | |||
| Age | 64.1 ± 0.5 | 64.6 ± 0.5 | 0.467 |
| % African ancestry | 77.8 ± 0.6 | 77.8 ± 0.5 | 0.996 |
| Pack-years smokedb | 32.9 ± 1.1 | 24.2 ± 1.3 | <0.001 |
at-Tests and chi-square tests were used to test for difference between cases and controls.
bAmong ever smokers.
Regional linkage by population
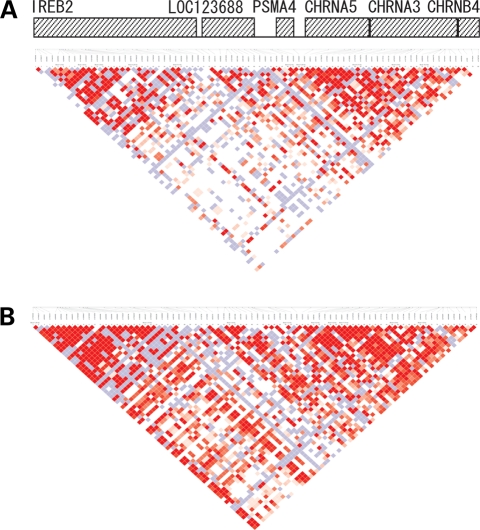
Linkage among SNPs in the 15q25.1 region was weaker among participants with ≥90% African ancestry than among those with ≤60% African ancestry (Fig. 1). Linkage between genotyped SNPs in the African-American population also was markedly lower than in the HapMap CEPH Utah residents with ancestry from northern and western Europe (CEU) population, but similar to that of the HapMap Yoruba in Ibadan, Nigeria (YRI) population (Supplementary Material, Fig. S1). Only 30 SNP pairs (1%) in African-Americans had r2 ≥ 0.80, compared with 336 SNP pairs (16%) in the HapMap CEU population and with 14 SNP pairs (0.6%) in the HapMap YRI population. This is consistent with our finding that the mean % African ancestry in both cases and controls was high, at 77.78% African ancestry.
Figure 1.
D′ linkage map of 77 chr15q25.1 SNPs in African-Americans from the Northern California Lung Cancer Study. (A) Individuals with an African ancestral percentage of ≥90%. (B) Individuals with an African ancestral percentage of ≤60%.
Single-SNP analysis
Associations with lung cancer risk: after eliminating SNPs with poor genotype cluster separation, MAF < 0.05, or control Hardy–Weinberg equilibrium (HWE) P < 0.01, 77 SNPs in 15q25.1 were analyzed with a log-additive model in relation to lung cancer risk adjusting for age, gender and % African ancestry. Four of 77 SNPs (rs7178270, P = 0.008; rs17486278, P = 0.008; rs2656068, P = 0.03; rs1504547 P = 0.04) had P < 0.05 for association with lung cancer (Supplementary Material, Table S1). However, only two of these had significant false-discovery rate (FDR) P-values (P < 0.05). These were CHRNB4 SNP rs7178270 (OR = 0.78, 95% CI 0.66–0.94, FDR P < 0.001) and CHRNA5 SNP rs17486278 (OR = 1.28, 95% CI 1.07–1.54, FDR P < 0.001). There was no statistically significant interaction between these two SNPs (P = 0.22). The association of rs17486278 with lung cancer risk was stronger using a dominant model; OR = 1.54, 95% CI 1.2–1.97, P < 5.6 × 10−4. The heterozygote T/G allele carried the highest risk (log-additive OR = 1.61 95% CI 1.24–2.08, P = 0.0003) (Supplementary Material, Table S2). Both rs17486278 and rs7178270 remained associated with lung cancer risk when log-additive analyses were pack-year adjusted (OR = 1.26, P = 0.022 and OR = 0.75, P = 0.003, respectively) (Supplementary Material, Table S3).
The analysis of single-SNP association with lung cancer risk was further stratified by family history of lung cancer, age at diagnosis, gender and ever smoking (Supplementary Material, Table S4). The minor allele of rs17486278 increased lung cancer risk most strongly in those with a family history of lung cancer and in women, whereas the minor allele of rs7178270 reduced lung cancer risk most strongly among ever smokers and women, among whom a statistically significant interaction with gender was found (P = 0.009) (Table 2 and Supplementary Material, Table S5). However, these results must be viewed cautiously because of differences in sample sizes in different risk strata. Testing for the combined effect of rs7178270 and rs17486278 by the number of risk alleles showed a statistically significant trend towards increased risk (P = 0.0016), with OR = 2.07, 95% CI 1.33–3.22, P = 0.001, for individuals with three risk alleles (Supplementary Material, Table S6).
Table 2.
Case/control associations of rs7178270 and rs17486278 with lung cancer in the Northern California Lung Cancer Study
| Cases | Controls | CHRNB4 | rs7178270 | P-valuea | P-value for gender interaction | CHRNA5 | rs17486278 | P-value | P-value for gender interaction | |
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |||||||
| SNP | C/G | G/T | ||||||||
| Minor allele | G | G | ||||||||
| MAF | 0.40 | 0.29 | ||||||||
| All | 448 | 611 | 0.78 | 0.66–0.94 | 0.008 | 1.28 | 1.07–1.54 | 0.008 | ||
| Age <50 years | 39 | 59 | 0.56 | 0.30–1.05 | 0.073 | 1.03 | 0.55–1.94 | 0.921 | ||
| Age 50 years and over | 409 | 552 | 0.81 | 0.66–1.00 | 0.045 | 1.35 | 1.09–1.67 | 0.005 | ||
| Ever smoked | 416 | 389 | 0.75 | 0.61–0.92 | 0.006 | 1.24 | 1.00–1.53 | 0.049 | ||
| History of familial LC | 86 | 73 | 0.66 | 0.40–1.10 | 0.109 | 1.80 | 1.06–3.03 | 0.028 | ||
| No history of familial LC | 338 | 510 | 0.81 | 0.66–0.98 | 0.033 | 1.23 | 1.00–1.51 | 0.048 | ||
| Female | 243 | 332 | 0.62 | 0.48–0.80 | 0.0002 | 0.009 | 1.44 | 1.12–1.86 | 0.005 | 0.192 |
| Male | 205 | 279 | 1.01 | 0.78–1.31 | 0.941 | 1.13 | 0.86–1.47 | 0.386 |
aLog-additive analysis corrected for age, gender and % African ancestry; gender-stratified analysis corrected for age and % African ancestry. Bold values indicate P ≤ 0.05 and a 95% CI range which does not overlap 1.00.
Linkage of lung cancer risk SNPs
SNPs associated with lung cancer in the single-SNP analysis were linked to a small number of SNPs in the region with a moderate r2 ≥ 0.5 (Supplementary Material, Table S7). Rs17486278 and rs2036527, just upstream of CHRNA5, have r2 = 0.59 and had an equal effect size among participants with a family history of lung cancer (OR = 1.82, CI 1.05–3.15, P = 0.032 and OR = 1.80, CI 1.06–3.03, P = 0.028, respectively). Of the 77 SNPs analyzed here, 72 were very weakly linked to CHRNB4 rs7178270 with r2 < 0.20. In the single-SNP analysis, rs578776 in CHRNA3 had a statistically significant effect on lung cancer risk OR = 1.46, CI 1.10–1.95, P = 0.008 when analyzed with a dominant model (data not shown). However, although rs578776 and rs6495307 have r2 = 0.67, rs6495307 was not associated with lung cancer risk (dominant P = 0.23, OR = 1.17).
Rs17486278 and rs16969968, a non-synonymous SNP often associated with lung cancer and nicotine dependence, were only slightly linked in African-Americans (r2 = 0.19). This contrasts sharply with linkage in the HapMap CEU population for which 24 of the 77 SNPs used in our study have r2 ≥ 0.60 with rs16969968 (and notably linkage with rs17486278 is r2 = 0.96) (Supplementary Material, Table S8). Furthermore, in HapMap CEU population, these same 24 SNPs have r2 ≥ 0.60 with rs17486278. In the African-American population, in contrast, only six SNPs have r2 ≥ 0.60 with rs16969968. None of these six SNPs shared a similar effect size for lung cancer risk as rs16969968 in the unstratified analyses.
Sliding window haplotype analysis
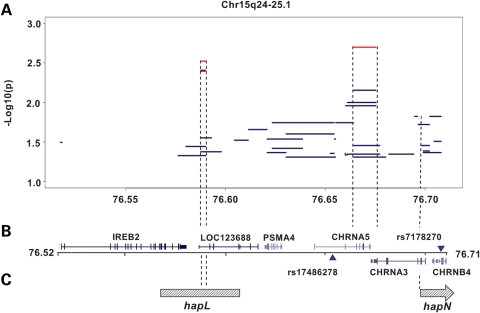
Two haplotypes determined by sliding window analyses showed statistically significant associations with lung cancer (Fig. 2 and Table 3). A two-SNP haplotype (rs7168796 and rs7164594) decreased lung cancer risk with global P = 0.004. These SNPs are between the first two exons of LOC123688, spanning chr15 at 76 587 548–76 590 111 bp. Of the four haplotype variations with frequencies >5%, GG had the lowest OR (OR = 0.56, 95% CI 0.38–0.82, P = 0.003) relative to AG, and haplotype AA also was inversely associated with lung cancer risk (P = 0.004, OR = 0.73, 95% CI 0.59–0.90). Notably, 45% of controls but only 35% of cases had one of these two haplotypes.
Figure 2.
Map of the associated 15q24–25.1 region in lung cancer. (A) −Log10(P) of significant sliding window haplotypes in the region. Global haplotypes are represented by blue lines, whereas individual haplotypes are represented by red lines. (B) Map of the 15q24–25.1 locus with rs17486278 and rs7178270; UCSC gene and SNP locations based on RefSeq, UniProt, GenBank, CCDS and comparative genomics. (C) Location of sliding window haplotypes in the region previously associated with familial lung cancer in Caucasians (10).
Table 3.
Two haplotypes significantly associated with lung cancer in African-Americans Northern California Lung Cancer Study
| Haplotypes | Percentage | OR (95% CI) | P-value | |
|---|---|---|---|---|
| Case | Control | |||
| Two-SNP haplotype | ||||
| Global | — | — | — | 0.004 |
| A-G | 56.1 | 49.3 | — | Referent |
| A-A | 27.8 | 33.4 | 0.73 (0.59–0.90) | 0.004 |
| G-A | 8.7 | 6.1 | 1.23 (0.84–1.81) | 0.293 |
| G-G | 7.4 | 11.2 | 0.56 (0.38–0.82) | 0.003 |
| Four-SNP haplotype | ||||
| Global | — | — | — | 0.007 |
| C-T-G-A | 49.3 | 53 | — | Referent |
| C-T-A-G | 8.9 | 7.2 | 1.36 (0.98–1.90) | 0.074 |
| C-T-G-G | 15.3 | 10.8 | 1.55 (1.18–2.04) | 0.002 |
| T-C-G-G | 12.4 | 13.7 | 0.98 (0.75–1.29) | 0.900 |
| T-T-G-G | 13.8 | 15.2 | 1 (0.78–1.30) | 0.950 |
The two-SNP haplotype includes rs7168796 and rs7164594, spanning 76 587 548–76 590 111 bp. The four-SNP haplotype includes rs11637635, rs17408276, rs16969968 and rs578776, spanning 76 664 204–76 675 454 bp. Bold values indicate P ≤ 0.05 and a 95% CI range which does not overlap 1.00.
There also was a four-SNP haplotype with (rs11637635, rs17408276, rs16969968 and rs578776) global P = 0.007 for lung cancer risk. The SNPs encompass the last four exons of CHRNA5 and the first two exons of CHRNA3: chr15 76 664 204–76 675 454. Five haplotype variations were detected, the most deleterious being CTGG (OR = 1.55, 95% CI 1.18–2.04, P = 0.002) compared with CTGA (Table 3).
Analyses of haplotype associations of the two- and four-SNP haplotypes with lung cancer risk were also stratified by gender, and a test for significant haplotype interaction was performed. No significant interaction with gender was found with either the two- or four-SNP haplotypes (P = 0.40 and P = 0.93, respectively) (Supplementary Material, Table S5).
Bioinformatic analysis of associated single SNPs and haplotypes identified by sliding window technique
Within the 2.6 kb region in LOC123688 bracketed by our two-SNP sliding window haplotype, two SNPs are predicted by Delta-MATCH to create transcription factor-binding sites (Table 4). Rs11852372 (difference z-score = 0.73) and rs9672189 (difference z-score = 1.0) lie just 2 bp apart and are each predicted to create binding sites for three transcription factors (FALZ, SRY and BRCA1 and FALZ, SRY and HMGA1, respectively). Rs17486278 was also predicted to lie within a CEBPA-binding site, but to have no effect on the binding of CEBPA (difference z-score = 0). Within the 11 kb region bracketed by the significant four-SNP sliding window haplotype, two SNPs were predicted by Patrocles and PolymiRTS to affect miRNA binding. In particular, rs8029939 near the 3′ end of CHRNA3 destroys a binding site for miRNA 584, whereas rs564585 is predicted to create binding sites for miRNA 23a, 23b and 130a* in the overlapping 3′ ends of both CHRNA3 and CHRNA5. Rs8029939 is specific to Yoruban and African-American populations.
Table 4.
Epigenetic candidate SNPs in proximity to lung cancer-associated loci
| Loci | bp location | Predicted SNP effect | Predicted regulatory element | NCBIa | ||
|---|---|---|---|---|---|---|
| AA MAF | YRI MAF | CEU MAF | ||||
| Two-SNP haplotype: LOC123688 | ||||||
| rs7168796b | 76587548 | 0.11 | 0.20 | 0 | ||
| rs11852372 | 76588448 | Creates TFBSc | SRY, FALZ, BRCA1 (dm_difZ = 0.73) | ndd | nd | nd |
| rs9672189 | 76588450 | Creates TFBS | SRY, FALZ, HMGA1 (dm_difZ = 1.0) | nd | nd | nd |
| rs7164594 | 76590111 | 0.50 | 0.51 | 0.21 | ||
| Top-hit SNP | ||||||
| rs17486278 | 766554536 | Lies within TFBS | CEBPA (dm_difZ = 0) | 0.24 | nd | 0.41 |
| Four-SNP haplotype: CHRNA5 and CHRNA3 | ||||||
| rs11637635 | 76664204 | 0.33 | 0.23 | 0.33 | ||
| rs17408276 | 76668672 | 0.17 | 0.05 | 0.33 | ||
| rs16969968 | 76669979 | 0.04 | 0 | 0.43 | ||
| rs564585 | 76673282 | Creates miRNAe binding site | hsa-miR-23a hsa-miR23b hsa-miR130a* | nd | nd | nd |
| rs8029939 | 76675404 | Disrupts miRNAf binding site | hsa-miR-584 | nd | 0.20 | 0 |
| rs578776 | 76675454 | 0.41 | 0.35 | 0.76 | ||
aAA MAF from the Coriell Cell Repository AFD_AFR_PANEL, YRI and CEU MAFs from the HapMAP_YRI and HapMap_CEU panels.
bUnderlined SNPs were genotyped in the current study.
cAll Transcription factor effects were found using the Delta-MATCH Database, df_difZ is a score from −1 to 1 indicating the predicted degree of change to a transcription factor-binding site, with −1 being the complete destruction of a binding site and 1 being the creation of a binding site.
dnd, no genotype data available for this population.
eAs predicted by the Patroclese Database.
fAs predicted by the Poly miRTS Database.
Single-SNP association with smoking behavior
No association was found between lung cancer-associated SNP rs7178270 and smoking behavior, as measured by pack-years smoked, cigarette consumption per day, maximum cigarette consumption per day or nicotine dependence as measured by the Fagerström Test for Nicotine Dependence (FTND). Lung cancer-associated SNP rs17486278 showed an association (P = 0.03) with mean FTND scores in ever smoker cases which were 1.1 point higher in those with two minor alleles than in those with no minor alleles (Supplementary Material, Table S9). Each of the SNPs in the four-SNP lung cancer risk haplotype was also associated with at least one measure of smoking behavior. SNP rs578776 was most associated with an increase in cigarettes per day (P = 0.008) in ever smoker cases; individuals with two minor G alleles had a mean cigarette consumption of 18.4 cigarettes per day relative to those with no minor alleles who smoked 14.8 cigarettes per day. SNP rs16969968 was most associated with maximum cigarettes smoked per day (P = 0.002) in ever smoker cases; individuals with two minor alleles smoked 52.6 cigarettes per day at their highest rate of cigarette consumption relative to those with no minor alleles who smoked 18.1 cigarettes per day (data not shown). SNP rs17480276 was most associated with increased pack-years in ever smoker cases (P = 0.01); individuals with two minor alleles showed a mean pack-year consumption of 59.5 pack-years relative to those with no minor alleles who smoked a mean of 32.4 pack-years. SNP rs11637635 was associated with nicotine dependence as measured by FTND scores in ever smoker cases, but the effect on FTND scores was inconsistent. No significant association was found between smoking behavior and SNPs in the two SNP lung cancer risk haplotype (rs7168796 and rs7164594).
Haplotype association with smoking behavior
The two- and four-SNP haplotypes were also tested for haplotype associations with smoking behavior. An association was found between the four-SNP haplotype and cigarettes per day in ever smoker cases (global haplotype P = 0.05). Ever smoker cases with the referent CTGA haplotype had a lower adjusted mean cigarette per day consumption than individuals with one of the other four common haplotype variants (data not shown). No association was found between the two-SNP lung cancer risk haplotype and smoking behavior (Supplementary Material, Tables S10).
DISCUSSION
As expected, SNPs in the 15q25.1 region were less likely to be linked in African-Americans compared with Caucasian populations, and linkage in the region decreased according to the % African ancestry estimated by an extensive panel of ancestry markers. This lower linkage provides the potential to finely map the multiple biological variations associated with lung cancer risk in the Chr15q25.1 region. We found a significant lung cancer association with rs17486278, which is in the first intron of CHRNA5. Despite the association of rs17486278 with increased nicotine dependence, the association with lung cancer risk remained significant after adjustment for smoking behavior, consistent with a previously published study that found that significant lung cancer associations with polymorphisms in the 15q25.1 region were no longer statistically significant in Caucasians after correction for smoking behavior, but remained significant in African-Americans (11).
Although rs17486278 has previously been reported to be associated with lung cancer (7), it is also tightly linked with another lung cancer risk SNP rs16969968 in these Caucasian populations. In the present study of African-Americans, rs17486278 was not linked to rs16969968 (r2 = 0.19), enabling specification of rs17486278 as an independent lung cancer association within the first intron of CHRNA5. Although rs16969968 was not significantly associated with lung cancer in African-Americans, the OR = 1.27 was very comparable to previous studies in Caucasians and the lack of statistical significance could be due to the low frequency of the risk variant in African-Americans (MAF = 0.07).
The consistent association of rs17486278 with lung cancer risk in African-Americans, despite its decreased linkage with rs16969968, is especially intriguing. Previous functional studies have attributed risk associated with the rs16969968 linkage group to the non-synonymous aspartic acid to asparagine amino acid substitution in the α5-nicotinic receptor subunit caused by rs16969968. In vitro studies have found that α4β2α5-nicotinic receptors which incorporate a normal aspartic acid containing an α5-subunit are more than twice as responsive to the nicotinic agonist epibatidine compared with the mutant asparagine containing α5-subunit. Notably, however, the expression of the α4β2α5 receptor remains unchanged (12). Interestingly, the risk-associated A allele of rs16969968, which results in the aspartic acid substitution, has also been associated with decreased in vivo expression of CHRNA5 in normal lung tissue of adenocarcinoma patients (13). Because the expression of CHRNA5 is significantly associated with the variation of rs16969968 in vivo in Caucasian populations, but not in vitro, a mutation linked to rs16969968 in Caucasians is likely to affect the expression of CHRNA5. Studies of CHRNA5 expression in Caucasians have been complicated by the presence of three separate linkage blocks in the region tagged by rs16969968, rs588765 and rs578776 and by interactions between these groups (13,14). Rs588765 is nominally linked (r2 = 0.41) with both rs16969968 and rs17486278 in Caucasian populations. The rs588765 linkage group contains rs3841324, a 22 bp in-del in the upstream CpG island of CHRNA5. The rs3841324 in-del was most significantly associated with CHRNA5 expression in a study of Caucasian brain tissue (14). The rs588765 linkage group was further associated with an increased risk of lung cancer and nicotine dependence when rs16969968 was controlled for, showing that the association of rs16969968 with the expression of CHRNA5 was a result of its linkage to rs588765 (14). In African-Americans, in contrast, the linkage group containing rs588765 has been shown to reduce nicotine dependence when rs16969968 is controlled for (14), suggesting that rs588765 is not linked to the same variations in African-Americans populations as it is in Caucasian populations and may no longer be associated with CHRNA5 expression. Indeed, in our population, rs588765 is not linked to either rs16969968 (r2 = 0.03) or rs17486278 (r2 = 0.19) and is not associated with either smoking behavior or lung cancer risk. Regardless, rs17486278 and the linked SNP rs2036527 are both associated with familial lung cancer in our study and bracket a 16 kb region which includes both rs3841324 and rs588765, indicating that variation impacting expression lies in this region. This 16 kb region is further implicated by the presence of a strong lung cancer association in Chinese populations (rs667282C>T, P = 2.0 × 10−12) (15).
The second top-hit SNP for lung cancer in African-Americans, rs7178270, is located between the first and second exons of CHRNB4 and decreases lung cancer risk in women but not in men (OR = 0.62 and 1.01, respectively). The interaction between polymorphism and gender found here corroborates previous SNP/gender effects seen in the 15q25.1 region in Caucasian and Japanese populations (16,17). Rs7178270 is also located in a sliding window haplotype region (HapN) significantly associated with lung cancer in Caucasians (10). The relationship between the Caucasian lung cancer risk haplotype to identified risk SNPs and haplotypes in the present study of African-Americans is shown in Figure 2.
In the present study, the combined risk associated with the most deleterious genotype combinations of rs7178270 and rs17486278 was estimated to be approximately 2-fold and occurred in 17% of controls and 24% of cases, a relatively large portion of the African-American population.
The sliding window haplotype analysis located two additional areas associated with lung cancer risk. A protective two-SNP haplotype (rs7168796 and rs7164594) spans 2.6 kb between the first and second exons of LOC123688, which is 30 kb upstream of PSMA4. Neither SNP was associated with lung cancer or smoking behaviors in the single-SNP analysis. SNP rs7168796 is not found in Caucasian populations and rs7164594 occurs at a much higher frequency in African-Americans than in Caucasians. Interestingly, the two-SNP rs7168796–rs7164594 haplotype identified by our study lies within the region covered by a larger familial lung cancer risk haplotype in Caucasians identified by Liu et al. (10) (Fig. 2). HapL spans 57 kb, covering the last five exons of IREB2 and the first three exons of LOC123688. Thus, the two-SNP haplotype identified here for African-Americans considerably narrows a candidate region previously defined in Caucasians.
To explore potential functional variation in the lung cancer risk regions, we employed several bioinformatic tools that revealed possible transcription factor and miRNA-binding sites that may mediate the effects of SNPs within our candidate regions. Within the 2.6 kb region involving the two-SNP haplotype, we found a pair of SNPs, rs11852372 and rs9672189, which are predicted to create binding sites for a number of transcription factors including FALZ and BRCA1.
The four-SNP haplotype associated with lung cancer including rs11637635, rs17408276, rs16969968 and rs578776 spans 11.3 kb, including the last four exons of CHRNA5 and the first two exons of CHRNA3. These four SNPs also had individual associations with smoking behavior. For SNP rs11637635, the T allele was associated with FTND score in ever smoker cases (P = 0.03), but not with other measures of smoking behavior, and the effect on FTND scores was inconsistent. In our data, the C allele of rs11637635 is tightly linked (r2 = 0.90) with the major G allele of rs588765 which has been shown in a previously published study, although not in our data, to be associated with an increased risk of nicotine dependence in African-Americans (9). This C allele appears in both the top lung cancer risk haplotype CTGG and the borderline risk CTAG haplotype. SNP rs16969968 was associated with nicotine dependence in African-Americans and Caucasians (9) and was associated with maximum cigarettes per day in this study. Our study also found a borderline significant association with lung cancer risk for haplotype CTAG (P = 0.074, OR = 1.36, 95% CI 0.98–1.90), which includes the risk allele A of rs16969968. The borderline significant association of this haplotype with lung cancer may be due to its low frequency in the African-American population; only 7.2% in controls. In the single-SNP analysis, rs578776 was associated with increased lung cancer risk using a dominant model. The rs578776 A allele has previously been associated with a decreased risk for nicotine dependence in Caucasians. Interestingly, although many SNPs differ in allele frequency between Caucasian and African-American populations, rs578776 is the only SNP in our study where the minor A allele in Caucasians becomes the major allele in African-Americans, consistent with allele frequencies for this SNP in other studies (9). The minor G allele for rs578776 in African-Americans is associated with an increase in cigarettes smoked per day in our study, and a change in this risk-associated G allele from the A allele in the referent haplotype creates the CTGG lung cancer risk haplotype (OR = 1.55, P = 0.002). Additional analysis of haplotype associations with smoking behavior in ever smoker cases found that all common haplotype variations of the four-SNP lung cancer risk haplotype were associated with an increase in mean cigarette per day consumption relative to individuals who carried the referent haplotype.
Bioinformatic analysis of SNPs in the 11.6 kb region spanned by the four-SNP haplotype found that rs8029939 and rs564585 lie between rs578776 and rs16969968. Both SNPs are predicted to change the binding of miRNAs in the regions 3′ to CHRNA5 and CHRNA3. Thus, rs8029939 and rs564858 are attractive candidate SNPs because of their location. Rs8029939 is especially interesting given that it is only polymorphic in African and African-American populations.
Our study found that not all variations in the 15q25.1 region associated with lung cancer risk are associated with smoking behaviors but, because of variability in reported nicotine intake among participants due to differences in cigarette brands, the depth of inhalation while smoking and environmental exposures, conclusively determining which lung cancer-associated SNPs are also associated with nicotine exposure needs to be determined through functional and animal studies. Moreover, because of the multiple comparisons made in our smoking behavior analyses, caution is required in interpreting the importance of associations that were not consistent in both cases and controls and those with marginally significant P-values. A strength of the current study is the population-based accrual of lung cancer cases and the in-person interviews that collected personal information. The very close similarity of the overall ancestry estimates of cases and controls indicates that the groups were drawn from the same base population with respect to African ancestry. We also believe that our candidate gene associations are robust because we carefully controlled for the potential effects of population stratification. Our study provides further evidence that multiple genetic alterations within the 15q25.1 region contribute to lung cancer risk, implicates rs7178270 as a polymorphism associated with a decreased risk of lung cancer specific to women and refines the locations of candidate loci. We also further confirm the significant association of this genomic region with lung carcinoma in the African-American population, which is impacted disproportionately by this disease.
MATERIALS AND METHODS
Study participants
The Northern California Lung Cancer Study is described in detail elsewhere (18) and was approved by the Committee on Human Research of the University of California, San Francisco, and by the Institutional Review Boards of all collaborating institutions. African-American cases and controls older than 18 years of age were identified during two collection periods spanning September 1998–March 2003 and July 2005–March 2008.
Cases were Northern California residents presenting with previously untreated, histologically confirmed lung cancer. Cases in the first accrual period were identified primarily through the Northern California Cancer Center (NCCC) rapid case ascertainment program. Cases in the second accrual period were identified through both the NCCC and the Kaiser Permanente Medical Care Program (KPMCP). Census data demonstrate that the patient population of KPMCP is closely representative of the general population in a number of demographic and economic areas. Case recruitment was performed as previously described (18).
Control participants ascertained in the first accrual period were recruited through three sources: random-digit dialing, Health Care Financing Administration records and community-based recruitment (e.g. health fairs, churches and senior centers). Controls in the second accrual period were recruited through the KPMCP. All controls were frequency matched to cases on age, gender and race/ethnicity with a control-to-case ratio of ∼2:1. Control participants completed in-person interviews and donated a blood and/or buccal specimen. In all, 467 cases and 625 controls with sufficient blood or buccal specimens were genotyped in this study.
SNP selection
SNPs were selected in the Chr15q25.1 lung cancer candidate region spanning 76 517 368–76 711 042 bp for the YRI and CEU International HapMap Project Populations (19). A combination of 69 tag (r2 ≥ 0.80) and non-tag SNPs with MAF ≥ 0.05 were selected to capture the variability present across the region in both parental populations. Linkage information for an African-American population was not available in the HapMap data sets at the time of SNP selection. SNPs previously found to be linked to lung cancer in Central European populations were also included (n = 13) (7). In total, 82 SNPs in the chr15q25.1 region were genotyped. As presented in more detail below, 3869 of 4222 African-American admixture SNPs compiled and previously published by Tian et al. (20) were used as the source of our admixture panel.
Genotyping platform
Illumina Golden Gate genotyping was performed at the University of California, San Francisco Genome Center with a Custom panel of 4608 SNPs. Unamplified genomic DNA samples extracted from whole blood (n = 940) were genotyped along with whole genome-amplified (WGA) blood or buccal DNA samples (n = 150) prepared as previously described (21). Genotypes for unamplified DNA and WGA DNA samples were clustered separately. A GenCall genotype quality threshold of 0.25 was used. Genotype reproducibility was verified with duplicates. Cluster accuracy was verified with CEPH trios from the Coriell Institute for Medical Research (Camden, NJ, USA) which were genotyped along with study samples. Reproducibility averaged 100% for seven unamplified DNA duplicate pairs (range 99.9–100%) and 99.96% for three WGA duplicate pairs (range 99.89–100%). Parent–parent–child heritability averaged 99.96% in 10 unamplified CEPH trios (range 99.93–100%) and 99.99% in two WGA CEPH trios (range 99.99–100%). Seven cases and 14 controls with an overall genotype rate <0.98 were excluded, along with 12 cases subsequently found to have a cancer other than primary lung cancer. In all, 448 lung cancer cases and 611 controls were included in the statistical analysis.
HWE calculations
The SAS software was used (22) to test the frequency distribution in controls at each SNP locus against HWE under the allele Mendelian biallelic expectation using the chi-square goodness-of-fit test; SNPs with an HWE P < 0.01 in control participants were excluded from case–control comparisons.
Calculations of % African ancestry
Selection of ancestry informative markers for African-American admixture mapping was based on a previously published panel of 3289 of 4222 SNPs (20). Five hundred and forty-five SNPs were excluded due to high LD (r2 ≥ 0.8) with at least one other SNP based on the genotyping data of European and African ancestral populations described in detail previously (20). Owing to an SNP call rate <90% in the ancestral populations, 110 SNPs were excluded. Two hundred and seventy-eight SNPs failed pre-genotyping quality control scoring for the Illumina genotyping platform. Among the 3289 SNPs genotyped, 153 failed to successfully genotype in both the genomic and WGA samples. Fifty-nine SNPs were excluded due to a call rate <90%. In addition, 130 SNPs on the X chromosome were excluded from the analysis. A total of 2947 SNPs were used to estimate the % African genetic ancestry for each study subject. STRUCTURE Version 2.2 (23,24) was used to estimate the % African genetic ancestry using 10 000 iterations of burn in period and 10 000 iterations of the Markov Chain Monte Carlo algorithms.
Linkage analysis
Haploview 4.1 was used to calculate r2 values between the genotyped SNPs and to generate D′ and r2 maps with genotype data from the present study and HapMap Phases 1 and 2 data sets (25). The % African ancestry statistic was used to create participant groups with ≥90% (n = 168) and ≤60% (n = 110) African ancestral contributions. Genotype data from the YRI and CEU HapMap cohorts were used to generate r2 maps for the 77 candidate SNPs analyzed in the present study (19).
Single-SNP association analyses
The effect of individual SNPs on lung cancer risk was assessed using unconditional logistic regression assuming log-additive (zero, one or two copies of variant alleles) and dominant models. Lung cancer risk was then assessed in the stratified groups: those with a family history of lung cancer, no family history of lung cancer, <50 years old, ≥50 years old, women, men, ever smokers (≥20 packs smoked total) and never smokers (data not shown due to the small number of never smoker cases). In addition to age, gender and % African ancestry, we included smoking (measured as pack-years, mean cigarettes per day and maximum cigarettes per day) in the model to assess confounding by smoking. Unconditional logistic regression assuming a log-additive model, and including a product term for each SNP and gender, was performed to test for the SNP × gender interaction for those SNPs with P < 0.05 in the gender-stratified analysis. Unconditional logistic regression was also used to assess whether the two SNPs with statistically significant lung cancer associations had independent effects by modeling them together with their product term. In addition, the effect of the total number of risk alleles from either of the two significantly associated SNPs was assessed.
Association of SNPs and haplotypes with smoking behavior
Participants with outlier smoking behavior were removed from both single-SNP and haplotype analysis: those who smoked ≥110 pack-years (n = 9); those who smoked ≥50 cigarettes per day (n = 6) and those who smoked ≥100 maximum cigarettes per day (n = 10).
Unconditional logistic regression and analyses of variants adjusted for age, gender and % African genetic ancestry were used to assess the association of individual SNPs with ever/never smoking status as well as pack-years, cigarettes per day, nicotine dependence and maximum cigarettes per day (data not shown). Nicotine dependence was measured using the items and scoring for the FTND suggested by Heatherton et al. (26). Cases and controls were analyzed separately.
Analysis of the association between haplotypes and smoking behavior was the r-project link is current and correct conducted using haplo.stats in R v2.8.1 (www.r-project.org) which uses an estimation–maximization (EM) algorithm to infer unphased haplotype probabilities. Haplotype effects were determined separately for lung cancer patients and controls and further stratified within these subgroups by current and ever smokers and were estimated using an adjusted weighted logistic regression in a two-step EM algorithm. All haplotype models assumed an additive mode of inheritance and were adjusted for age (continuous), gender and % African ancestry. Adjusted global score statistics were computed to ascertain the overall association between the haplotype and the smoking behavior (ever/never smoking status, pack-years, mean cigarettes per day, maximum cigarettes per day and FTND scores). In each analysis, the most common haplotype was the referent group and haplotypes with a frequency of <5% were designated as ‘rare’ and pooled into one group.
FDR calculations
A permutation-based procedure controlling for FDR (27) was used to account for multiple testing. Permutation-based procedures utilize the empirical dependency structure of the data to construct more powerful FDR controlling procedures. To allow for the calculation of age, gender and % African genetic ancestry adjusted P-values, case–control status and these three covariates were permuted together to preserve relationship. Assuming, from the original data, that the log-additive association P-value for the ith SNP is Pi (i = 1, … , S = 2947), we computed 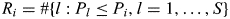 for i = 1, … , S. For each permutation b (b= 1, … , B = 1000), log-additive P-values (P1,b, … , PS,b) were calculated for all SNPs, and we computed
for i = 1, … , S. For each permutation b (b= 1, … , B = 1000), log-additive P-values (P1,b, … , PS,b) were calculated for all SNPs, and we computed 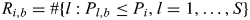 for i = 1, … , S. To calculate FDR at Pi (i = 1, … , S), the true positive (TP) number was estimated as
for i = 1, … , S. To calculate FDR at Pi (i = 1, … , S), the true positive (TP) number was estimated as 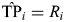 , and the false positive (FP) number was estimated as
, and the false positive (FP) number was estimated as 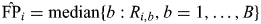 . We conservatively assumed the proportion of non-associated SNPs to be 1, then based on the FDR estimation defined as in Storey and Tibshirani (27), the FDR-adjusted P-value for the ith SNP was:
. We conservatively assumed the proportion of non-associated SNPs to be 1, then based on the FDR estimation defined as in Storey and Tibshirani (27), the FDR-adjusted P-value for the ith SNP was:
Sliding window haplotype analysis
Haplotype analyses were performed using the haplo.stats R package (http://cran.r-project.org/) to capture the information potentially missed by the single-SNP analyses by increasing the statistical power to tag causal variants and accounting for cis-interactions between two or more SNPs (28). Haplotype analyses were performed using the sliding window approach with haplotype windows of two to six SNPs. Global P-values were calculated for each haplotype window to evaluate whether the distribution of haplotypes was significantly different between cases and controls.
Bioinformatic analysis
An investigation of predicted epigenetic SNP effects utilized the Patrocles (http://www.patrocles.org/) (29), PolymiRTS (http://compbio.uthsc.edu/miRSNP/home.php) (30) and Delta-MATCH (http://snplogic.org/) (31) databases. Delta-MATCH provides a difference z-score ranging from −1 to 1 for the change of predicted binding affinities at polymorphic transcription factor-binding sites throughout the human genome, with −1 indicating the predicted destruction of a transcription factor-binding site and 1 indicating the creation. A difference z-score near an absolute value of 1 thus points to SNPs that may dramatically alter gene expression. Patrocles and PolymiRTS databases calculate the effect of SNPs on predicted miRNA-binding sites.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institute of Environmental Health Sciences (R01 ES06717) and the National Cancer Institute (R25 CA112355 to J.S.C. and R01 CA 52689 to M.R.W.).
Supplementary Material
REFERENCES
- 1.Group U.S.C.S.W. United States Cancer Statistics: 1999–2005 Incidence and Mortality Web-based Report. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2009. [Google Scholar]
- 2.Alberg A.J., Brock M.V., Samet J.M. Epidemiology of lung cancer: looking to the future. J. Clin. Oncol. 2005;23:3175–3185. doi: 10.1200/JCO.2005.10.462. doi:10.1200/JCO.2005.10.462. [DOI] [PubMed] [Google Scholar]
- 3.Higgins R.S., Lewis C., Warren W.H. Lung cancer in African Americans. Ann. Thorac. Surg. 2003;76:S1363–S1366. doi: 10.1016/s0003-4975(03)01208-6. doi:10.1016/S0003-4975(03)01208-6. [DOI] [PubMed] [Google Scholar]
- 4.Stewart J.H.t. Lung carcinoma in African Americans: a review of the current literature. Cancer. 2001;91:2476–2482. doi:10.1002/1097-0142(20010615)91:12<2476::AID-CNCR1283>3.0.CO;2-Z. [PubMed] [Google Scholar]
- 5.Cote M.L., Kardia S.L., Wenzlaff A.S., Ruckdeschel J.C., Schwartz A.G. Risk of lung cancer among white and black relatives of individuals with early-onset lung cancer. JAMA. 2005;293:3036–3042. doi: 10.1001/jama.293.24.3036. doi:10.1001/jama.293.24.3036. [DOI] [PubMed] [Google Scholar]
- 6.Amos C.I., Wu X., Broderick P., Gorlov I.P., Gu J., Eisen T., Dong Q., Zhang Q., Gu X., Vijayakrishnan J., et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. doi:10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P., et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. doi:10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 8.Thorgeirsson T.E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K.P., Manolescu A., Thorleifsson G., Stefansson H., Ingason A., et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. doi:10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saccone N.L., Wang J.C., Breslau N., Johnson E.O., Hatsukami D., Saccone S.F., Grucza R.A., Sun L., Duan W., Budde J., et al. The CHRNA5–CHRNA3–CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. doi:10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Liu P., Wen W., James M.A., Wang Y., Bailey-Wilson J.E., Amos C.I., Pinney S.M., Yang P., de Andrade M., et al. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. Cancer Res. 2009;69:7844–7850. doi: 10.1158/0008-5472.CAN-09-1833. doi:10.1158/0008-5472.CAN-09-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz A.G., Cote M.L., Wenzlaff A.S., Land S., Amos C.I. Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J. Thorac. Oncol. 2009;4:1195–1201. doi: 10.1097/JTO.0b013e3181b244ef. doi:10.1097/JTO.0b013e3181b244ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierut L.J., Stitzel J.A., Wang J.C., Hinrichs A.L., Grucza R.A., Xuei X., Saccone N.L., Saccone S.F., Bertelsen S., Fox L., et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. doi:10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falvella F.S., Galvan A., Frullanti E., Spinola M., Calabro E., Carbone A., Incarbone M., Santambrogio L., Pastorino U., Dragani T.A. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin. Cancer Res. 2009;15:1837–1842. doi: 10.1158/1078-0432.CCR-08-2107. doi:10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- 14.Wang J.C., Cruchaga C., Saccone N.L., Bertelsen S., Liu P., Budde J.P., Duan W., Fox L., Grucza R.A., Kern J., et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum. Mol. Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. doi:10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C., Hu Z., Yu D., Huang L., Jin G., Liang J., Guo H., Tan W., Zhang M., Qian J., et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. doi:10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 16.Lips E.H., Gaborieau V., McKay J.D., Chabrier A., Hung R.J., Boffetta P., Hashibe M., Zaridze D., Szeszenia-Dabrowska N., Lissowska J., et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int. J. Epidemiol. 39:563–577. doi: 10.1093/ije/dyp288. doi:10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiraishi K., Kohno T., Kunitoh H., Watanabe S., Goto K., Nishiwaki Y., Shimada Y., Hirose H., Saito I., Kuchiba A., et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. doi:10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- 18.Cabral D.N., Napoles-Springer A.M., Miike R., McMillan A., Sison J.D., Wrensch M.R., Perez-Stable E.J., Wiencke J.K. Population- and community-based recruitment of African Americans and Latinos: the San Francisco Bay Area Lung Cancer Study. Am. J. Epidemiol. 2003;158:272–279. doi: 10.1093/aje/kwg138. doi:10.1093/aje/kwg138. [DOI] [PubMed] [Google Scholar]
- 19.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. doi:10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.Tian C., Hinds D.A., Shigeta R., Kittles R., Ballinger D.G., Seldin M.F. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am. J. Hum. Genet. 2006;79:640–649. doi: 10.1086/507954. doi:10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen H.M., Wiemels J.L., Wrensch M., Wiencke J.K. DNA quantification of whole genome amplified samples for genotyping on a multiplexed bead array platform. Cancer Epidemiol. Biomarkers Prev. 2007;16:1686–1690. doi: 10.1158/1055-9965.EPI-06-1024. [DOI] [PubMed] [Google Scholar]
- 22.SAS Institute Inc. SAS 9.2 Help and Documentation. Cary, NC: SAS Institute Inc; (2002–2008) [Google Scholar]
- 23.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Heatherton T.F., Kozlowski L.T., Frecker R.C., Fagerstrom K.O. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br. J. Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. doi:10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 27.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. doi:10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N., Zhang K., Zhao H. Haplotype-association analysis. Adv. Genet. 2008;60:335–405. doi: 10.1016/S0065-2660(07)00414-2. doi:10.1016/S0065-2660(07)00414-2. [DOI] [PubMed] [Google Scholar]
- 29.Hiard S., Charlier C., Coppieters W., Georges M., Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2009;38:D640–D651. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao L., Zhou M., Wu L., Lu L., Goldowitz D., Williams R.W., Cui Y. PolymiRTS database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–D54. doi: 10.1093/nar/gkl797. doi:10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pico A.R., Smirnov I.V., Chang J.S., Yeh R.F., Wiemels J.L., Wiencke J.K., Tihan T., Conklin B.R., Wrensch M. SNPLogic: an interactive single nucleotide polymorphism selection, annotation, and prioritization system. Nucleic Acids Res. 2009;37:D803–D809. doi: 10.1093/nar/gkn756. doi:10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.