Abstract
Malformations of cortical development are characteristic of a plethora of diseases that includes polymicrogyria, periventricular and subcortical heterotopia and lissencephaly. Mutations in TUBA1A and TUBB2B, each a member of the multigene families that encode α- and β-tubulins, have recently been implicated in these diseases. Here we examine the defects that result from nine disease-causing mutations (I188L, I238V, P263T, L286F, V303G, L397P, R402C, 402H, S419L) in TUBA1A. We show that the expression of all the mutant proteins in vitro results in the generation of tubulin heterodimers in varying yield and that these can co-polymerize with microtubules in vitro. We identify several kinds of defects that result from these mutations. Among these are various defects in the chaperone-dependent pathway leading to de novo tubulin heterodimer formation. These include a defective interaction with the chaperone prefoldin, a reduced efficiency in the generation of productive folding intermediates as a result of inefficient interaction with the cytosolic chaperonin, CCT, and, in several cases, a failure to stably interact with TBCB, one of five tubulin-specific chaperones that act downstream of CCT in the tubulin heterodimer assembly pathway. Other defects include structural instability in vitro, diminished stability in vivo, a compromised ability to co-assemble with microtubules in vivo and a suppression of microtubule growth rate in the neurites (but not the soma) of cultured neurons. Our data are consistent with the notion that some mutations in TUBA1A result in tubulin deficit, whereas others reflect compromised interactions with one or more MAPs that are essential to proper neuronal migration.
INTRODUCTION
The organization of the mammalian brain depends on events involving extensive neuronal migration during development. Following proliferation in the ventricular zone, neurons migrate to their final destination in an orchestrated fashion (1). These migration events are influenced by a spectrum of genes that regulate the advancement of the leading neurite, translocation of the nucleus and retraction of the trailing process. Not surprisingly, mutations in these genes result in a range of neurodevelopmental diseases that are characterized by severe cortical malformations. In most cases, there are profound accompanying disabilities, including severe mental retardation and epilepsy (2).
Malformations of cortical development have been classified into several subcategories. These include polymicrogyria, periventricular heterotopia, subcortical band heterotopia and lissencephaly. The pathological features of these diseases have been associated with mutations in a number of genes, including SPRX2 (3), KIAA1279 (4), GPR56 (5,6), PAX6 (7), TBR2 (8), COL18A1 (9), FLNA1 (10), RAB3GAP1 (11), DCX (12,13) and LIS1 (14,15). Both DCX and LIS1 have been implicated in the regulation of cytoskeletal function via modulation of microtubule polymerization. LIS1 binds to dynein via a number of distinct sites (16,17), as well as to several proteins (mNudC, mNudE, NudEL) that are involved in nuclear distribution (18,19). These observations imply a link between LIS1 and nucleokinesis. DCX co-assembles with brain microtubules, both nucleating and stabilizing them (20), consistent with an important role in influencing microtubule function (21,22).
It is becoming increasingly clear that nuclear migration and growth cone extension are key components of neuronal migration and that both are dependent on a dynamic microtubule network (23). The subunit from which microtubules are assembled is the α/β-tubulin heterodimer, consisting of one α- and one β-tubulin polypeptide. Each microtubule is a polarized polymer consisting of 13 protofilaments; these are formed by the head-to-tail juxtaposition of tubulin heterodimers and surround a hollow core. In mammals, a small multigene family encodes the α- and β-tubulins, and the pattern of expression of tubulin-encoding genes shows variation among different tissues as well as distinctive patterns of regulation during development (24,25). The dynamic behavior of microtubules is characterized by the entry or release of tubulin heterodimers from the plus ends of the microtubule polymer such that the microtubule either grows or shrinks, a process termed ‘dynamic instability’ (26). A key feature of dynamic instability is that incorporation of tubulin heterodimers containing GTP bound to the β-subunit results in a so-called ‘GTP cap’, which stabilizes the microtubule end, and that the size of this cap determines whether a given microtubule will grow or transition to a rapidly depolymerizing phase. The importance of microtubules and the proper regulation of their dynamic behavior for neuronal migration are emphasized by our discovery that mutations in a α-tubulin subunit encoded by TUBA1A cause lissencephaly (27). Indeed, mutations in TUBA1A (28,29) cause a variety of complex brain disorders. Moreover, there is an association between bilateral asymmetrical polymicrogyria and de novo mutations in TUBB2B (15), and heterozygous missense mutations in TUBB3 result in a spectrum of nervous system disorders referred to as TUBB3 syndromes (30).
In principle, mutations in tubulins could result in defects in neuronal migration and differentiation via a number of different mechanisms. In one scenario, there could be a defect in microtubule dynamics conferred, for example, by compromised binding of guanine nucleotide or by a disruption of one or more critical interactions with a microtubule effector such as a molecular motor (kinesin or dynein) responsible for intracellular trafficking in neurons. In a second and unrelated potential mechanism, a mutation in tubulin could result in a reduced abundance of functional tubulin heterodimers. In this context, it is important to note that the tubulin heterodimer cannot assemble spontaneously. Rather, newly translated α- and β-tubulin polypeptides must first interact with several chaperone proteins. These include prefoldin (PFD) (31), the cytosolic chaperonin CCT (32) and a series of five chaperone proteins termed TBCA-TBCE (33) that function in concert to integrate α- and β-tubulins (neither of which exist as a stable independent entity) into the heterodimer (summarized in Supplementary Material, Fig. S1). Genetic experiments with model organisms have shown that CCT as well as TBCB, TBCD, TBCC and TBCE are essential for life: consequently, defective interaction of a mutant form of tubulin with any one of these chaperones could result in haploinsufficiency [defined as a situation in which one copy of a wild-type gene is insufficient to provide wild-type function (34,35); note that this is not limited to null mutations, but also extends to missense mutations that result in compromised function (36,37)]. Indeed, analysis of the mechanism underlying a pachygyria-causing mutation in the TUBA1A mutation R264C has revealed a number of defects in the overall tubulin heterodimer assembly pathway, the most striking of which is a failure of quasi-native α-tubulin molecules to interact efficiently with TBCB (38). Here we present an analysis of the biochemical and biological consequences of nine disease-causing mutations in TUBA1A [I188L, P263T, L286F, R402C, R402H, S419L (28); I238V (39); L397P (29) and V303G]. Our data reveal the existence of a class of mutations that fail to interact efficiently with TBCB, as well as other defects that compromise the biosynthetic pathway leading to heterodimer assembly or that affect microtubule dynamics.
RESULTS
Location of disease-associated residues within the structure of the α-tubulin polypeptide
We mapped the location of disease-associated mutated residues onto the structure of the α-tubulin polypeptide (Fig. 1). This analysis showed that the mutations are widely scattered: seven mutated residues (P263, L286, V303, L397, R402, S419) are located on the surface, whereas two (I188 and I238) are buried in the molecule's interior. Two of the mutated residues (R402 and L397) lie close to points of contact with β-tubulin, whereas two others (L286 and V303) lie in fairly close proximity to the GTP-binding pocket. The fact that the mutations show no evidence of clustering suggested that several different mechanisms might contribute to the clinical pathology. We therefore proceeded with a comprehensive biochemical and biological analysis of the effects of the mutations on tubulin folding, heterodimer assembly, microtubule dynamics and stability.
Figure 1.

Location of mutated residues within the structure of the α-tubulin polypeptide. The figure shows the surface location of seven amino acids [R264 (38), P263, L286, V303, L397, R402 and S419 (this study)] on the α-tubulin polypeptide. Note that two different mutations (R402C and R402H) are located at R402. Two other disease-associated mutations located at I188 and I238 are not shown in the figure because they are buried in the interior of the molecule.
Expression of disease-causing mutations by transcription/translation in vitro
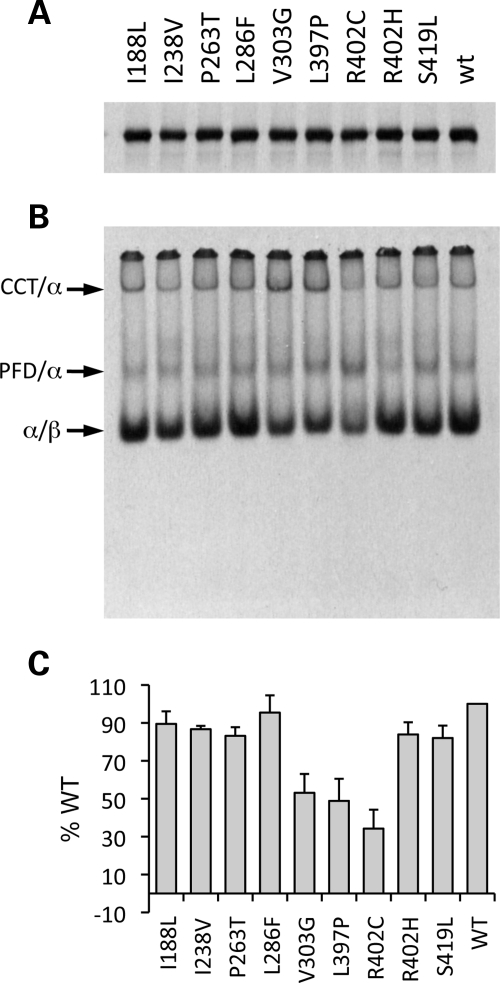
We first examined the effects of the disease-associated mutations in a translational context. To do this, we analyzed the products of in vitro transcription/translation generated in rabbit reticuloctye lysate. The reaction products were analyzed in two ways: under denaturing conditions (to assess the overall efficiency of translation) and under native conditions (to assess the relative efficiency of formation of native heterodimer). Analysis of the reaction products under denaturing conditions showed that the wild-type control and all the various disease-associated mutations were translated in the cell-free cocktail with essentially the same efficiency (Fig. 2A). In contrast, analysis of the identical reaction products under native conditions revealed that although all of the mutations resulted in the generation of a product that migrated as native heterodimer, the yield of this product varied across a continuum from an amount similar to the wild-type control (in the case of L286F) to slightly reduced amounts (in the case of mutants I188L, I238V, P263T, R402H and S419L) to significantly diminished amounts (in the case of mutants, V303G, L397P and, most conspicuously, R402C) (Fig. 2B and C). We also noted a discernible enhancement in the yield of CCT/α-tubulin binary complexes in reactions done with V303G and L397P, as well as an increase in the yield of the band corresponding to the α-tubulin/PFD co-complex (31) in the case of R402C (Fig. 2B).
Figure 2.
Expression of TUBA1A mutations upon transcription/translation in a cell-free system. Wild-type and mutant sequences were expressed as [35S]methionine-labeled polypeptides by transcription/translation in rabbit reticulocyte lysate. Equal aliquots of the reaction products were analyzed under either denaturing (SDS–PAGE) (A) or native conditions (B). Arrows in (B) (top to bottom) show the migration positions of the α-tubulin/CCT binary complex, the PFD/α-tubulin co-complex and tubulin heterodimers, respectively. (C) Quantification (with standard deviation) of the yield of tubulin heterodimer derived from four independent experiments such as that shown in (B). For each mutation, the amount of heterodimer generated is normalized with respect to the translational efficiency as shown by SDS–PAGE, and the data compared with wild-type (taken as 100%).
We sought to determine whether the relatively low yield of heterodimer in the case of several disease-associated mutants might be ascribable to a compromised rate of transit through the assembly pathway. To do this, we examined the products of transcription/translation reactions in a time-dependent manner (Supplementary Material, Fig. S2). Several of the mutations (I188L, I238V, P263T, L286F, R402H, S419L) were generally similar in the kinetics of intermediate and product formation compared with the wild-type control, while others showed clear differences. For example, analysis of R402C confirmed an enhanced abundance of the PFD/α-tubulin binary complex and showed a delayed decline in label associated with this species relative to the wild-type control. In the case of V303G and L397P, there was a delayed decline in label associated with the CCT/α-tubulin binary complex. These data are consistent with the results presented in Figure 2 and suggested the existence of one or more defects in the tubulin heterodimer assembly pathway in the case of V303G, L397P and R402C (see below).
Mutant-containing tubulins are universally competent for microtubule co-polymerization in vitro
To test whether mutant-containing tubulin heterodimers are competent to assemble into microtubules, we mixed the products of in vitro transcription/translation reactions with native, depolymerized bovine brain microtubules. These were then taken through three successive cycles of GTP-dependent polymerization and depolymerization. At the end of each cycle, an aliquot containing the same amount of total protein was removed and analyzed by SDS–PAGE. The result of this experiment showed that all mutant proteins co-cycled with authentic brain microtubules with undiminished efficiency compared with a wild-type control (Fig. 3). We conclude that all the disease-causing TUBA1A mutations under study are competent for assembly into microtubules in vitro.
Figure 3.
Co-cycling of heterodimers containing TUBA1A mutant α-tubulins with bovine brain microtubules. Wild-type and mutant sequences expressed as [35S]methione-labeled proteins by transcription/translation in rabbit reticulocyte lysate were tested for their ability to co-cycle with added native bovine brain microtubules through successive cycles of polymerization and depolymerization. At the end of each cycle, aliquots containing identical, amounts of total protein were removed and analyzed by SDS–PAGE.
Deficiencies in the production of CCT-mediated IQ folding intermediates
In principle, a reduced yield of assembly competent heterodimers could be a consequence of one or more defects at any step in on the complex pathway leading to de novo tubulin heterodimer assembly (Supplementary Material, Fig. S1). An early and critical event in this pathway is the generation of productive folding intermediates via ATP-dependent cycling of tubulin polypeptides with the cytosolic chaperonin CCT. This results in the generation of chaperonin-bound quasi-native folding intermediates termed IQ, defined by their acquisition of a non-exchangeable (i.e. N-site) GTP-binding pocket (40). We used this criterion to measure the relative efficiency with which CCT can cycle mutant α-tubulins relative to a wild-type control. In each case, equal amounts of unlabeled, urea-denatured target proteins made via expression in Escherichia coli were presented to CCT and the reactions incubated in the presence of ATP and α-32P-labeled GTP. The reaction products were then analyzed on native gels. The results of this experiment showed that mutants I188L, I238V, L397P and (most conspicuously) R402C all generated lower yields of 32P-labeled IQ intermediates compared with the wild-type control (Fig. 4A and B). We also monitored the kinetics of IQ formation in the case of those mutations (V303G, L397P and R402C) that gave a compromised yield of heterodimers upon transcription/translation in a cell-free system (Fig. 2). The data showed a diminished rate (relative to the control) of IQ formation for L397P and R402C and (in the case of R402C) a time-dependent decay of IQ intermediates (Fig. 4C). We conclude that several disease-associated TUBA1A mutations result in a diminished efficiency whereby they are cycled by CCT so as to produce potentially productive folding intermediates; in addition, IQ intermediates are unstable in the case of R402C.
Figure 4.
Generation of quasi-native (IQ) folding intermediates via ATP-dependent interaction with CCT. Wild-type and mutant TUBA1A sequences were expressed as unlabeled insoluble inclusion bodies in E. coli, unfolded in 8 m urea, and equal amounts used as target proteins in folding reactions containing CCT, ATP and 32P-labeled GTP. Following incubation to allow the generation of IQ intermediates (see text), the reaction products were resolved on native polyacrylamide gels. (A) IQ formation for wild-type and mutant TUBA1A target proteins. (B) Bar diagram showing the quantification (with standard deviation) of data based on four experiments done in the same manner as those shown in (A). (C) Kinetics of IQ formation for those disease-associated mutations (L397P, V303G and R402C) that produce a compromised yield of heterodimers upon transcription/translation in vitro (Fig. 2). Note the time-dependent decay of CCT-bound [32P]GTP in the case of R402C.
Defective interactions with TBCB revealed by in vitro CCT-driven folding reactions and TBCD-driven heterodimer disruption reactions
To explore the existence of defects in interactions that occur downstream of CCT in the tubulin heterodimer assembly pathway (Supplementary Material, Fig. S1), we did in vitro folding reactions using fully purified components (i.e. CCT and the downstream chaperones TBCB-E). Wild-type and mutant α-tubulins were expressed as [35S]methionine-labeled proteins in E. coli. The resulting insoluble inclusion bodies were purified, unfolded in 8 m urea and used as target proteins in reconstituted folding reactions in the presence of the various components required for productive folding (33,41). In each case, the reaction products were resolved by native PAGE.
We first did fully reconstituted folding reactions containing CCT, ATP, GTP, TBCB, TBCC, TBCD, TBCE and native tubulin (provided as a source of β-tubulin). In these experiments, we found that, consistent with our transcription/translation data (Fig. 2), all the disease-associated mutations were capable of generating native heterodimers, albeit in varying yield (Fig. 5A). We also did folding reactions with CCT, ATP, GTP and TBCB alone. This approach was based on our recent observation that the TUBA1A pachygyria-causing mutation R264C is characterized by a low yield of de novo assembled heterodimers as a result of a defect in the ability of CCT-generated α-tubulin folding intermediates to stably bind to TBCB (38). We found that seven of the mutations under study (I188L, I238V, P263T, L286F, R402C, R402H and S419L) produced a product that co-migrated with the TBCB/α-tubulin intermediate generated in a parallel reaction done with the wild-type control, although the yield of this intermediate varied significantly among these mutants. Conspicuously, however, in common with R264C, mutants V303G and L397P both failed to yield any detectable amount of this product (Fig. 5B). These data define a new class of disease-causing mutations in TUBA1A that shares a defective or inefficient binding of TBCB to CCT-generated α-tubulin folding intermediates.
Figure 5.
Defective interactions with TBCB in CCT-driven folding reactions and in TBCD-mediated tubulin destruction reactions. Wild-type and mutant α-tubulin polypeptides were expressed as 35S-labeled insoluble inclusion bodies in host E. coli cells. These were purified, unfolded in 8 m urea and used as probes in in vitro CCT-mediated folding reactions containing CCT, ATP, GTP and either the full complement of chaperones (TBCB, TBCC, TBCD and TBCE) required for the de novo assembly of the tubulin heterodimer (A) or in parallel folding reactions containing CCT, ATP, GTP and TBCB alone (B). In each case, the reaction products were resolved on native polyacrylamide gels. (C) De novo assembled native tubulin heterodimers 35S-labeled in their α-subunits were generated by transcription/translation in vitro. Following purification, the labeled heterodimers were incubated with a molar excess of TBCD (to disrupt them) in the presence of TBCB (present as a capturing agent for α-tubulin subunits free of β-subunits). The reaction products were analyzed on non-denaturing polyacrylamide gels. Arrows in (A) and (B) (top to bottom) show the migration positions of the α-tubulin/CCT binary complex, the TBCB/α-tubulin co-complex and tubulin heterodimers, respectively.
We sought further insight into the nature of defective interactions with TBCB by taking advantage of the fact that the tubulin heterodimer assembly reaction can be driven in reverse: the action of TBCD on native tubulin heterodimers disrupts them, sequestering β-tubulin as a co-complex with TBCD (Fig. S1). Under these conditions, the α-tubulin subunit is released and rapidly decays to a non-native state unless it is captured by interaction with TBCB to form a stable TBCB/α-tubulin co-complex. To examine the efficiency of TBCB/α-tubulin co-complex formation, purified wild-type and mutant heterodimers 35S-labeled in their α-tubulin subunits were incubated with a 2-fold stoichiometric excess of TBCD (to disrupt the heterodimers) and a 10-fold molar excess of TBCB (to capture labeled, released α-tubulin subunits). The reaction products were resolved by native PAGE. These experiments showed that TBCD-mediated disruption of heterodimers containing 35S-labeled I188L, I238V, P263T, L286F, R402C, R402H and S419L resulted in the production of TBCB/α-tubulin that was comparable to wild-type levels. On the other hand, L397P and V303G yielded only very low levels of this co-complex (Fig. 5C). These data are broadly consistent with the results of our forward (CCT-driven) folding reactions (Fig. 5A and B). We interpret the barely detectable presence of a TBCB/α-tubulin product in either forward or back reactions in the case of L397P as reflecting a severely compromised ability of this mutant to stably interact with TBCB. In the case of R402C, the modest yield of TBCB/α-tubulin in forward folding reactions (Fig. 5B), but relatively abundant yield of this intermediate in back reactions (Fig. 5C), is consistent with the compromised yield and instability of CCT-generated IQ intermediates in the case of this mutant (Fig. 4). On the other hand, V303G produces very little or no detectable TBCB/α-tubulin co-complex in CCT-driven (i.e. forward) folding reactions (Fig. 5B), and only a modest quantity of the same intermediate in TBCD-mediated tubulin heterodimer disruption reactions (Fig. 5C). The generation of a modest level of heterodimer in fully reconstituted folding reactions done with V303G (Fig. 5A) therefore appears paradoxical.
To resolve this paradox, we did an experiment to directly test the stability of the TBCB/α-tubulin co-complex in heterodimer assembly reactions done with V303G. Labeled, urea-unfolded wild-type or V303G probes were presented to CCT in in vitro folding reactions containing ATP and GTP; these were incubated for 1 h in order to generate IQ intermediates. TBCB was added (in order to allow the formation of the TBCB/α-tubulin co-complex) and the reactions divided into three equal aliquots. In one case, the incubation was continued without further addition. In a second case, TBCC, TBCD and TBCE were immediately added in order to promote the generation of de novo assembled tubulin heterodimers. In the third case, TBCC, TBCD and TBCE were added after a 10 min delay, introduced so as to provide an opportunity for potentially unstable α-tubulin/TBCB intermediates to decay. Consistent with the experiments shown in Figure 5, analysis of the reaction products by native PAGE showed that reactions containing the full complement of TBCs each resulted in the formation of tubulin heterodimer (Fig. 6, compare lanes 2 and 6), whereas reactions done with TBCB alone resulted in little or no TBCB/α-tubulin product in the case of V303G (Fig. 6, compare lanes 1 and 5). Most importantly, when wild-type reactions containing TBCB were incubated for 10 min prior to the addition of TBCC, TBCD and TBCE, there was little discernible effect on the yield of de novo assembled heterodimers (Fig. 6, compare lanes 6 and 7); on the other hand, the yield of de novo assembled heterodimers in a parallel reaction done with V303G was dramatically reduced (Fig. 6, compare lanes 3 and 7). It follows that in the case of V303G, the TBCB/α-tubulin co-complex can form transiently and can contribute to functional de novo heterodimer assembly, but it cannot persist as a stable entity. We conclude that the V303G mutation results in a weakened interaction of CCT-generated IQ intermediates with TBCB such that the formation of a stable TBCB/α-tubulin co-complex is compromised.
Figure 6.
The TUBA1A V303G mutation results in the formation of an unstable TBCB/α-tubulin co-complex. 35S-labeled urea-denatured wild-type and V303G mutant tubulins were used in CCT-driven in vitro folding assays containing ATP and GTP. After 1 h of incubation at 30° to form IQ intermediates (see text), either TBCB alone was added (lanes marked B) (included so as to generate the TBCB/α-tubulin co-complex) or TBCB, TBCC, TBCD and TBCE (lanes marked BCDE) were added (included so as to generate heterodimers). Lanes marked B*CDE show the products of parallel reactions done with TBCB alone and incubated for 10 min prior to the addition of TBCC, TBCD and TBCE. Note that when TBCB, TBCC, TBCD and TBCE were added contemporaneously, the yield of heterodimers was similar in reactions done with βV303G and wild-type target proteins. In contrast, when a 10 min delay was introduced between the addition of TBCB and the remaining TBCs, there was a greatly diminished yield of heterodimers in the case of V303G. Lanes marked CDE show control reactions done in the absence of TBCB, showing the requirement for this component for heterodimer formation. Arrows (top to bottom) show the migration positions of the α-tubulin/CCT binary complex, the TBCB/α-tubulin co-complex and tubulin heterodimers, respectively.
Effect of TUBA1A mutations on heterodimer stability in vitro and in vivo
One possible consequence of amino acid substitution mutations is a change in secondary or tertiary structure that could lead to instability. We tested this possibility in two ways. First, we analyzed the susceptibility of the purified mutant proteins to digestion with low levels of the non-specific protease proteinase K as a measure of their structural integrity (40). At various intervals, aliquots were withdrawn from the reactions, the proteolytic activity quenched by addition of phenyl methyl sulfonyl fluoride and the reaction products analyzed by SDS–PAGE. Most of the mutant proteins were as resistant to proteolytic digestion as the wild-type control. However, a conspicuous exception was V303G. In this case, the mutant protein was more extensively degraded than the wild-type control: ∼90% of the intact protein was no longer evident after 15 min of digestion, and the progressive disappearance of intact protein was accompanied by the appearance of a discrete degradation product that migrated at about 35 kDa (Fig. 7A). We conclude that the V303G mutation leads to a reduction in the structural integrity of the folded polypeptide. Second, we measured the stability of the various mutant tubulins in vivo by pulse-chase experiments using FLAG-tagged constructs to transfect HEK293 cells. We found that some mutant tubulins (I188L, I238V, L286F, R402H and S419L) behaved in a manner similar to the wild-type control. In contrast, two mutations (P263T and V303G) showed a reduced stability, whereas L397P and R402C were highly unstable (Fig. 7B). The rapid decay of R402C is in agreement with the short half-life of IQ intermediates in the case of this mutation (Fig. 4C), whereas the extreme instability of L397P is consistent with our observation that this mutation does not appear competent for assembly into microtubules in vivo (see below).
Figure 7.
Structural instability of heterodimers containing V303G and stability of TUBA1A mutant proteins in vivo. (A) Wild-type and V303G mutant TUBA1A sequences were expressed as [35S]methione-labeled proteins in rabbit reticulocyte lysate, purified and tested for their resistance to proteolysis by the non-specific protease proteinase K. At increasing time intervals, the proteolytic reaction was quenched and the reaction products analyzed by SDS–PAGE. Note the diminished abundance of intact α-tubulin (relative to the wild-type control) with increasing times of proteinase K digestion in the case of V303G, and the appearance of a degradation product that migrates at about 35 kDa. All other disease-associated mutants showed a proteolytic resistance pattern similar to the wild-type control (data not shown). (B) Relative stability of wild-type and mutant proteins determined by pulse-chase experiments in vivo. Bars show the percentage (relative to t = 0) of the radiolabel remaining in FLAG immunoprecipitable α-tubulin after a 5 h chase.
Expression of disease-associated α-tubulin mutants in vivo
To assess the competence of mutant-bearing heterodimers to assemble into microtubules in vivo, we generated constructs designed for their expression by transfection in mammalian cells. To distinguish the transgene from endogenously expressed α-tubulins, each mutant construct was engineered in such a way as to include a FLAG epitope substituted in place of the encoded C-terminal amino acid (38). In each case, transfected cells were examined by immunofluorescence using an anti-FLAG antibody (to detect the expression of the transgene) and an anti-α-tubulin antibody (to detect the overall microtubule population). In these experiments, we found that most of the mutant proteins (I188L, I238V, P263T, L286F, V303G, R402C, R402H and S419L) were capable of incorporation into the interphase microtubule network in a manner that was indistinguishable from the wild-type control (Supplementary Material, Fig. S3). In contrast, the expression of L397P resulted in a distinctive phenotype in which the transfected protein appeared as a diffuse signal throughout the cytoplasm, with little or no detectable incorporation into microtubules (Supplementary Material, Fig. S4A). We reasoned that this phenomenon might be influenced by the placement of the FLAG epitope, as previously described in the case of R264C (38). To test this notion, we engineered a set of alternative constructs in which the FLAG tag was either placed following the C-terminally encoded tyrosine residue or substituted for the 5, 8 or 11 C-terminal amino acids of the authentic protein. The 12 C-terminal amino acids of α-tubulin are not required for incorporation into HeLa cell microtubules in vivo (42), while the removal of multiple acidic residues from the C-terminus counterbalances the acidic charge conferred by the FLAG epitope. We found that, as with our original FLAG-tagged construct, transfection of these constructs into HeLa cells all resulted in proteins that failed to incorporate into microtubules (Supplementary Material, Fig. S4B). We conclude that the L397P mutant is most probably compromised in its ability to co-assemble into microtubules in vivo.
We did several kinds of experiment to examine the effect on microtubule dynamics of those mutants that were competent for assembly into microtubules in vivo. First, HeLa cells transfected with FLAG-tagged constructs were treated with nocodazole to depolymerize their microtubules. Following depolymerization, the cells were restored to drug-free medium and the regrowth of microtubules from the centrosome was monitored at various times thereafter. We found no significant difference in the rate of microtubule growth from the centrosome in the case of I188L, I238V, L286F, V303G, R402C, R402H and S419L compared with a wild-type control (data not shown). However, in the case of P263T, we noted a distinctive phenotype: between 5 and 20 min of recovery, there was a conspicuous persistence (relative to the wild-type control) of the diffuse high background of label that reflects unpolymerized cytosolic tubulin heterodimers (Fig. 8). This diffuse background signal eventually subsided as the cytosol became repopulated with microtubules, but much more slowly compared with the wild-type control. Similar data (not shown) were obtained in parallel experiments in COS-7 cells in which microtubule depolymerization was effected by exposure to cold rather than nocodazole. These observations could reflect either P263T heterodimer instability outside the context of the microtubule polymer or a compromised rate at which mutant heterodimers can assemble into the microtubule lattice. Second, we examined microtubule dynamics in vivo via co-transfection of COS-7 cells with GFP-tagged EB3 and FLAG-tagged wild-type P263T or V303G TUBA1A, the latter selected as an example of a mutant form that had a similar stability in vivo and behaved indistinguishably from the wild-type in our microtubule regrowth experiments. We found that expression of P263T (but not V303G) resulted in a significant damping (relative to the control) of microtubule velocity in a dose-dependent manner (Fig. 9A and accompanying movies in Supplementary Material, Figs S5 and S6). These data are consistent with the delayed cytosolic repopulation of microtubules following nocodazole or cold-induced depolymerization in the case of cells overexpressing P263T. We conclude that the expression of P263T most likely results in the assembly of heterodimers that have a deleterious effect on normal microtubule dynamics.
Figure 8.
Effect of TUBA1A mutant P263T on microtubule regrowth following nocodazole-induced depolymerization. HeLa cells were transfected with constructs engineered for the expression of FLAG-tagged wild-type and P263T mutant forms of TUBA1A. Thirty-six hours post-transfection, the cells were treated with nocodazole and restored to drug-free medium. At the times indicated in the figure, cells were fixed and examined by immunofluorescence using a rabbit anti-FLAG antibody (to detect the transgene, shown in red) and a mouse monoclonal anti-α-tubulin antibody (to detect the overall microtubule network, shown in green). Note the extended persistence of diffuse cytosolic labeling (present in all cases at t = 0 min) during early recovery times in the case of P263T. Bar = 10 µm.
Figure 9.
Effect of expression of P263T and V303G on microtubule dynamics in COS-7 cells and in cultured cortical neurons. (A and B) Constructs engineered for the expression of C-terminally FLAG-tagged wild-type, P263T and V303G TUBA1A were co-transfected into either COS-7 cells (A) or electroporated into E15.5 cultured mouse cortical neurons (B) in combination with a construct driving the expression of EB3-GFP. Two different molar ratios (1:0.5 and 1:2) of EB3-GFP versus TUBA1A plasmid DNA were used (see Materials and Methods). Representative illustrations are shown of the comet-like labeling of microtubule plus ends by EB3-GFP in a WT TUBA1A and EB3-GFP co-transfected COS-7 cell (A) and a 1DIV cortical neuron (B), together with the quantification of microtubule plus end velocities for each condition. The area delineated by the white rectangle in (A) is reproduced in colour below, together with corresponding images from COS-7 cells co-expressing EB3-GFP and either P263T or V303G. Plus ends are color-coded at each fourth frame (i.e. every 4 s). The first frame is shown in red, the second in green and the third in blue. Note the decrease in plus end velocities upon overexpression of the P263T mutant in COS-7 cells as illustrated by the closer proximity of color-coded tips in comparison to wild-type (WT) and V303G. Mean velocities of five different COS-7 cells per condition (n = 5, representing approximately 100 comets in total) or of six to eight different cortical neurons per condition (n = 6–8, representing 43–89 comets in total), respectively, were averaged to obtain mean of velocities for each condition. A Mann–Whitney non-parametric test showed that the decreased velocities apparent in COS-7 cells expressing P263T are significant when the EB3-GFP: P263T ratio is 1:2 [P = 0.0317; marked by a single asterisk in (A)], but not significant at a ratio of 1:0.5, consistent with a quantitative effect of the mutant protein. In contrast, expression of the V303G mutation does not significantly affect microtubule growth. Similarly, in primary cultured cortical neurons transfected with EB3-GFP:P263T at a ratio of 1:2, a Mann–Whitney non-parametric test revealed that the decreased microtubule growth velocities are significant in neurites, but not in the soma [P = 0.0093; marked by a double asterisk in (B)]. (C) Imaged explanted cortical cells, assessed as neurons on the basis of their morphologies, were subsequently fixed and processed for immunofluorescence using antibodies directed against the FLAG epitope (shown in red), as well as an antibody directed against the neuron-specific βIII-tubulin isotype (shown in blue). Scale bars = 10 µm.
In view of the compromised dynamic behavior conferred on microtubules in HeLa and COS-7 cells by the expression of the P263T mutation, we extended our in vivo expression experiments to include an analysis of the corresponding effect on microtubule growth in primary cultures of mouse cortical neurons. In these experiments, we used cells that had reached the 1 DIV (days in vitro) stage; this corresponds to stage 2 of neuronal polarization in vitro and is characterized by the extension of multiple neurites, with a single neurite ultimately becoming the axon. We selected this time window to assess microtubule growth because this period depends critically on the proper regulation of microtubule dynamics (43). As in the case of experiments done in COS-7 cells, we found a significant damping of microtubule growth rate in the case of neurons expressing P263T, but not V303G. Surprisingly, however, this decrease in growth rate occurred only in microtubules in neurites, and not in microtubules in the soma (Fig. 9B and accompanying movies in Supplementary Material, Figs S7 and S8). These intriguing data could reflect either a difference in isotype composition between microtubules in these different cell compartments or a compartment-specific presence of one or more MAPs that affect microtubule dynamics.
DISCUSSION
The discovery of mutations in genes encoding both α- and β-tubulin that are associated with neuronal migration disorders (27) reinforces the importance of proper microtubule function as a critical condition for normal neuronal migration during brain development (15). Here we establish the nature of the defects in a spectrum of neuronal migration disorders caused by mutations in TUBA1A. In several cases, these defects can be ascribed to specific interactive events in the de novo tubulin heterodimer assembly pathway. This pathway is required for several reasons. First, newly synthesized tubulin polypeptides are particularly prone to misfolding and aggregation under the conditions of high protein concentration that prevail in eukaryotic cells. Second, because neither subunit, on its own, is stable under physiological conditions (33), specific molecular machinery has evolved whose function is to lock the α- and β-tubulin subunits together. Excluding regulatory effectors such as Arl2 (44), the pathway involves at least seven chaperone proteins, the majority of which play an indispensible role in heterodimer formation (Supplementary Material, Fig. S1). The essential nature of this pathway and its many components offers a multitude of opportunities for genetic changes that could result in compromised heterodimer assembly.
A range of defects in the tubulin folding pathway affect heterodimer assembly
We found that all of the mutations we analyzed were expressed with essentially the same overall efficiency in a cell-free system (Fig. 2A). This result is consistent with the fact that all are single base substitutions within the exons of the TUBA1A gene and would therefore not be predicted to affect the translational machinery. On the other hand, three of the mutations (V303G, L3497P, R402C) had a significant impact on the efficiency of tubulin heterodimer formation (Fig. 2B and C). We identified a number of defects in the assembly pathway that are likely to contribute to the observed reduction in heterodimer formation in the case of these mutants. First, we noted an enhanced yield of the PFD/α-tubulin complex in cell-free translation reactions done with R402C (Fig. 2B). PFD functions by presenting newly synthesized tubulin polypeptides to CCT and by promoting productive folding in the presence of competing but non-productive pathways (31). The enhanced level of PFD/α-tubulin complexes in the case of R402C implies a compromised efficiency with which PFD is able to discharge its target protein upon docking with CCT. This conclusion is consistent with the reduced level (compared with wild-type) of CCT/α-tubulin binary complexes in transcription/translation reactions done with this mutant (Fig. 2B). Second, we found a reduced efficiency whereby CCT-generated quasi-native (IQ) intermediates in the case of several mutations (I188L, I238V and especially L397P and R402C) (Fig. 4), implying a compromised ability of these polypeptides to acquire a native N-site GTP-binding pocket as a result of multiple ATP-dependent iterations with the chaperonin. Third, in the case of V303G and L397P, we identified defective interaction of CCT-generated intermediates with TBCB [one of the tubulin-specific chaperones that capture CCT-generated α-tubulin folding intermediates (Supplementary Material, Fig. S1)] (Figs 5 and 6). Consistent with these observations, transcription/translation reactions done with V303G and L397P show enhanced relative yields of CCT/α-tubulin binary complexes (Fig. 2B and Supplementary Material, Fig. S2), presumably reflecting accumulation of chaperonin-bound intermediates in the absence of efficient capture by TBCB. A summary of these defects in tubulin heterodimer biogenesis is presented in Table 1.
Table 1.
Defective interactions in the tubulin heterodimer assembly pathway resulting from TUBA1A disease-associated mutations
| I188La | I238Va | R264Cb | V303Ga | L397Pa | R402Ca | |
|---|---|---|---|---|---|---|
| Defective discharge from PFD | ++ | |||||
| Defective production of CCT-bound IQ intermediates | + | + | ++ | ++ | +++ | |
| Defective interaction with TBCB | +++ | +++ | +++ |
aThis paper.
bFrom Tian et al. (38).
Role of TBCB
With the exception of TBCA, all of the chaperone proteins that capture α- and β-tubulin folding intermediates and assemble them into functional tubulin heterodimers are essential for life in higher eukaryotes (45,46). Defective interactions with these chaperones as a result of mutations in any α- or β-tubulin isotype might therefore be expected to result in microtubule phenotypes. The same is true of mutations in TBCs themselves. In the case of TBCE, for example, deletion mutations are responsible for a devastating condition known as HRD (hypoparathyroidism, mental retardation and facial dysmorphism) (47,48). Although there are no known cases of inherited mutations in TBCB, the data presented here define a new class of mutations in TUBA1A, members of which are characterized by a diminished capacity of CCT-generated IQ intermediates to stably interact with TBCB (Table 1). The importance of proper TBCB function in neuronal development is highlighted by the fact that the modulation of the level of expression of this protein leads to abnormalities including axonal length and abnormalities in growth cone microtubules (49).
Effect of TUBA1A mutations on microtubule behavior in vivo
Although several α-tubulin isotypes are expressed in the mammalian brain, TUBA1A is dominant. The mouse homolog, Tuba1a, whose amino acid sequence is identical to its human counterpart, mimics the overall level of α-tubulin expression during development, rising to a peak during late embryogenesis, and then decreasing in the first 10–15 days of post-natal life (50). Expression of this isotype is therefore likely to be a major contributor to the pool of tubulin heterodimers required for incorporation into microtubules during the critical period of neuronal migration and differentiation that determines correct cortical lamination. It follows that any mutation resulting in a diminished supply of TUBA1A could impact on the overall abundance of tubulin heterodimers, with a resulting paucity of microtubules that might compromise proper neuronal migration and differentiation. Indeed, haploinsufficiency of any tissue-specific tubulin isotype [such as those expressed exclusively in brain (51), testis (24) or hematopoietic tissue (25)] might be expected to lead to disease.
None of the nine disease-associated TUBA1A mutations we examined resulted in a complete failure to yield tubulin heterodimers upon coupled transcription/translation, implying that heterozygous null mutations in this gene are likely to be lethal. For those mutations that generated tubulin heterodimers in significantly reduced yield (V303G, L397P and R402C, Fig. 2), tubulin deficit may be considered as a likely contributing factor to the disease phenotype. Two of these mutant proteins (V303G and R402C) as well as those that gave a relatively uncompromised yield of heterodimers upon transcription/translation (I188L, I238V, P263T, L286F, R402H, S419L; Fig. 2) were capable of efficient incorporation into microtubules in vivo (Supplementary Material, Fig. S3). In contrast, the expression of FLAG-tagged L397P failed to result in incorporation of the mutant protein into microtubules (Supplementary Material, Fig. S4A). We considered the possibility that the addition of the C-terminal FLAG tag to the L397P mutant protein (required in order to distinguish mutant proteins from endogenously expressed α-tubulin isotypes in transfection experiments) might have compromised its functionality. However, changing the location of the tag had little or no effect on the observed phenotype (Supplementary Material, Fig. S4). One possibility is that the failure of heterodimers carrying the L397P mutation to incorporate into microtubules in vivo is a result of a compromised interaction with one or more of the many microtubule tip interacting proteins (+TIPS) that influence microtubule dynamics by modulating the efficiency of incorporation of heterodimers at microtubule plus ends (26). This explanation is particularly attractive in light of the fact that the L397P mutation had no detectable effect on the ability of mutant-containing heterodimers to co-cycle with native brain microtubules in vitro (Fig. 3). On the other hand, functional compromise of the L397P mutation in vivo as a result of addition of the FLAG tag cannot be formally ruled out.
Pulse chase experiments done with the various disease-associated mutations in vivo showed that several (P263T, V303G, R402C and L397P in particular) are significantly less stable than wild-type TUBA1A (Fig. 7B). Contributing factors could include instability of the assembled heterodimer as well as instability of intermediate complexes (including, but not limited to, the PFD/α-tubulin, CCT/α-tubulin and TBCB/α-tubulin co-complexes) that precede the heterodimer in the assembly pathway. The increased vulnerability of these mutants to degradation in vivo could be a contributing factor to the disease phenotype.
For those mutations that were assembly competent in vivo, with the exception of the P263T mutation, we found no clear evidence of any effect on the growth rate of microtubules from the centrosome following either nocodazole or cold-induced depolymerization. In the case of P263T, however, we observed an abnormal phenotype consisting of a persistent diffuse cytosolic signal that eventually subsided (though much more slowly compared with the wild-type control) as the cytoplasm became repopulated with microtubules (Fig. 8). This diffuse signal is indistinguishable from that seen in cells expressing the wild-type protein that have been exposed to the drug and is presumably ascribable to unpolymerized heterodimers. The altered kinetics of microtubule regrowth in the case of P263T is consistent with our observation of damped microtubule velocity in experiments in which we measured microtubule growth in COS-7 cells transfected with the P263T mutant in vivo (Fig. 9A and accompanying movies, Supplementary Material, Figs S5 and S6). The damping effect of P263T expression on microtubule growth rate was also evident in dissociated neuronal cells in culture, although only in the case of microtubules in neurites and not in microtubules in the cell body (Fig. 9B and accompanying movies, Supplementary Material, Figs S7 and S8). This remarkable finding could reflect a difference in microtubule isotype composition in these two cellular compartments that influences microtubule growth rate. Alternatively, the damping effect of P263T in neurites could be attributable to the compartment-specific expression of a MAP [such as a member of the Dis1/XMAP215 family (52,53)] that stimulates microtubule growth rate. In either event, our data support the view that the expression of P263T mutant-containing heterodimers results in kinetically constrained assembly into the microtubule polymer. This could reflect a dominant-negative effect of this mutation.
For those mutations (I188L, I238V, L286F, R402H, S419L) for which we found either no obvious effect on de novo heterodimer assembly or microtubule dynamics, the disease phenotype is likely to be caused by an effect on one or more microtubule-dependent processes such as binding of associated proteins, including microtubule motors, that are critical during cortical development. Indeed, a precedent for such a mechanism exists in the recent description of a newly discovered class of mutations in the β-tubulin-encoding gene TUBB3 (30). These mutations result in the ocular motility disorder CFEOM3, in some cases together with intellectual and behavioral impairments or other neurological deficits. We note that residue S419 in α-tubulin resides in helix H12 and that this region has recently been shown to contain amino acids that are crucial for kinesin motility (54). The S419L mutation is therefore a plausible candidate for compromised interaction with kinesin.
Disease-causing mutations in tubulin genes: implications for the multitubulin hypothesis
Since the original discovery that in vertebrates, α- and β-tubulins are each encoded by a multigene family (55), it has been speculated that the various isotypes contribute to significant functional differences among different populations of microtubules. This notion has been termed the ‘multitubulin hypothesis’ (56,57). Evidence relating to the validity of this hypothesis has been equivocal. On the one hand, in cultured cells, microtubules are assembled as co-polymers of whatever isotypes are expressed (58). Indeed, the engineered expression of tissue-specific tubulins (e.g. brain, testis or hematopoietic specific α- or β-tubulin isotypes) in cultured cells leads to their free intermingling with those tubulins that are expressed endogenously (59,60). These observations are puzzling in light of the universal interspecies conservation of amino acid sequences (located almost exclusively within the acidic C-termini) that distinguish the various tubulin isotypes in vertebrate species. On the other hand, genetic experiments in Drosophila suggest that the architecture and organization of microtubules can be influenced by isotype composition via interactions with extrinsic proteins (61,62). Moreover, in humans, disease-causing mutations in genes encoding individual tubulin isotypes [TUBA1A (27,33), TUBA8 (63), TUBB2B (15), TUBB3 (30)] result in different phenotypes, some involving defects in neuronal migration (TUBA1A, TUBA8, TUBB2B) and others not (TUBB3). Taken together, these latter observations argue in favor of variations in tubulin isotype composition conferring subtle differences on dynamics and other microtubule properties that are required for critical events in the context of the developing brain.
MATERIALS AND METHODS
Computational methods
Computer-generated visualization of the placement of the mutations under study on the surface of the α-tubulin polypeptide (RCSB PDB code: 1TUB) was rendered using PyMOL (http://www.pymol.org).
Cloning and in vitro translation
A construct containing a full-length cDNA encoding TUBA1A (38) was cloned into the pET23 (Novagen Inc.) and pcDNA 3.1+ (Invitrogen, Carlsbad, CA, USA) vectors. Mutations corresponding to I188L, I238V, P263T, L286F, V303G, L397P, R402C, R402H and S419L were introduced into these constructs by site-directed mutagenesis using a QuikChange II kit (Stratagene, La Jolla, CA, USA). A tag encoding the FLAG epitope (DYKDDDDK) was incorporated into the TUBA1A wild-type and mutant sequences such that it substituted for the C-terminal tyrosine residue. In the case of L397P, additional FLAG-tagged constructs were generated in which the epitope was positioned such that it substituted for the 5, 8 or 11 C-terminal residues of TUBA1A. All constructs were checked by DNA sequencing. Transcription/translation reactions in rabbit reticulocyte lysate (TNT; Promega, Madison, WI, USA) were done in the presence of [35S]methionine as described previously (38). Tubulin heterodimers 35S-labeled in their α-subunit were generated and purified as described (33). Labeled reaction products were resolved by either SDS–PAGE or on 4.5% native polyacrylamide gels as described previously (64).
Microtubule co-polymerization experiments
Products of in vitro translation reactions were mixed with depolymerized bovine brain microtubules and taken through successive cycles of polymerization and depolymerization as described (33). At the end of each of these cycles, aliquots containing equal amounts of depolymerized material were removed and analyzed by SDS–PAGE.
In vitro folding reactions
In vitro folding assays were done in folding buffer containing CCT, ATP, GTP and tubulin chaperones (TBCB, TBCC, TBCD, TBCE) as described previously (65). Target proteins (i.e. wild-type or mutant TUBA1A tubulins) were expressed as either unlabeled or 35S-labeled proteins in E. coli and the inclusion bodies purified and unfolded in 8 m urea as described (32). Experiments to determine the efficiency of formation of quasi-native folding intermediates (40) were done as described previously (38). In all cases, reaction products were analyzed by electrophoresis on native polyacrylamide gels as described (66). Back reactions (see text) were done by incubating purified wild-type or mutant tubulin heterodimers [35S]methionine-labeled by in vitro transcription/translation in their α-subunit with a 2-fold molar excess of TBCD [so as to disrupt the heterodimer (44,64) for 30 min at 30°C as described (33)] and a 10-fold stoichiometric excess of TBCB [included in order to capture the released α-subunit as a TBCB/α-tubulin co-complex (33)].
Determination of assembly competence in vivo and measurement of microtubule regrowth following nocodazole or cold-induced depolymerization
FLAG-tagged constructs were transfected into HeLa cells and processed for immunofluoresence using anti-FLAG and anti-β-tubulin antibodies as described previously (38). To monitor microtubule growth following induced depolymerization, cells were either treated with nocodazole and allowed to recover as described (38) or incubated on ice for 30 min before restoring to 37°C. In either case, microtubule regrowth from the centrosome was monitored at various intervals following fixation with paraformaldehyde. Images were captured using a Zeiss Axiophot epifluorescence microscope.
Measurement of microtubule dynamics in vivo
Transfection of COS-7 cells
COS-7 cells seeded in micro-dishes designed for live imaging (µ-Dish 35 mm; Ibidi, München, Germany) were co-transfected using Nanofectin (PAA Laboratories, Pasching, Austria) with a combination of EB3-GFP (generously provided by F. Niedergang, Institut Cochin, Paris, France) and a pRK5 vector containing either wild-type or mutant sequences encoding TUBA1A. Transfections were done using two different amounts of TUBA1A plasmid DNA, with a constant amount of EB3-GFP DNA such that the ratio of EB3-GFP:TUBA1A was either 1:0.5 or 1:2.0. Transfected cells were cultured for 24 h prior to imaging.
Preparation and electroporation of primary cultures of cortical neurons
E15.5 embryos from pregnant OF1 mice were used for the preparation of dissociated cortical cell cultures. Brains were dissected, cortices were isolated from the ventral telencephalon and the tissue was mechanically dissociated following incubation at 37°C for 10 min in HBSS supplemented with 20 mm HEPES and 0.25% trypsin. Dissociated cells were resuspended in DMEM with 10% FBS. A total of 5 × 106 cells were electroporated with a combination of EB3-GFP and pRK5 vectors (the latter containing sequences encoding either wild-type or mutant TUBA1A) at a ratio of 1:2, using the Amaxa Nucleofector Technology and Mouse Neuron Nucleofector kit (Lonza, Basel, Switzerland) following the guidelines supplied by the manufacturer, and further seeded on Ibidi micro-dishes with grids allowing the subsequent relocation of imaged cells.
Live cell imaging and analysis
Prior to imaging, cells were transferred to video medium (COS-7 cells: DMEM without phenol red containing 10% FBS, 10 mm HEPES; neuronal cells: DMEM: F12 without phenol red supplemented with 0.3% glucose, 1× N2, 1× B27 and 1 mm pyruvic acid). Cells were allowed to adapt to the new medium in ambient air for at least 30 min at 37°C. Image acquisition was done using a thermostated spinning disk confocal microscope equipped with a piezo electric z controller. The acquisition sequence was 2 min long, with scans performed every 1 s. Scans were performed at 3 z positions (spaced by 0.2 µm) at every time point. Stacks were built using Metamorph v7.5 (Molecular Devices, Silicon Valey, CA, USA) and analyzed using the ImageJ plugin ‘Manual Tracking’ developed by Fabrice P. Cordelières (Institut Curie, Paris, France). About 20 comets were tracked in each of five different cells for each condition (n = 5). Instant velocities of comets were measured between each time point and further averaged over their time courses.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). The efficiency of co-transfection in imaged cells was verified following confocal data acquisition by immunofluorescence using an anti-FLAG (rabbit polyclonal; Sigma-Aldrich Inc., St Louis, MO, USA) and either an anti-α-tubulin (mouse monoclonal DM1A; Sigma-Aldrich Inc., St Louis, MO, USA) antibody or an anti-βIII-tubulin (Tuj1) (mouse monoclonal Tuj1; Covance, Princeton, NJ, USA) antibody. The proportion of EB3-GFP and TUBA1A-FLAG co-transfected cells was assessed and estimated to be ∼95%.
Stability measurements in vitro and in vivo
For the measurement of sensitivity to proteolysis in vitro, tubulin heterodimers containing [35S]methionine-labeled wild-type or mutant TUBA1A sequences were generated by in vitro transcription/translation and purified as described (33). The reaction products were subjected to digestion with proteinase K and analyzed by SDS–PAGE as described previously (64). Determination of the stability of wild-type and mutant proteins in vivo was done following transfection of FLAG-tagged constructs into HEK293 cells. Twenty-eight hours post-transfection, the cells were labeled for 1.5 h with [35S]methionine, chased by growth in unlabelled medium and FLAG-tagged material immunoprecipitated from a cytosolic extract with a rabbit anti-FLAG antibody (Santa Cruz Biologicals, Inc.) at t = 0 and t = 5 h as described (67). Immunoprecipitated material was resolved by SDS–PAGE and the level of FLAG-tagged proteins was determined using a phosphorimager.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from the National Institutes of Health (HD057028 to N.J.C.), the Inserm Avenir program and a Coup d'Elan from the Fondation Bettencourt Schueller (to F.F.), grants from Agence National pour la Recherche (ANR-NEURO-2005; ANR-MNP 2008 to J.C.) and Fondation pour la Recherche Médicale (FRM 2007 to J.C.) and a PhD grant from the French Ministry of Research and an EMBO short-term fellowship (ASTF 66.00-2008 to X.H.J.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Victoria Cabot for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ayala R., Shu T., Tsai L.H. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. doi:10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Francis F., Meyer G., Fallet-Bianco C., Moreno S., Kappeler C., Socorro A.C., Tuy F.P., Beldjord C., Chelly J. Human disorders of cortical development: from past to present. Eur. J. Neurosci. 2006;23:877–893. doi: 10.1111/j.1460-9568.2006.04649.x. doi:10.1111/j.1460-9568.2006.04649.x. [DOI] [PubMed] [Google Scholar]
- 3.Roll P., Rudolf G., Pereira S., Royer B., Scheffer I.E., Massacrier A., Valenti M.P., Roeckel-Trevisiol N., Jamali S., Beclin C., et al. SRPX2 mutations in disorders of language cortex and cognition. Hum. Mol. Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. doi:10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 4.Brooks A.S., Bertoli-Avella A.M., Burzynski G.M., Breedveld G.J., Osinga J., Boven L.G., Hurst J.A., Mancini G.M., Lequin M.H., de Coo R.F., et al. Homozygous nonsense mutations in KIAA1279 are associated with malformations of the central and enteric nervous systems. Am. J. Hum. Genet. 2005;77:120–126. doi: 10.1086/431244. doi:10.1086/431244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piao X., Hill R.S., Bodell A., Chang B.S., Basel-Vanagaite L., Straussberg R., Dobyns W.B., Qasrawi B., Winter R.M., Innes A.M., et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. doi:10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 6.Piao X., Chang B.S., Bodell A., Woods K., Benzeev B., Topcu M., Guerrini R., Goldberg-Stern H., Sztriha L., Dobyns W.B., et al. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann. Neurol. 2005;58:680–687. doi: 10.1002/ana.20616. doi:10.1002/ana.20616. [DOI] [PubMed] [Google Scholar]
- 7.Glaser T., Jepeal L., Edwards J.G., Young S.R., Favor J., Maas R.L. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. doi:10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 8.Baala L., Briault S., Etchevers H.C., Laumonnier F., Natiq A., Amiel J., Boddaert N., Picard C., Sbiti A., Asermouh A., et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat. Genet. 2007;39:454–456. doi: 10.1038/ng1993. doi:10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- 9.Paisan-Ruiz C., Scopes G., Lee P., Houlden H. Homozygosity mapping through whole genome analysis identifies a COL18A1 mutation in an Indian family presenting with an autosomal recessive neurological disorder. Am. J. Med. Genet. B Neuropsychiatric Genet. 2009;150B:993–997. doi: 10.1002/ajmg.b.30929. [DOI] [PubMed] [Google Scholar]
- 10.Fox J.W., Lamperti E.D., Eksioglu Y.Z., Hong S.E., Feng Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. doi:10.1016/S0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 11.Aligianis I.A., Johnson C.A., Gissen P., Chen D., Hampshire D., Hoffmann K., Maina E.N., Morgan N.V., Tee L., Morton J., et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg micro syndrome. Nat. Genet. 2005;37:221–223. doi: 10.1038/ng1517. doi:10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 12.des Portes V., Pinard J.M., Smadja D., Motte J., Boespflug-Tanguy O., Moutard M.L., Desguerre I., Billuart P., Carrie A., Bienvenu T., et al. Dominant X linked subcortical laminar heterotopia and lissencephaly syndrome (XSCLH/LIS): evidence for the occurrence of mutation in males and mapping of a potential locus in Xq22. J. Med. Genet. 1997;34:177–183. doi: 10.1136/jmg.34.3.177. doi:10.1136/jmg.34.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.des Portes V., Pinard J.M., Billuart P., Vinet M.C., Koulakoff A., Carrie A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 14.Dobyns W.B., Truwit C.L. Lissencephaly and other malformations of cortical development: 1995 update. Neuropediatrics. 1995;26:132–147. doi: 10.1055/s-2007-979744. doi:10.1055/s-2007-979744. [DOI] [PubMed] [Google Scholar]
- 15.Jaglin X.H., Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555–566. doi: 10.1016/j.tig.2009.10.003. doi:10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Faulkner N.E., Dujardin D.L., Tai C.Y., Vaughan K.T., O'Connell C.B., Wang Y., Vallee R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 17.Tai C.Y., Dujardin D.L., Faulkner N.E., Vallee R.B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. doi:10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efimov V.P., Morris N.R. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. doi:10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitagawa M., Umezu M., Aoki J., Koizumi H., Arai H., Inoue K. Direct association of LIS1, the lissencephaly gene product, with a mammalian homologue of a fungal nuclear distribution protein, rNUDE. FEBS Lett. 2000;479:57–62. doi: 10.1016/s0014-5793(00)01856-1. doi:10.1016/S0014-5793(00)01856-1. [DOI] [PubMed] [Google Scholar]
- 20.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.C., Friocourt G., McDonnell N., Reiner O., Kahn A., et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. doi:10.1016/S0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 21.Moores C.A., Perderiset M., Francis F., Chelly J., Houdusse A., Milligan R.A. Mechanism of microtubule stabilization by doublecortin. Mol. Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. doi:10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Moores C.A., Perderiset M., Kappeler C., Kain S., Drummond D., Perkins S.J., Chelly J., Cross R., Houdusse A., Francis F. Distinct roles of doublecortin modulating the microtubule cytoskeleton. EMBO J. 2006;25:4448–4457. doi: 10.1038/sj.emboj.7601335. doi:10.1038/sj.emboj.7601335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallee R.B., Seale G.E., Tsai J.W. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19:347–355. doi: 10.1016/j.tcb.2009.03.009. doi:10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villasante A., Wang D., Dobner P., Dolph P., Lewis S.A., Cowan N.J. Six mouse alpha-tubulin mRNAs encode five distinct isotypes: testis-specific expression of two sister genes. Mol. Cell. Biol. 1986;6:2409–2419. doi: 10.1128/mcb.6.7.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D., Villasante A., Lewis S.A., Cowan N.J. The mammalian beta-tubulin repertoire: hematopoietic expression of a novel, heterologous beta-tubulin isotype. J. Cell Biol. 1986;103:1903–1910. doi: 10.1083/jcb.103.5.1903. doi:10.1083/jcb.103.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhmanova A., Steinmetz M.O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. doi:10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 27.Keays D.A., Tian G., Poirier K., Huang G.J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnar Z., et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. doi:10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirier K., Keays D.A., Francis F., Saillour Y., Bahi N., Manouvrier S., Fallet-Bianco C., Pasquier L., Toutain A., Tuy F.P., et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum. Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. doi:10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- 29.Bahi-Buisson N., Poirier K., Boddaert N., Saillour Y., Castelnau L., Philip N., Buyse G., Villard L., Joriot S., Marret S., et al. Refinement of cortical dysgeneses spectrum associated with TUBA1A mutations. J. Med. Genet. 2008;45:647–653. doi: 10.1136/jmg.2008.058073. doi:10.1136/jmg.2008.058073. [DOI] [PubMed] [Google Scholar]
- 30.Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L., et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 140:74–87. doi: 10.1016/j.cell.2009.12.011. doi:10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vainberg I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., Cowan N.J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. doi:10.1016/S0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y., Thomas J.O., Chow R.L., Lee G.H., Cowan N.J. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. doi:10.1016/0092-8674(92)90622-J. [DOI] [PubMed] [Google Scholar]
- 33.Tian G., Lewis S.A., Feierbach B., Stearns T., Rommelaere H., Ampe C., Cowan N.J. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J. Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. doi:10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rédei G.P. Encyclopedic Dictionary of Genetics, Genomics, and Proteomics. Hoboken, NJ: Wiley-Liss; 2003. [Google Scholar]
- 35.Lodish H.F. Molecular Cell Biology. New York: W.H. Freeman; 2008. [Google Scholar]
- 36.Pece-Barbara N., Cymerman U., Vera S., Marchuk D.A., Letarte M. Expression analysis of four endoglin missense mutations suggests that haploinsufficiency is the predominant mechanism for hereditary hemorrhagic telangiectasia type 1. Hum. Mol. Genet. 1999;8:2171–2181. doi: 10.1093/hmg/8.12.2171. doi:10.1093/hmg/8.12.2171. [DOI] [PubMed] [Google Scholar]
- 37.Saitsu H., Kato M., Mizuguchi T., Hamada K., Osaka H., Tohyama J., Uruno K., Kumada S., Nishiyama K., Nishimura A., et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 2008;40:782–788. doi: 10.1038/ng.150. doi:10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 38.Tian G., Kong X.P., Jaglin X.H., Chelly J., Keays D., Cowan N.J. A pachygyria-causing alpha-tubulin mutation results in inefficient cycling with CCT and a deficient interaction with TBCB. Mol. Biol. Cell. 2008;19:1152–1161. doi: 10.1091/mbc.E07-09-0861. doi:10.1091/mbc.E07-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fallet-Bianco C., Loeuillet L., Poirier K., Loget P., Chapon F., Pasquier L., Saillour Y., Beldjord C., Chelly J., Francis F. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain. 2008;131:2304–2320. doi: 10.1093/brain/awn155. doi:10.1093/brain/awn155. [DOI] [PubMed] [Google Scholar]
- 40.Tian G., Vainberg I.E., Tap W.D., Lewis S.A., Cowan N.J. Quasi-native chaperonin-bound intermediates in facilitated protein folding. J. Biol. Chem. 1995;270:23910–23913. doi: 10.1074/jbc.270.41.23910. [DOI] [PubMed] [Google Scholar]
- 41.Cowan N.J., Lewis S.A. Type II chaperonins, prefoldin and the tubulin-specific chaperones. Adv. Protein Chem. 2002;59:73–104. doi: 10.1016/s0065-3233(01)59003-8. doi:10.1016/S0065-3233(01)59003-8. [DOI] [PubMed] [Google Scholar]
- 42.Gu W., Cowan N.J. Assembly properties of altered beta-tubulin polypeptides containing disrupted autoregulatory domains. Mol. Cell. Biol. 1989;9:3418–3428. doi: 10.1128/mcb.9.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes A.P., Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu. Rev. Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. doi:10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhamidipati A., Lewis S.A., Cowan N.J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. doi:10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grishchuk E.L., McIntosh J.R. Sto1p, a fission yeast protein similar to tubulin folding cofactor E, plays an essential role in mitotic microtubule assembly. J. Cell Sci. 1999;112:1979–1988. doi: 10.1242/jcs.112.12.1979. [DOI] [PubMed] [Google Scholar]
- 46.Steinborn K., Maulbetsch C., Priester B., Trautmann S., Pacher T., Geiges B., Kuttner F., Lepiniec L., Stierhof Y.D., Schwarz H., et al. The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 2002;16:959–971. doi: 10.1101/gad.221702. doi:10.1101/gad.221702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parvari R., Hershkovitz E., Grossman N., Gorodischer R., Loeys B., Zecic A., Mortier G., Gregory S., Sharony R., Kambouris M., et al. Mutation of TBCE causes hypoparathyroidism–retardation–dysmorphism and autosomal recessive Kenny–Caffey syndrome. Nat. Genet. 2002;32:448–452. doi: 10.1038/ng1012. doi:10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 48.Tian G., Huang M.C., Parvari R., Diaz G.A., Cowan N.J. Cryptic out-of-frame translational initiation of TBCE rescues tubulin formation in compound heterozygous HRD. Proc. Natl Acad. Sci. USA. 2006;103:13491–13496. doi: 10.1073/pnas.0602798103. doi:10.1073/pnas.0602798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Fanarraga M., Carranza G., Bellido J., Kortazar D., Villegas J.C., Zabala J.C. Tubulin cofactor B plays a role in the neuronal growth cone. J. Neurochem. 2007;100:1680–1687. doi: 10.1111/j.1471-4159.2006.04328.x. [DOI] [PubMed] [Google Scholar]
- 50.Lewis S.A., Lee M.G., Cowan N.J. Five mouse tubulin isotypes and their regulated expression during development. J. Cell Biol. 1985;101:852–861. doi: 10.1083/jcb.101.3.852. doi:10.1083/jcb.101.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgoyne R.D., Cambray-Deakin M.A., Lewis S.A., Sarkar S., Cowan N.J. Differential distribution of beta-tubulin isotypes in cerebellum. EMBO J. 1988;7:2311–2319. doi: 10.1002/j.1460-2075.1988.tb03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinoshita K., Habermann B., Hyman A.A. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 2002;12:267–273. doi: 10.1016/s0962-8924(02)02295-x. doi:10.1016/S0962-8924(02)02295-X. [DOI] [PubMed] [Google Scholar]
- 53.Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. doi:10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uchimura S., Oguchi Y., Hachikubo Y., Ishiwata S., Muto E. Key residues on microtubule responsible for activation of kinesin ATPase. EMBO J. 29:1167–1175. doi: 10.1038/emboj.2010.25. doi:10.1038/emboj.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cleveland D.W., Lopata M.A., MacDonald R.J., Cowan N.J., Rutter W.J., Kirschner M.W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980;20:95–105. doi: 10.1016/0092-8674(80)90238-x. doi:10.1016/0092-8674(80)90238-X. [DOI] [PubMed] [Google Scholar]
- 56.Cleveland D.W. The multitubulin hypothesis revisited: what have we learned? J. Cell Biol. 1987;104:381–383. doi: 10.1083/jcb.104.3.381. doi:10.1083/jcb.104.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luduena R.F. Are tubulin isotypes functionally significant. Mol. Biol. Cell. 1993;4:445–457. doi: 10.1091/mbc.4.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopata M.A., Cleveland D.W. In vivo microtubules are copolymers of available beta-tubulin isotypes: localization of each of six vertebrate beta-tubulin isotypes using polyclonal antibodies elicited by synthetic peptide antigens. J. Cell Biol. 1987;105:1707–1720. doi: 10.1083/jcb.105.4.1707. doi:10.1083/jcb.105.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis S.A., Gu W., Cowan N.J. Free intermingling of mammalian beta-tubulin isotypes among functionally distinct microtubules. Cell. 1987;49:539–548. doi: 10.1016/0092-8674(87)90456-9. doi:10.1016/0092-8674(87)90456-9. [DOI] [PubMed] [Google Scholar]
- 60.Gu W., Lewis S.A., Cowan N.J. Generation of antisera that discriminate among mammalian alpha- tubulins: introduction of specialized isotypes into cultured cells results in their coassembly without disruption of normal microtubule function. J. Cell Biol. 1988;106:2011–2022. doi: 10.1083/jcb.106.6.2011. doi:10.1083/jcb.106.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raff E.C., Fackenthal J.D., Hutchens J.A., Hoyle H.D., Turner F.R. Microtubule architecture specified by a beta-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. doi:10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- 62.Wilson P.G., Borisy G.G. Evolution of the multi-tubulin hypothesis. Bioessays. 1997;19:451–454. doi: 10.1002/bies.950190603. doi:10.1002/bies.950190603. [DOI] [PubMed] [Google Scholar]
- 63.Abdollahi M.R., Morrison E., Sirey T., Molnar Z., Hayward B.E., Carr I.M., Springell K., Woods C.G., Ahmed M., Hattingh L., et al. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am. J. Hum. Genet. 2009;85:737–744. doi: 10.1016/j.ajhg.2009.10.007. doi:10.1016/j.ajhg.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian G., Huang Y., Rommelaere H., Vandekerckhove J., Ampe C., Cowan N.J. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. doi:10.1016/S0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 65.Tian G., Vainberg I.E., Tap W.D., Lewis S.A., Cowan N.J. Specificity in chaperonin-mediated protein folding. Nature. 1995;375:250–253. doi: 10.1038/375250a0. doi:10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
- 66.Zabala J.C., Cowan N.J. Tubulin dimer formation via the release of alpha- and beta-tubulin monomers from multimolecular complexes. Cell. Motil. Cytoskeleton. 1992;23:222–230. doi: 10.1002/cm.970230306. doi:10.1002/cm.970230306. [DOI] [PubMed] [Google Scholar]
- 67.Jansens A., Braakman I. Pulse-chase labeling techniques for the analysis of protein maturation and degradation. Methods Mol. Biol. 2003;232:133–145. doi: 10.1385/1-59259-394-1:133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.