Abstract
Heart rate (HR) has been identified as a risk factor for cardiovascular disease (CVD), yet little is known regarding genetic factors influencing this phenotype. Previous research in American Indians (AIs) from the Strong Heart Family Study (SHFS) identified a significant quantitative trait locus (QTL) for HR on chromosome 9p21. Genetic association on HR was conducted in the SHFS. HR was measured from electrocardiogram (ECG) and echocardiograph (Echo) Doppler recordings. We examined 2248 single-nucleotide polymorphisms (SNPs) on chromosome 9p21 for association using a gene-centric statistical test. We replicated the aforementioned QTL [logarithm of odds (LOD) = 4.83; genome-wide P= 0.0003] on chromosome 9p21 in one SHFS population using joint linkage of ECG and Echo HR. After correcting for effective number of SNPs using a gene-centric test, six SNPs (rs7875153, rs7848524, rs4446809, rs10964759, rs1125488 and rs7853123) remained significant. We applied a novel bivariate association method, which was a joint test of association of a single locus to two traits using a standard additive genetic model. The SNP, rs7875153, provided the strongest evidence for association (P = 7.14 × 10−6). This SNP (rs7875153) is rare (minor allele frequency = 0.02) in AIs and is located within intron 9 of the gene KIAA1797. To support this association, we applied lymphocyte RNA expression data from the San Antonio Family Heart Study, a longitudinal study of CVD in Mexican Americans. Expression levels of KIAA1797 were significantly associated (P = 0.012) with HR. These findings in independent populations support that KIAA1797 genetic variation may be associated with HR but elucidation of a functional relationship requires additional study.
INTRODUCTION
Several studies identify elevated heart rate (HR) as a risk factor for cardiovascular disease (CVD) independent of hypertension, cholesterol level, or tobacco and alcohol consumption (1–3). HR is regulated by the autonomic nervous system. Sympathetic and parasympathetic inputs are integrated within the specialized myocytes (pacemaker cells) in the sinoatrial node, where the electrical impulse is propagated to produce a heart beat. The underlying genetic factors influencing the complex control of HR are largely unknown.
Presently, case–control genome-wide association studies (GWASs) are the most widely used approach to associate genetic variation with phenotypic diversity. These studies have identified a number of common variants, increasing our understanding of molecular pathways underlying common chronic diseases. However, this research is limited by its ability to determine genetic association across populations, account for the majority of the heritable components in complex traits, and often overlooks rare genetic variants (4). Alternatively, family-based linkage or association studies are powered to detect underlying rare variants that may have a larger effect on phenotype diversity (5). Additionally, joint analysis of two related phenotypes can theoretically extract more information while simultaneously providing greater statistical power to detect association (6).
Family-based studies demonstrate genes influence HR regulation. Heritability estimates vary between populations, ranging from 0.21 in the Framingham Heart Study (FHS) (7) to 0.42 in the Netherlands Twins Register (8). Recent studies also attempted to localize genes impacting HR. Laramie et al. (9) found a significant quantitative trait locus (QTL) on chromosome 10 [logarithm of odds (LOD) = 4.2] for participants from the HyperGEN Study and a suggestive QTL on chromosome 5 (LOD = 1.92) across different ethnic groups in a meta-analysis of the NHLBI Family Blood Pressure Program. On chromosome 4, Wilk et al. (10) found significant evidence for an HR QTL in African and European American hypertension patients (LOD = 3.18), and Martin et al. (11) detected significant linkage (LOD = 3.9) within Metabolic Risk Complication of Obesity Genes participants. A GWAS conducted on HR variability in FHS using 70 987 common genetic variants did not find any single-nucleotide polymorphisms (SNPs) that attained statistical significance after Bonferroni correction (12). A second GWAS on HR variability of three isolated European populations detected significant association for HR measured as the RR interval (HR = 60 000/RR) within GPR133, a G-protein coupled receptor (rs885489, P = 3.9 × 10−8) on chromosome 12 and replication of an earlier association for QT interval with the NOS1AP gene (rs2880058, P = 2.00 × 10−10) (13). Potential explanations for this result may be that HR is influenced by several genetic variants each contributing a small effect, that alleles influencing variation in HR are rare or that HR candidate genes are not significantly represented on the analyzed genotyped arrays. Given these varied linkage and GWAS results, it is likely that HR is influenced by multiple genes and their effects may differ between populations.
The genetic components of HR in American Indians (AIs) are unknown. In automated genome-wide linkage screens conducted in the Strong Heart Family Study (SHFS), significant evidence of linkage (LOD = 3.3, P = 0.016) was observed for a QTL influencing HR levels on chromosome 9p21 in joint analysis of data from all three SHFS centers (14). Previous GWASs have demonstrated that several SNPs located on chromosome 9p21 are associated with risk for coronary heart disease (CHD) and/or myocardial infarction (MI) (15,16), indicating that this was an interesting chromosomal region for future research. In addition, the aforementioned GWAS on HR variability in isolated European populations reported two SNPs (rs13300284 and rs12552736) associated with RR interval on chromosome 9p21 in the same region (13). The primary objective of this study was to conduct linkage and association for HR using phenotypic data from echocardiograph (Echo) and electrocardiogram (ECG) HR measures in each of the three SHFS field centers (Arizona, Oklahoma, Dakotas) and 2248 SNPs on chromosome 9p21 in AI participants.
RESULTS
Table 1 displays descriptive statistics of HR measures and covariates that were included in our analysis of genetic factors related to Echo and ECG HR. Participants from Oklahoma had the highest Echo HR (68.1 ± 10.7 b.p.m.) and those from Arizona had the lowest (66.5 ± 10.5 b.p.m.). The only significant covariates of Echo HR at P < 0.05 were age in females and height for both sexes in Arizona and adipose body mass in the combined analysis. Heritability estimates for Echo and ECG HR (Table 2) were statistically significant and were similar across populations, ranging from 0.30 (±0.06) in Oklahoma to 0.26 (±0.08) in Arizona. Heritability estimates for ECG HR ranged from 0.35 (±0.05) in the Dakotas to 0.27 (±0.05) in Arizona and Oklahoma.
Table 1.
Characteristics of SHFS participants stratified by field center
| Arizona | Dakota | Oklahoma | Combined | |
|---|---|---|---|---|
| Echo (ECG) sample size | 1204 (1187) | 1173 (1186) | 1193 (1148) | 3567 (3521) |
| Female, n (%) | 752 (62) | 695 (59) | 698 (59) | 2145 (60) |
| Age (years), mean (±SD) | 37.09 (15.9) | 38.91 (17.10) | 43.58 (17.29) | 39.85 (16.99) |
| Echo HR (b.p.m.), mean (±SD) | 66.49 (10.47) | 66.65 (11.03) | 68.06 (10.71) | 67.06 (10.76) |
| ECG HR (b.p.m.), mean (±SD) | 65.15 (10.51) | 64.04 (10.47) | 65.04 (10.25) | 64.80 (10.53) |
| Height (m), mean (±SD) | 1.65 (8.7)* | 1.69 (8.75) | 1.67 (9.14) | 1.67 (9.07) |
| Body mass index, mean (±SD) | 35.12 (8.17) | 30.14 (6.82) | 31.18 (6.90) | 32.14 (7.62) |
| Hypertensive medication, current n (%) | 268 (22) | 180 (15) | 274 (23) | 722 (20) |
| Lean body mass (kg) (±SD) | 54.93 (12.37) | 56.04 (12.38) | 54.77 (12.50) | 55.24 (12.43) |
| Fat mass (kg) (±SD) | 41.43 (9.7) | 34.17 (10.02) | 36.33 (9.41) | 37.33 (10.19)** |
| HOMAIR, mean (±SD) | 4.57 (4.29) | 4.12 (5.18) | 3.16 (3.08) | 3.92 (4.32) |
| Fasting insulin (µU/ml), mean (±SD) | 19.69 (14.26) | 17.01 (14.89) | 15.52 (13.49) | 17.42 (14.32) |
| Fasting glucose(mg/dl), mean (±SD) | 121.68 (55.48) | 104.20 (33.66) | 106.59 (37.78) | 110.83 (44.05) |
| HBA1C (%) (±SD) | 7.26 (2.30) | 6.09 (1.61) | 6.62 (1.89) | 6.71 (2.05) |
| Diastolic blood pressure (mmHg), mean (±SD) | 76.48 (11.33) | 75.13 (10.48) | 76.83 (11.52) | 76.15 (11.14) |
| Systolic blood pressure (mmHg), mean (±SD) | 120.75 (16.50) | 119.71 (15.32) | 126.52 (16.86) | 122.33 (16.51) |
| Mean arterial blood pressure (mmHg), mean (±SD) | 91.29 (11.53) | 90.01 (10.71) | 93.46 (11.66) | 91.60 (11.40) |
| Pulse pressure mean (mmHg) (±SD) | 44.05 (13.87) | 44.40 (12.51) | 49.53 (14.43) | 45.99 (13.85) |
| Alcohol, current n (%) | 709 (59) | 777 (66) | 566 (47) | 2053 (58) |
| Smokers, current n (%) | 307 (25) | 500 (43) | 397 (33) | 1204 (34) |
NS, not statistically significant; Echo, echocardiograph; ECG, electrocardiogram; HOMAIR, homeostasis model assessment of insulin resistance; HBA1C, glycosylated hemoglobin.
*Statistically significant with Echo HR levels at P-value <0.05.
**Statistically significant with Echo HR levels at P-value <0.01.
Table 2.
Heritability and linkage results for two HR phenotypes in AIs
| Arizona | Dakota | Oklahoma | Combined | |
|---|---|---|---|---|
| Heritability of Echo HR, (±SD) | 0.26 (0.08)** | 0.29 (0.06)** | 0.30 (0.06)** | 0.28 (0.03)** |
| Heritability of ECG HR, (±SD) | 0.27 (0.06)** | 0.35 (0.05)** | 0.27 (0.05)** | 0.30 (0.05)** |
| Echo linkage results on chromosome 9p21 | ||||
| Echo HR LOD | 0.032 | 0.000 | 3.67** | 1.37 |
| ECG/Echo bivariate linkage results on chromosome 9p21a | ||||
| ECG/Echo LOD | 0.000 | 0.000 | 4.83** | 0.98 |
aBivariate linkage model with simultaneous analysis of Echo and ECG HR modeling the correlations between them and testing the null hypothesis that variance components relating to major gene effects on both traits are equal to 0 (17).
**Statistically significant with Echo HR levels at P < 0.01.
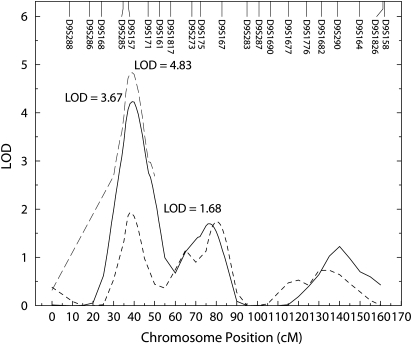
Results of linkage analyses in the previously implicated region of chromosome 9 for each of three field centers and the combined sample are presented in Table 2. After accounting for covariates, the only center to maintain a significant LOD score was Oklahoma (3.67, P = 0.006) on chromosome 9p21 between markers D9S185 and D9S157 for Echo HR. A suggestive LOD score of 1.68 was also detected in the Oklahoma cohort at the same position for ECG HR. Further investigation of the Oklahoma Echo HR LOD score indicated that the majority of support for the observed QTL was from two pedigrees (LODs = 2.8 and 0.92, respectively). In the combined sample, this region yielded an Echo HR LOD score of 1.37 and no evidence of linkage in the Arizona (LOD = 0.032) and Dakota (LOD = 0.000) cohorts. The effects of additional covariates aside from age*sex are the reasons that the combined Echo HR QTL was reduced from the preliminary LOD score of 3.3 to 1.4 in the combined sample.
The ECG is a standard method for measuring HR and provided a suggestive LOD score at the same position on chromosome 9p21 as Echo. However, a number of individuals in the Oklahoma pedigree that contributed the majority of evidence for the Echo QTL were missing ECG measurements. Based on statistical evidence demonstrating that ECG and Echo HR are significantly genetically correlated (rhoG = 0.88, P = 4.82 × 10−9), we conducted linkage analysis (17) for both ECG and Echo HR phenotypes simultaneously, modeling the genetic and environmental correlations between them along with effects of age, sex and their interactions in Sequential Oliogenic Linkage Analysis Routines (SOLAR) (18). Such joint or bivariate linkage facilitates the testing of whether the same QTLs in a region influence two correlated phenotypes and can improve power to detect QTLs under a standard additive genetic linkage model (17). This linkage resulted in increased evidence for a QTL (LOD = 4.83, P = 0.0003) at the same position on chromosome 9p21 (Fig. 1), indicating that these two phenotypes are influenced by the same gene in this region within Oklahoma participants. Bivariate analysis in Arizona, Dakota and the combined field centers did not result in significant linkage results (Table 2).
Figure 1.
Multipoint linkage plot of QTLs observed on chromosome 9p21 in the Oklahoma cohort from the SFHS. Light gray dashed line indicates HR; solid gray line, Echo HR; dotted light gray line, ECG HR.
To identify a gene(s) responsible for the HR QTL on chromosome 9p21 in Oklahoma, we conducted a gene-centric statistical analysis (19) for Echo HR using genotypes from 2248 heterozygous SNPs. Association analysis was done on Echo measurements of HR because ECG data were missing in key families demonstrating evidence for a QTL influencing HR. Power analysis indicates that with a Bonferroni correction for the testing of 2248 SNPs (i.e. a P-value threshold of 2.23 × 10−5), which is conservative given linkage disequilibrium (LD) between markers, we reach 80% power in Oklahoma to detect variants accounting for ≥2.1% of the trait variance. Forty genes were investigated within the 1-LOD region of the chromosome 9p21 QTL. Twenty-one of these genes contained SNPs showing nominal association with HR (i.e. measured genotype P-values <0.05). There were 555 genotyped SNPs within 10 kb of one of these 21 genes. Four genes [KIAA1797, interferon-epsilon-1 (IFNE1), LOC729983 and LOC402359] showed evidence of association (P < 0.05) in the gene-centric test (Table 3). The genes LOC4022359 and LOC729983 are pseudogenes, KIAA1797 is a hypothetical protein and IFNE1 belongs to a family of cytokines with pleiotropic effects that involve inhibition of viral replication and cell proliferation (20). Six SNPs (rs7875153, rs7848524, rs4446809, rs10964759, rs1125488 and rs7853123) within these four genes were still significant after correction for the effective number of SNPs. One of these significant SNPs (rs7848524) showed deviation from Hardy–Weinberg equilibrium (HWE = 0.02). Previous research on HR had identified two SNPs (rs13300284 and rs12552736) on chromosome 9p21 demonstrating suggestive association with the RR interval measure of HR. These two intergenic SNPs were included in our association analysis of Echo HR and were not significant in the Dakota, Oklahoma or combined AI centers. However, both SNPs (rs13300284 and rs12552736) demonstrated marginal evidence of association (P = 0.01) in the Arizona center.
Table 3.
Oklahoma Center SHFS gene-centric statistical analysis with 2248 heterozygous SNPs on chromosome 9p21
| Gene | Best SNP | SNP P-value | Number of SNPs | Effective number of SNPs | Gene-centric test | HWEa |
|---|---|---|---|---|---|---|
| KIAA1797 | rs7875153 | 0.00008 | 69 | 25 | 0.002 | 0.47 |
| — | rs4446809 | 0.00014 | — | — | 0.003 | 0.46 |
| — | rs10964759 | 0.00045 | — | — | 0.010 | 0.61 |
| IFNE1 | rs1125488 | 0.02503 | 3 | 1 | 0.025 | 0.10 |
| LOC729983 | rs7853123 | 0.00369 | 11 | 8 | 0.029 | 0.08 |
| LOC402359 | rs7848524 | 0.00098 | 8 | 5 | 0.049 | 0.02 |
| LOC392298 | rs3913107 | 0.00545 | 17 | 12 | 0.065 | 0.15 |
| LOC646505 | rs4977348 | 0.00535 | 15 | 12 | 0.066 | 0.49 |
| IFNA16 | rs12337907 | 0.02288 | 3 | 3 | 0.068 | 0.03 |
| PLAA | rs12003612 | 0.02711 | 12 | 4 | 0.108 | 0.49 |
| SLC24A2 | rs10757082 | 0.00295 | 63 | 38 | 0.112 | 0.12 |
| LOC402360 | rs1036107 | 0.00657 | 30 | 20 | 0.131 | 0.20 |
| MTAP | rs10117507 | 0.01581 | 28 | 9 | 0.142 | 0.03 |
| ELAVL2 | rs4977886 | 0.00785 | 50 | 30 | 0.236 | 0.53 |
| LOC392288 | rs11792752 | 0.01486 | 22 | 17 | 0.253 | 0.99 |
| C9orf82 | rs10511793 | 0.04801 | 18 | 6 | 0.288 | 0.30 |
| C9orf11 | rs911602 | 0.04525 | 14 | 8 | 0.362 | 0.89 |
| MOBKL2B | rs4879507 | 0.01262 | 68 | 41 | 0.517 | 0.48 |
| TEK | rs7035592 | 0.01786 | 71 | 42 | 0.750 | 0.56 |
| MLLT3 | rs7868612 | 0.02751 | 50 | 29 | 0.798 | 0.35 |
| IFNA1 | rs7864960 | 0.03402 | 1 | — | — | 0.93 |
| IFNA6 | rs660675 | 0.02922 | 1 | — | — | 0.83 |
| LOC729932 | rs7853123 | 0.00369 | 1 | — | — | 0.08 |
Significant SNPs in bold.
aHWE, Hardy–Weinberg equilibrium P-value for the best SNP.
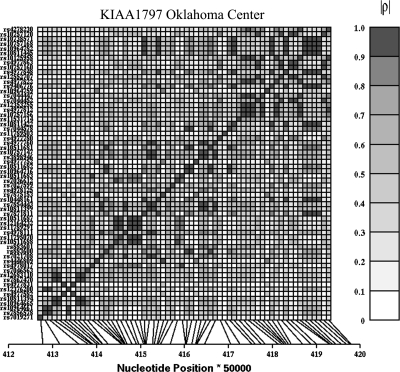
After correcting for effective number of SNPs in each gene accounting for the LD between markers, three SNPs within KIAA1797 met the threshold for inclusion (0.0021): rs7875153, rs4446809 and rs1096479. The KIAA1797 SNP, rs7875153, had the best measured genotype P-value (7.95 × 10−5). A plot of the LD between genotyped SNPs in KIAA1797 demonstrated rs7875153 was in high LD (r2 = 0.97) with rs1096479 as shown in Figure 2. This indicates that the significant association demonstrated by rs1096479 is due to its high LD with rs7875153. In addition, the KIAA1797 SNP rs4446809 is also in LD with the LOC402359 SNP rs7848524 (r2 = 0.84), indicating that LD may be why it is demonstrating statistical significance. The SNP rs7875153 is located in intron 9 of KIAA1797. Allele frequencies for rs7875153 in all three centers and the combined sample are presented in Table 4. The minor allele frequency (MAF) ranged from a high of 0.0433 in the Dakotas to a low of 0.006 in Arizona. Additionally, the same two families carrying the rare A allele in Oklahoma are the ones that contributed substantially to the 9p21 linkage signal. LOC729983 (rs7853123) and IFNE1 (rs1125488) each contained a single significant SNP. No evidence of association was found for HR with the best SNP, rs7875153, in the two other centers investigated for this research.
Figure 2.
LD plot of 69 SNPs within 10 kb of KIAA1797 within the Oklahoma cohort from the SHFS. Darker colors indicate higher r2 values.
Table 4.
Allele frequencies for SNP rs7875153 in the three Strong Heart centers and all centers combined
| Allele frequencya | ||
|---|---|---|
| Center | G | A |
| Oklahoma | 0.9720 | 0.0280 |
| Dakotas | 0.9567 | 0.0433 |
| Arizona | 0.9935 | 0.0065 |
| Combined | 0.9800 | 0.0200 |
aAllele frequencies from large pedigrees have been calculated using standard maximum likelihood methods accounting for family structure.
Given that only KIAA1797 remains significant in the gene centric test after Bonferroni correcting for the testing of 21 genes, we focused our subsequent analyses on this gene. The fact that Echo and ECG are measures of the same HR phenotype allowed us to conduct a novel joint association analysis of both traits simultaneously with the SNP, rs7875153, which had provided the best evidence for association. In this type of joint or bivariate association analysis, we use a standard additive genetic model, testing whether trait mean varies by genotype at the SNP. The fact that the Echo and ECG phenotypes are measurements of the same underlying trait, and are thus on the same scale, allows a single degree of freedom test as we can constrain the effect of genotype on the trait mean to be equal for both HR phenotypes. The difference between the log-likelihoods of a model in which the SNP effect is estimated versus one in which it is constrained to zero is then distributed as a χ2 distribution with 1 degree of freedom. We applied this novel type of bivariate association analysis to Echo and ECG HR with the SNP, rs7875153. This bivariate association analysis strengthened evidence for association of HR with rs7875153 (P = 7.14 × 10−6) in Oklahoma participants.
To independently support these association results in a separate ethnic population, we used lymphocyte RNA expression data on 1240 individuals from the San Antonio Family Heart Study (SAFHS), a longitudinal genetic epidemiological study of CVD in Mexican Americans residing in San Antonio, Texas (21). The expression profiling methodology has been described in detail by Göring et al. (22). We used phenotypic data on HR measured from ECG in SAFHS participants and linear regression to test the correlation between HR and KIAA1797 RNA levels. This test utilizes a χ2 test with one degree of freedom to determine whether the regression beta-coefficient is different from zero. Tests were conducted within a variance component framework to account for non-independence among family members. Expression data for KIAA1797 were correlated with ECG HR in Mexican Americans (P = 0.0115) and the regression coefficient (−0.0038) was negative, indicating that individuals with higher levels of KIAA1797 expression have lower HRs. The SNP showing association with HR in our Oklahoma sample, rs7875153, is not currently available in the SAFHS.
DISCUSSION
HR is an important clinical phenotype that is associated with CVD (1–3). Individuals characterized by increased HR show elevated levels of total cholesterol, triglycerides, fasting insulin and other known risk factors for hypertension (23). The relationship between elevated HR-associated disease and mortality is not completely understood. Researchers suggest that elevated HR is an indicator of irregular behavior in the autonomic nervous system and modification that lowers HR through surgical (24) or pharmaceutical means (25) in animals reduces development of coronary atherosclerosis. However, owing to evidence that HR is an independent predictor of CVD, identification of underlying genetic factors influencing HR may allow for the advancement in clinical treatments that aid in lowering risk of atherosclerosis and CVD. Our results indicate HR is a significantly heritable trait in AIs, and our findings are consistent with previous estimates from other population-based studies. Heritability estimated in our study ranged from 0.26 (±0.08) in Arizona to 0.30 (±0.06) in Oklahoma for Echo HR and 0.27 (±0.05) in Arizona and Oklahoma to 0.35 (±0.05) in the Dakotas for ECG HR. Both of these results fit within the range of 0.21 in FHS (7) to 0.41 in twin studies from the Netherlands (8).
Results from the AI bivariate linkage of HR detected a significant QTL on chromosome 9p21 in Oklahoma SHFS participants. Further investigation of 2248 SNPs within this region demonstrated genetic association with three SNPs in KIAA1797. The best SNP, rs7875153, is rare in these AI groups with MAFs <0.05. However, in other HapMap populations, the MAF of this SNP ranges from 0.138 in the Japanese to 0.198 in Chinese and is intermediate at 0.158 in Europeans and Africans (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=7875153). Given that AI participants investigated for this study come from a wide geographic range in North America, it is unlikely that isolation of a specific population group is the reason for the lower MAF in the SHFS. To further support this observed association, we used RNA expression data from the SAFHS (22) and identified a significant correlation between levels of KIAA1797 RNA and HR measured by ECG in Mexican Americans (21). This evidence points to either a yet-undefined role of KIAA1797 in regulating HR or some other function within the autonomic nervous system and suggests that variants influencing inter-individual variation in HR may act through the regulation of KIAA1797 expression.
The gene KIAA1797 codes for a hypothetical protein with an undefined function. However, it is known to interact with the gene vinculin (VCL), a gene involved in the development of dilated cardiomypathy (26,27). One group of researchers suggests that KIAA1797 is involved in mitotic chromosome condensation; however, they offer little evidence in this regard (28). Mitotic chromosome condensation refers to the biological process where the chromatin is unraveled during the interphase stage of mitosis and becomes invisible. The chromosome becomes visible once again during mitosis when chromatin is bundled into a more condensed state (29). Until recently, it was believed that cardiac myocytes did not undergo cellular replication, making the relationship between a gene potentially involved in mitosis and HR tenuous. However, a recent study suggested that cardiomyocytes renew at an age-dependent rate of 1% starting at the age of 25 to 0.45% by the age of 75 (30). This raises the possibility that if KIAA1797 is influencing HR via myocyte turnover, the effect of KIAA1797 may change with age, either as a consequence or as a cause of the changing rate of cardiomyocyte renewal with age. However, there was no correlation between KIAA1797 RNA levels and age in the SAFHS, suggesting that gene expression is stable across the adult lifespan, and there was no indication that the effect of associated SNPs in KIAA1797 on HR changed with age in the SHFS (data not shown). This indicates that further research is necessary to better understand the relationship between KIAA1797 and HR.
A number of recent GWAS studies have implicated chromosome 9p21 in CVD (15,31), ischemic stroke (32,33), MI (16,33–35), and type II diabetes (36). These studies identified four SNPs (rs10757274, rs2383206, rs2383207 and rs10757278) that are related to MI and CHD (37–39). All four of these SNPs were included in our association analysis for Echo HR and are located in the gene, LOC729983. However, none was significantly associated with Echo or ECG HR. In addition, all of these SNPs are located approximately 1 million base pairs from the SNP that shows the strongest association with Echo HR. This does not imply that these SNPs are not involved in CVD but they do not appear to be associated with HR. In addition, two previous intergenic SNPs (rs13300284 and rs12552736) on chromosome 9p21 demonstrated suggestive association with HR measured as the RR interval (13). Both of these SNPs were included in our analysis and were not significant in the Dakotas, Oklahoma or combined AI center for Echo HR. Thus, it is unlikely that they contribute to the chromosome 9 HR linkage signal observed in the Oklahoma sample. They were, however, marginally significant (P = 0.01 for both rs13300284 and rs12552736) for the association with HR within the Arizona center.
To our knowledge, this is the first study to demonstrate genetic association for HR in the AI population. CVD accounts for a high percentage of mortality and morbidity in AI communities, yet the underlying causes and associated risk factors are still not fully understood (40,41). Therefore, understanding differences between populations and their underlying genetic factors that contribute to risk factors of CVD is an important consideration for further studies. One interesting finding is that ECG did not provide as robust a QTL as Echo when detecting a QTL for HR in Oklahoma participants. This result is due to the evidence that individuals in the two Oklahoma families that contributed to the Echo QTL were missing ECG values. However, there are no significant differences in phenotypic measurements in those participants with Echo but missing ECG values when compared with participants with both measures available, and there was no systematic exclusion of those individuals in obtaining ECG results. Other potential reasons for differences between HR measurements could be due to the different positions that the participants are placed in during the examination (slightly on left side for Echo HR versus flat on back for ECG) time of day the examination was conducted or that Echo HR measurement increases sympathetic activity. In addition, previous cardiology research in non-diabetic Strong Heart Study (SHS) participants demonstrated left ventricular and aortic size differences in these three AI communities and there is the potential that these differences may be indicative of the observed phenotypic differences in HR (42,43).
The strength of the current study involves the combined use of linkage, genetic association analysis and RNA expression data to account for potential variation within a given phenotype. Successful genetic association studies result in the localization of a gene or genetic region involved with the investigated phenotype. Therefore, various sources of independent data can be used to augment evidence for the involvement of a particular gene and improve assumptions regarding causal associations. In this study, we used family-based genetic data from three geographically distinct AI groups to identify a region and gene of interest and RNA expression data from a longitudinal study of Mexican Americans in order to find independent confirmation connecting the gene KIAA1797 and HR. We also applied a bivariate association method, conducting joint association analyses for two measures of HR, for a better understanding of the relationship between two related phenotypic traits and a single genetic locus. This joint analysis of both HR measures simultaneously was important to tie together the significant evidence of a QTL on chromosome 9p21 and association of KIAA1797 markers with Echo HR in the SHFS and the significant correlation of ECG HR with KIAA1797 expression levels in the SAFHS. Additionally, combining these two phenotypes (Echo and ECG HR) in a single association analysis should theoretically increase power to detect association. While this study combined information from two different population studies, both cohorts were ascertained without regard to phenotype or disease status. There is the potential that the relationship between RNA expression level and investigated phenotypes may vary across populations. However, we feel that this combining of different sources of information allowed for greater resolution in understanding the genetic factors that may be involved in HR.
In summary, we identified a novel QTL on chromosome 9p21 from the SFHS Oklahoma center that influences HR in AIs. Through association analysis of 2248 SNPs in this region, we identified significant association with SNPs in KIAA1797. The best SNP, rs7875153, was significant in Oklahoma but not in the other two centers or in the combined sample. However, we demonstrated further support for the relationship between KIAA1797 and HR through the use of RNA expression data from the SAFHS. Therefore, this lack of replication may imply that rs7875153 is in high LD with another relatively rare functional variant in KIAA1797 private to the two families in the Oklahoma sample or that statistical power was insufficient to detect it in the other centers. Given the significance of HR in understanding CVD, these results may provide important information for understanding the different ethnic populations. However, in order to better determine whether genetic variation within KIAA1797 is associated with HR, further research and deep sequencing of the gene are necessary with a focus on identifying potential regulatory variants that might explain the correlation between gene expression and HR.
MATERIALS AND METHODS
Study population
SHS investigates cardiovascular risk factors and heart disease in the AI population (41). Population-based samples of AI participants aged 45–74 years were recruited from three geographic areas, Arizona, Oklahoma, and North and South Dakota, to understand risk factors involved with CVD in this population. In 1998, the SHFS was initiated for additional genetic studies and participants aged 14 years and older were recruited without regard to disease status. Families were selected through sibships of at least five siblings, of whom at least three had participated in the original SHS. Parents, spouses, offspring, spouses of offspring and grandchildren of the original participants were then recruited to construct extended pedigrees. All participants provided informed consent. In addition, study approval was obtained from relevant AI tribes and institutional review boards.
Phenotype measurement
HR was measured from ECGs (ECG HR) and Echo Doppler recording (Echo HR) at SHS exam 4. Standard 12-lead ECGs were performed with MAC-PC or MAC-12 digital ECG systems (GE Medical Systems) similar to SHS exam 1, as described previously (44). ECG HR was calculated from standard 12-lead ECGs using computerized algorithm. Echocardiograms were performed using commercially available phased-array Echos using identical methods employed in SHS exam 2, as described previously (42,45). Echo HR was measured using a digitized computer station (Digisonics, Inc.). In families genotyped for the genome-wide linkage screen, Echo HR measurements are available for 3567 AI participants (1204 in Arizona, 1173 in the Dakotas and 1193 in Oklahoma) and ECG HR measurements for 3521 AI participants (1187 in Arizona, 1186 in the Dakotas, 1148 in Oklahoma).
Genotyping
SNP genotyping and association analysis were conducted using a set of 1122 ‘founders’ chosen from SHFS participants. These were SHFS participants genotyped for the original genome scan but did not have parents or grandparents with genotyping data. These individuals were therefore likely to be contributing most of the alleles informative for linkage and association analyses. A set of 2329 SNPs from the 8.2 Mb 1-LOD support interval on chromosome 9p21 were included on Illumina iSelect Custom 12-Sample BeadChips and genotyped using the Infinium II Assay with one bead type per SNP. Assays were performed according to manufacturer's protocol (Illumina, San Diego, CA, USA). Genomic DNA (200 ng) was whole-genome-amplified, enzymatically fragmented and hybridized to locus-specific polymers (50 bp) that are covalently linked to beads embedded on the BeadChips. Hybridization was followed by allele-specific primer extension and fluorescent staining. All steps were performed on a Tecan Freedom EVO 150 cm liquid handler (Tecan, Männedorf, Switzerland) with Illumina GTS Robot Control software (Illumina). Fluorescent intensities were detected using the Illumina BeadArray Reader and analyzed using BeadStudio software from Illumina. Cluster calls were checked for accuracy and genotypes were exported as text files for use in association analysis. Six samples were typed in replica as controls for genotyping and allele calling consistencies. Illumina included sample-dependent and -independent controls on their chips to test for accuracy. Of 2329 SNPs on chromosome 9p21, 2248 SNPs were genotyped and heterozygous, 2 SNPs were typed but homozygous and 79 SNPs failed.
Statistical analysis
Maximum likelihood methods, taking into account the relationships among family members, were used to estimate allelic frequencies for each marker with the computer program SOLAR (18). The computer program Loki was used to compute multipoint identity by descent (IBD) matrices, with marker map positions drawn from DeCode Genetics (46). IBD matrices were estimated separately for each field center and combined for joint analysis of all three centers to account for potential differences in allele frequency between populations. In order to identify pedigree misspecifications, pedigree relationship statistical tests (47) were used. The PEDSYS program, INFER, was used to determine unknown genotypes from close relatives, if they could be unambiguously inferred, and to resolve Mendelian inconsistencies (48).
Genetic association and linkage analyses, both for single phenotypes and for joint bivariate analyses of both measures of HR simultaneously, were all conducted using variance component methods, as implemented in SOLAR (18). Sex, sex-specific age and age (2) were included as covariates in all analyses. Bivariate multipoint linkage analyses of Echo and ECG HR were conducted for each of three field centers and for a combined analysis of all three centers. Bivariate linkage tests the linkage of two correlated phenotypes to a single genetic region simultaneously and tests the null hypothesis that variance components relating to major gene effects on both traits are equal to 0 (17). Covariates used in this analysis are shown in Table 1. Variables were kept within the final polygenic model, and carried forward to linkage and association models, if significantly associated with HR at P < 0.05. Since variance components are sensitive to kurtosis, HR was transformed using an inverse normalization procedure available in SOLAR (49). Genome-wide P-values were obtained using the Feingold et al. method (50). HWE was determined using maximum likelihood estimation, by comparing the likelihood of a genotype-based frequency model with that of allele-frequency model, which assumes HWE. Estimations for each of the measured SNPs were conducted in SOLAR (18).
Genetic association was determined through the application of a gene-centric statistical test that utilizes the effective number of SNPs, given the LD between them, and was better able to determine significance under multiple testing (19). We implemented this method by defining the physical location of each SNP within 10 000 base pairs (10 kb) of 40 genes in chromosome 9p21. Measured genotype analysis was conducted on each SNP in order to calculate a nominal P-value for association. The measured genotype analysis was conducted for each polymorphic SNP, in which the number of minor alleles is added to the quantitative polygenic genetic model as a covariate in order to assess the effect of the SNP genotype on the mean of the trait. This model was fitted to the data and compared with the null model of no difference in trait mean by genotype using a likelihood-ratio test. Twice the difference in log-likelihoods of these models was distributed as a χ2 random variable with 1 degree of freedom. The resulting likelihood-ratio test statistic was recorded for each SNP (19). We calculated the effective number of SNPs in each gene using Moskvina and Schmidt's (51) method implemented in SOLAR (18). The nominal P-value and the effective number of SNPs were used to determine an adjusted P-value for each gene to be used in multiple testing. This gene-centric P-value was calculated using the equation
where ‘corrected’ is the corrected P-value, ‘nominal’ is the uncorrected P-value and ‘effective’ is the effective number of SNPs. This approach allows for non-independence among family members and accounts for effects of other potential covariates (19).
We applied a novel bivariate association analysis, jointly testing association with both measures of HR simultaneously, based within the variance component framework using the SNP rs7875153 as this had provided the best measured genotype value. In this type of analysis, the effect of a SNP on the mean trait values of the two phenotypes is constrained to be equal for both measures and is included as a covariate in the bivariate analysis. The difference between the log-likelihoods of a standard additive genetic model in which the SNP effect is estimated versus one in which it is constrained to zero is then distributed as a χ2 distribution with 1 degree of freedom.
FUNDING
This research was funded by National Institute of Health Grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, U01 HL65521 and R01 HL45522. Development of SOLAR was supported by National Institute of Health grant MH59490. This investigation was conducted in part in facilities constructed with the support from the Research Facilities Improvement Program under grants numbered C06 RR013556 and C06 RR017515. The AT&T Genomics Computing Center supercomputing facilities used for statistical genetic analyses were supported in part by a gift from the SBC Foundation.
ACKNOWLEDGEMENTS
We thank the Strong Heart Family Study participants, the Indian Health Service hospitals and clinical staff and the staff of the SHS field centers. The views expressed in this paper are those of the authors and do not necessarily reflect those of the Indian Health Service. All authors read and approved the final manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Greenland P., Daviglus M.L., Dyer A.R., Liu K., Huang C.F., Goldberger J.J., Stamler J. Resting heart rate is a risk factor for cardiovascular and noncardiovascular mortality: The Chicago Heart Association Detection Project in Industry. Am. J. Epidemiol. 1999;149:853–862. doi: 10.1093/oxfordjournals.aje.a009901. [DOI] [PubMed] [Google Scholar]
- 2.Cook S., Togni M., Schaub M.C., Wenaweser P., Hess O.M. High heart rate: a cardiovascular risk factor? Eur. Heart J. 2006;27:2387–2393. doi: 10.1093/eurheartj/ehl259. doi:10.1093/eurheartj/ehl259. [DOI] [PubMed] [Google Scholar]
- 3.King D.E., Everett C.J., Mainous A.G., III, Liszka H.A. Long-term prognostic value of resting heart rate in subjects with prehypertension. Am. J. Hypertens. 2006;19:796–800. doi: 10.1016/j.amjhyper.2006.01.019. doi:10.1016/j.amjhyper.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Frazer K.A., Murray S.S., Schork N.J., Topol E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. doi:10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 5.Iles M.M. What can genome-wide association studies tell us about the genetics of common disease? PLoS Genet. 2008;4:e33. doi: 10.1371/journal.pgen.0040033. doi:10.1371/journal.pgen.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Bonham A.J., Li J., Pei Y.F., Chen J., Papasian C.J., Deng H.W. Family-based bivariate association tests for quantitative traits. PLoS ONE. 2009;4:e8133. doi: 10.1371/journal.pone.0008133. doi:10.1371/journal.pone.0008133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh J.P., Larson M.G., O'Donnell C.J., Tsuji H., Evans J.C., Levy D. Heritability of heart rate variability: the Framingham Heart Study. Circulation. 1999;99:2251–2254. doi: 10.1161/01.cir.99.17.2251. [DOI] [PubMed] [Google Scholar]
- 8.Kupper N.H., Willemsen G., van den Berg M., de Boer D., Posthuma D., Boomsma D.I., de Geus E.J. Heritability of ambulatory heart rate variability. Circulation. 2004;110:2792–2796. doi: 10.1161/01.CIR.0000146334.96820.6E. doi:10.1161/01.CIR.0000146334.96820.6E. [DOI] [PubMed] [Google Scholar]
- 9.Laramie J.M., Wilk J.B., Hunt S.C., Ellison R.C., Chakravarti A., Boerwinkle E., Myers R.H. Evidence for a gene influencing heart rate on chromosome 5p13–14 in a meta-analysis of genome-wide scans from the NHLBI family blood pressure program. BMC Med. Genet. 2006;7:17. doi: 10.1186/1471-2350-7-17. doi:10.1186/1471-2350-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilk J.B., Myers R.H., Zhang Y., Lewis C.E., Atwood L., Hopkins P.N., Ellison R.C. Evidence for a gene influencing heart rate on chromosome 4 among hypertensives. Hum. Genet. 2002;111:207–213. doi: 10.1007/s00439-002-0780-9. doi:10.1007/s00439-002-0780-9. [DOI] [PubMed] [Google Scholar]
- 11.Martin L.J., Comuzzie A.G., Sonnenberg G.E., Myklebust J., James R., Marks J., Blangero J., Kissebah A.H. Major quantitative trait locus for resting heart rate maps to a region on chromosome 4. Hypertension. 2004;43:1146–1151. doi: 10.1161/01.HYP.0000122873.42047.17. doi:10.1161/01.HYP.0000122873.42047.17. [DOI] [PubMed] [Google Scholar]
- 12.Newton-Cheh C., Guo C.Y., Wang T.J., O'Donnell C.J., Levy D., Larson M.G. Genome-wide association study of electrocardiographic and heart rate variability traits: the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl. 1):S7. doi: 10.1186/1471-2350-8-S1-S7. doi:10.1186/1471-2350-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marroni F., Pfeufer A., Aulchennko Y.S., Franklin C.S., Isaacs A., Pichler I., Wild S.H., Oostra B.A., Wright A.F., Campbell H., et al. A genome-wide association scan of RR and QT interval duration in three European genetically isolated populations. The EUROSPAN project. Circ. Cardiovasc. Genet. 2009;2:322–328. doi: 10.1161/CIRCGENETICS.108.833806. doi:10.1161/CIRCGENETICS.108.833806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutherford S., Voruganti V.S., Goring H.H., Laston S.L., Haack K., Almasy L., Comuzzie A., Lee E.T., Best L.G., Fabsitz R.R., et al. A heart rate genetic locus on chromosome 9p21 in the Strong Heart Family Study. Hypertension. 2008;52:e101. [Google Scholar]
- 15.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. doi:10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. doi:10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 17.Almasy L., Dyer T.D., Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet. Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. doi:10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Almasy L., Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. doi:10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlesworth J.C., Peralta J.M., Drigalenko E., Goring H.H., Almasy L., Dyer T.D., Blangero J. Toward the identification of causal genes in complex diseases: a gene-centric joint test of significance combining genomic and transcriptomic data. BMC Proc. 2009;3(Suppl. 7):S92. doi: 10.1186/1753-6561-3-s7-s92. doi:10.1186/1753-6561-3-s7-s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark G.R. How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev. 2007;18:419–423. doi: 10.1016/j.cytogfr.2007.06.013. doi:10.1016/j.cytogfr.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacCluer J.W., Stern M.P., Almasy L., Atwood L.A., Blangero J., Comuzzie A.G., Dyke B., Haffner S.M., Henkel R.D., Hixson J.E., et al. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr. Rev. 1999;57:S59–S65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 22.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. doi:10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 23.Fujiura Y., Adachi H., Tsuruta M., Jacobs D.R., Jr, Hirai Y., Imaizumi T. Heart rate and mortality in a Japanese general population: an 18-year follow-up study. J. Clin. Epidemiol. 2001;54:495–500. doi: 10.1016/s0895-4356(00)00323-1. doi:10.1016/S0895-4356(00)00323-1. [DOI] [PubMed] [Google Scholar]
- 24.Beere P.A., Glagov S., Zarins C.K. Retarding effect of lowered heart rate on coronary atherosclerosis. Science. 1984;226:180–182. doi: 10.1126/science.6484569. doi:10.1126/science.6484569. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan J.R., Manuck S.B., Adams M.R., Weingand K.W., Clarkson T.B. Inhibition of coronary atherosclerosis by propranolol in behaviorally predisposed monkeys fed an atherogenic diet. Circulation. 1987;76:1364–1372. doi: 10.1161/01.cir.76.6.1364. [DOI] [PubMed] [Google Scholar]
- 26.de Hoog C.L., Foster L.J., Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. doi:10.1016/S0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 27.Vasile V.C., Will M.L., Ommen S.R., Edwards W.D., Olson T.M., Ackerman M.J. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol. Genet. Metab. 2006;87:169–174. doi: 10.1016/j.ymgme.2005.08.006. doi:10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Matlik K., Redik K., Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koshland D., Strunnikov A. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. doi:10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 30.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabe-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H., et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. doi:10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., et al. Genome wide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. doi:10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu W.L., Li S.J., Liu D.T., Wang Y., Niu S.Q., Yang X.C., Zhang Q., Yu S.Z., Jin L., Wang X.F. Genetic variants on chromosome 9p21 and ischemic stroke in Chinese. Brain Res. Bull. 2009;79:431–435. doi: 10.1016/j.brainresbull.2009.04.001. doi:10.1016/j.brainresbull.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Wahlstrand B., Orho-Melander M., Delling L., Kjeldsen S., Narkiewicz K., Almgren P., Hedner T., Melander O. The myocardial infarction associated CDKN2A/CDKN2B locus on chromosome 9p21 is associated with stroke independently of coronary events in patients with hypertension. J. Hypertens. 2009;27:769–773. doi: 10.1097/HJH.0b013e328326f7eb. doi:10.1097/HJH.0b013e328326f7eb. [DOI] [PubMed] [Google Scholar]
- 34.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., et al. Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. doi:10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paynter N.P., Chasman D.I., Buring J.E., Shiffman D., Cook N.R., Ridker P.M. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann. Intern. Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doria A., Wojcik J., Xu R., Gervino E.V., Hauser T.H., Johnstone M.T., Nolan D., Hu F.B., Warram J.H. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. JAMA. 2008;300:2389–2397. doi: 10.1001/jama.2008.649. doi:10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdullah K.G., Li L., Shen G.Q., Hu Y., Yang Y., MacKinlay K.G., Topol E.J., Wang Q.K. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest) Ann. Hum. Genet. 2008;72:654–657. doi: 10.1111/j.1469-1809.2008.00454.x. doi:10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G.Q., Li L., Rao S., Abdullah K.G., Ban J.M., Lee B.S., Park J.E., Wang Q.K. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2008;28:360–365. doi: 10.1161/ATVBAHA.107.157248. doi:10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 39.Shen G.Q., Rao S., Martinelli N., Li L., Olivieri O., Corrocher R., Abdullah K.G., Hazen S.L., Smith J., Barnard J., et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J. Hum. Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. doi:10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 40.Lee E.T., Welty T.K., Fabsitz R., Cowan L.D., Le N.A., Oopik A.J., Cucchiara A.J., Savage P.J., Howard B.V. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am. J. Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 41.Howard B.V., Lee E.T., Cowan L.D., Devereux R.B., Galloway J.M., Go O.T., Howard W.J., Rhoades E.R., Robbins D.C., Sievers M.L., et al. Rising tide of cardiovascular disease in American Indians. The Strong Heart Study. Circulation. 1999;99:2389–2395. doi: 10.1161/01.cir.99.18.2389. [DOI] [PubMed] [Google Scholar]
- 42.Devereux R.B., Roman M.J., Paranicas M., O'Grady M.J., Lee E.T., Welty T.K., Fabsitz R.R., Robbins D., Rhoades E.R., Howard B.V. Impact of diabetes on cardiac structure and function: the Strong Heart Study. Circulation. 2000;101:2271–2276. doi: 10.1161/01.cir.101.19.2271. [DOI] [PubMed] [Google Scholar]
- 43.Devereux R.B., Roman M.J., O'Grady M.J., Fabsitz R.R., Rhoades E.R., Crawford A., Howard B.V., Lee E.T., Welty T.K. Differences in echocardiographic findings and systemic hemodynamics among non-diabetic American Indians in different regions: the Strong Heart Study. Ann. Epidemiol. 2000;10:324–332. doi: 10.1016/s1047-2797(00)00059-4. doi:10.1016/S1047-2797(00)00059-4. [DOI] [PubMed] [Google Scholar]
- 44.Okin P.M., Devereux R.B., Fabsitz R.R., Lee E.T., Galloway J.M., Howard B.V. Strong Heart Study. Quantitative assessment of electrocardiographic strain predicts increased left ventricular mass: the Strong Heart Study. J. Am. Coll. Cardiol. 2002;40:1395–1400. doi: 10.1016/s0735-1097(02)02171-x. doi:10.1016/S0735-1097(02)02171-X. [DOI] [PubMed] [Google Scholar]
- 45.Bella J.N., MacCluer J.W., Roman M.J., Almasy L., North K.E., Best L.G., Lee E.T., Fabsitz R.R., Howard B.V., Devereux R.B. Heritability of left ventricular dimensions and mass in American Indians: the Strong Heart Study. J. Hypertens. 2004;22:281–286. doi: 10.1097/00004872-200402000-00011. doi:10.1097/00004872-200402000-00011. [DOI] [PubMed] [Google Scholar]
- 46.Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 47.Sun L., Wilder K., McPeek M.S. Enhanced pedigree error detection. Hum. Hered. 2002;54:99–110. doi: 10.1159/000067666. doi:10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 48.Dyke B. PEDSYS: A Pedigree Data Management System User's Manual. PGL Technical Report 2. San Antonio: Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research; 1995. [Google Scholar]
- 49.Rutherford S., Cai G., Lopez-Alvarenga J.C., Kent J.W., Voruganti V.S., Proffitt J.M., Curran J.E., Johnson M.P., Dyer T.D., Jowett J.B., et al. A chromosome 11q quantitative-trait locus influences change of blood-pressure measurements over time in Mexican Americans of the San Antonio Family Heart Study. Am. J. Hum. Genet. 2007;81:744–755. doi: 10.1086/521151. doi:10.1086/521151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feingold E., Brown P.O., Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am. J. Hum. Genet. 1993;53:234–251. [PMC free article] [PubMed] [Google Scholar]
- 51.Moskvina V., Schmidt K.M. Individual SNP allele reconstruction from informative markers selected by a non-linear gauss-type algorithm. Hum. Hered. 2006;62:97–106. doi: 10.1159/000096097. doi:10.1159/000096097. [DOI] [PubMed] [Google Scholar]