Abstract
The neuromuscular disease myotonic dystrophy type I (DM1) affects multiple organ systems with the major symptoms being severe muscle weakness, progressive muscle wasting and myotonia. The causative mutation in DM1 is a CTG repeat expansion in the 3′-untranslated region of the DM protein kinase (DMPK) gene. RNA transcribed from the expanded allele contains the expanded CUG repeats and leads to the nuclear depletion of Muscleblind-like 1 (MBNL1) and to the increased steady-state levels of CUG-binding protein 1 (CUGBP1). The pathogenic effects of MBNL1 depletion have previously been tested by the generation of MBNL1 knockout mice, but the consequence of CUGBP1 overexpression in adult muscle is not known. In a DM1 mouse model expressing RNA containing 960 CUG repeats in skeletal muscle, CUGBP1 up-regulation is temporally correlated with severe muscle wasting. In this study, we generated transgenic mice with doxycycline-inducible and skeletal muscle-specific expression of CUGBP1. Adult mouse skeletal muscle overexpressing CUGBP1 reproduces molecular and physiological defects of DM1 tissue. The results from this study strongly suggest that CUGBP1 has a major role in DM1 skeletal muscle pathogenesis.
INTRODUCTION
The most common cause of adult-onset muscular dystrophy is myotonic dystrophy type I (DM1) with an estimated incidence of 1 in 8000 individuals worldwide. DM1 is highly variable both in the severity and in the type of symptoms affecting skeletal, cardiac and smooth muscle as well as the endocrine and central nervous systems. Involvement of the skeletal muscle is complex and includes myotonia, tissue insulin resistance, muscle weakness and atrophy/wasting that primarily affects distal limb muscles early in the disease with proximal progression (1,2). Respiratory failure resulting from progressive muscle weakness is the major cause of death in individuals affected by DM1 (3,4). The mechanisms underlying the process of muscle wasting are largely undefined.
DM1 is caused by a CTG trinucleotide repeat expansion in the 3′-untranslated region of the DM protein kinase (DMPK) gene (5). The mutation does not primarily cause disease through loss of DMPK function, but rather through the gain of RNA function. The CTG repeat expansion is transcribed and the CUG repeat RNA is retained in the nucleus as nuclear foci (6). Two pathways are affected by the accumulation of this toxic RNA. First, the RNA-binding protein Muscleblind-like 1 (MBNL1) binds to and is sequestered by the CUG repeat RNA, causing its depletion from the nucleus (7). Second, CUG-binding protein 1 (CUGBP1) is up-regulated in DM1 myoblasts, heart and skeletal muscle (8,9) through a PKC-mediated phosphorylation event which stabilizes the protein (10). MBNL1 and CUGBP1 antagonistically regulate alternative splicing transitions during normal heart and skeletal muscle development (11). CUGBP1 also plays roles in translational control and mRNA stability (12). The depletion of MBNL1 and increased expression of CUGBP1 results in widespread transcriptional and post-transcriptional changes, including alterations in mRNA stability and alternative splicing (11–14).
The best-characterized post-transcriptional change in DM1 is the misregulation of alternative splicing. Global mRNA profiling studies in DM1 mouse models using splicing-sensitive microarrays found alterations in more than 200 splicing events (13). In the majority of cases examined so far, the misregulation is due to the disruption of a program of developmentally regulated splicing such that embryonic splice variants are aberrantly expressed in adult tissues. The abnormal expression of specific genes is directly responsible for the onset of some disease symptoms. A well-established example is the muscle-specific chloride channel 1 (CLCN1) which is mis-spliced in DM1 tissue resulting in mRNA degradation. The lack of chloride channel function in adult muscle produces myotonia (15–18). Recent evidence suggests that aberrant expression of ryanodine receptor (RyR1) with exon 70 exclusion in adult DM1 skeletal muscle may alter excitation–contraction coupling and promote muscle degeneration (19,20). However, solid links between many symptoms in the skeletal muscle system and individual alternative splicing or transcriptional events remain to be established.
We previously generated a DM1 mouse model (EpA960/HSA-Cre-ERT2) with tamoxifen-inducible and skeletal muscle-specific expression of RNA containing 960 interrupted CUG repeats within the natural position of DMPK exon 15 (DMPK-CUG960 RNA) (21). Induction of DMPK-CUG960 RNA in skeletal muscle reproduced many disease features including characteristic histological abnormalities, myotonia, decreased muscle function, severe muscle wasting, MBNL1 co-localization with nuclear foci, CUGBP1 up-regulation and misregulated alternative splicing (21). The muscle wasting phenotype was temporally correlated with increased CUGBP1 expression, suggesting that CUGBP1 may contribute to, or be primarily responsible for, muscle wasting in DM1. Other data also support this correlation, as follows: (i) splicing events responsive to CUGBP1 but not MBNL1 reverted to embryonic patterns, demonstrating a downstream consequence of elevated CUGBP1; (ii) a DM1 mouse model expressing CUG250 RNA in the context of the human skeletal actin gene (HSALR) has neither increased CUGBP1 nor a strong muscle degeneration phenotype (22); (iii) MBNL1 knockout mice (MBNL1ΔE3/ΔE3) do not exhibit severe muscle wasting suggesting that MBNL1 depletion alone is not able to reproduce this disease feature (23); (iv) constitutive transgenic overexpression of CUGBP1 in developing mouse skeletal muscle results in impaired skeletal muscle differentiation and dystrophic muscle histology (24,25); (v) CUGBP1 overexpression in Drosophila somatic muscles generated loss of muscle integrity (26), and (vi) inducible expression of exogenous CUGBP1 in adult mouse cardiomyocytes reproduces DM1 cardiac symptoms (27). Cumulatively, these data provide a strong rationale for testing the hypothesis that CUGBP1 plays a pathogenic role in skeletal muscle wasting observed in DM1.
To determine whether CUGBP1 overexpression in adult mouse skeletal muscle is sufficient to reproduce features of DM1 including skeletal muscle wasting, we generated transgenic mice with doxycycline-inducible and skeletal muscle-specific expression of CUGBP1. Mice with an 8-fold induction of CUGBP1 in skeletal muscle exhibited decreased muscle weight, impaired muscle performance and dystrophic muscle histology characteristic of DM1, including a large fraction of myofibers containing central nuclei. Additionally, we show that many alternative splicing events misregulated in human DM1 skeletal muscle are also misregulated in mouse muscle overexpressing CUGBP1. These data show that CUGBP1 up-regulation in skeletal muscle is pathogenic and is sufficient to reproduce functional and molecular features of DM1.
RESULTS
CUGBP1 overexpression is sufficient to produce a muscle phenotype reminiscent of DM1
We previously generated a doxycycline-responsive mouse line, TRECUGBP1, which encodes an N-terminal Flag-tagged human CUGBP1 LYLQ isoform located downstream of a tetracycline-responsive element and a minimal CMV promoter (27). TRECUGBP1 mice were crossed to a mouse line with skeletal muscle-specific expression of a modified reverse tetracycline transactivator (rtTA), MDAFrtTA. These mice express rtTA2S-M2, a modified rtTA with an increased doxycycline sensitivity and lower basal activity, from the rat myosin light chain 1/3 promoter/enhancer (28). At 2–3 months of age, bitransgenic MDAFrtTA/TRECUGBP1 mice were induced by feeding doxycycline-containing food (+dox). Two groups of uninduced controls were used: (i) bitransgenic MDAFrtTA/TRECUGBP1 mice fed a normal diet (–dox) and (ii) monotransgenic TRECUGBP1 or MDAFrtTA (+dox) mice. Both control groups produced similar results in all experiments performed and were therefore grouped for statistical analysis.
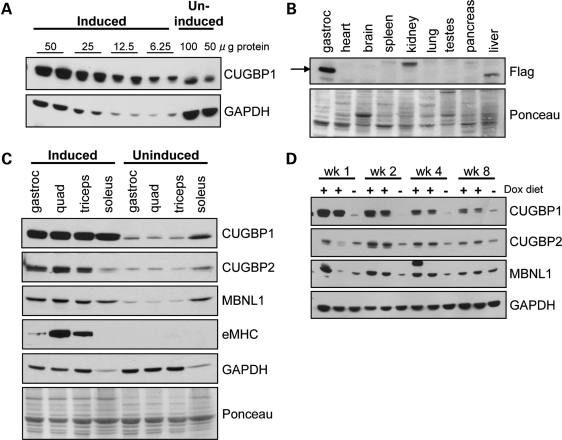
MDAFrtTA/TRECUGBP1 mice given food containing 2 g dox/kg food achieved short-term robust expression of CUGBP1. Serial dilution analysis conducted on gastrocnemius muscle protein lysate following 2 weeks of induction showed an 8-fold increase in CUGBP1 (Fig. 1A). Western blotting with an anti-Flag antibody determined CUGBP1 was expressed from the exogenous transgene specifically in the skeletal muscle and no bands of the correct molecular weight were detected in any other organ tissue tested (Fig. 1B). The bands in the liver and kidney protein lysates were deemed non-specific because they were not recognized by CUGBP1 antibody (data not shown). Additionally, CUGBP1 was up-regulated in the predominately fast twitch quadriceps and triceps, as well as the slow twitch soleus muscle (Fig. 1C). There was some induction of CUGBP2 and MBNL1, potentially due to the expression within regenerating myofibers, supported by the up-regulation of embryonic myosin heavy chain (eMHC), a marker of regeneration (Fig. 1C). CUGBP1 protein induction was not maintained over time, and by 8 weeks on dox diet, CUGBP1 was only up-regulated 2–3-fold (Fig. 1D).
Figure 1.
MDAFrtTA/TRECUGBP1 mice display CUGBP1 up-regulation in skeletal muscle upon dox administration. (A) Western blot for CUGBP1 following serial dilution of gastrocnemius muscle protein lysates from induced MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) mice show an 8-fold elevation in CUGBP1 expression compared with uninduced control mice. (B) Western blot using anti-Flag antibody to detect exogenous CUGBP1 shows that the expression is specific to the skeletal muscle of MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) mice. Ponceau staining of the blot was used as the loading control. (C) CUGBP1 is highly expressed in all MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) skeletal muscles tested including the gastrocnemius, quadriceps, triceps and soleus muscles. Western blots for CUGBP2 and MBNL1 and the regeneration marker eMHC are also shown. Both GAPDH and Ponceau stain were used as loading controls. (D) CUGBP1 western blot over the 8-week time course on dox diet shows short-term induction in MDAFrtTA/TRECUGBP1 mice. CUGBP2 and MBNL1 protein expression are also shown.
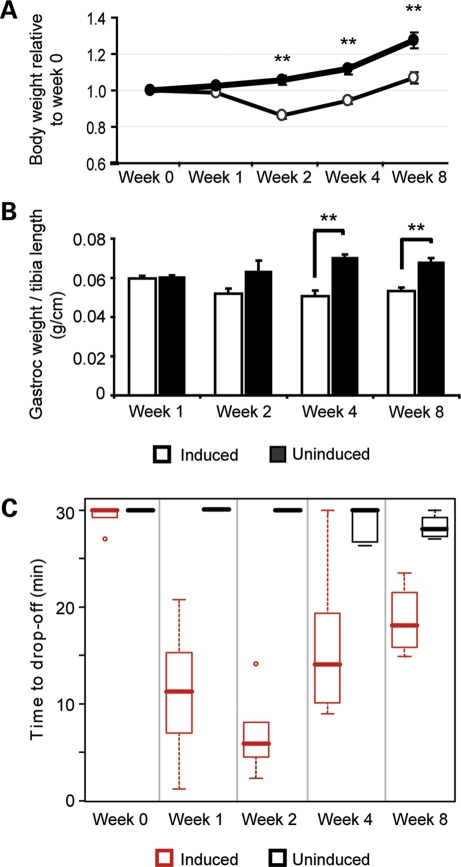
MDAFrtTA/TRECUGBP1 mice had a strong, observable phenotype by 2 weeks on dox diet exhibiting impaired movement, abnormal gait and an 18% reduction in total body weight compared with uninduced control mice (Supplementary Material Movie S1 and Fig. 2A). Furthermore, there was a 30% reduction in gastrocnemius muscle weight by 4 weeks on dox diet (Fig. 2B). Muscle function was assessed in MDAFrtTA/TRECUGBP1 mice by an acute graded treadmill assay over the 8-week time course on dox diet. During the time points of high CUGBP1 induction, namely 1–4 weeks on dox diet, MDAFrtTA/TRECUGBP1 mice exhibited a significant reduction in muscle function (Fig. 2C). However by 8 weeks of induction, when CUGBP1 expression levels are significantly reduced, muscle performance improved (Fig. 2C). These data demonstrate a very tight correlation between the level of CUGBP1 expression and the severity of muscle functional properties.
Figure 2.
Induced MDAFrtTA/TRECUGBP1 mice exhibit reduced muscle weight and muscle function. (A) Total mouse body weight at 1, 2, 4 and 8 weeks following CUGBP1 induction relative to the pre-induction weight. Induced mice (open circles, n ≥ 5) have a significant reduction in body weight when compared with uninduced control mice (closed circles, n ≥ 5), **P < 0.001. (B) Induced mice (open bars, n = 8 muscles) have a significant reduction in gastrocnemius muscle weight when compared with uninduced control mice (closed bars, n = 8 muscles), **P < 0.001. Only male mice were used for this assay and muscle weights were normalized to the length of the tibia. (C) Mice were subject to an acute treadmill exercise regime at each time point and the time to drop-off is represented in a box plot. The top and bottom of each box denote the 75th and 25th percentiles, respectively, and the median is shown by the bold middle line. Induced mice (red boxes, n ≥ 4) have a significant reduction in muscle function at 1–4 weeks on dox diet compared with uninduced control mice (black boxes, n ≥ 4), P < 0.001.
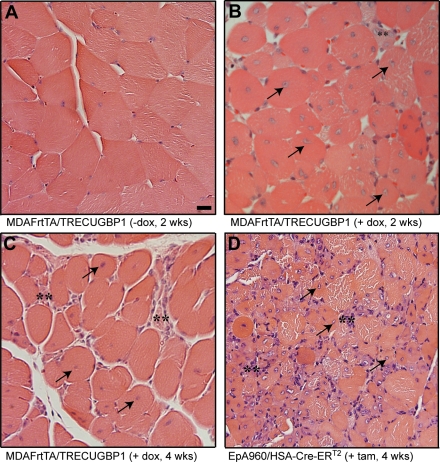
Histological examination of uninduced MDAFrtTA/TRECUGBP1 gastrocnemius muscle by hematoxylin and eosin staining showed normal muscle histology with peripherally located nuclei and no evidence of myofiber atrophy or regeneration (Fig. 3A). By 2 weeks on dox diet, the muscle histology was strikingly abnormal with an abundance of myofibers with centrally located nuclei (>10% of the sample) (Fig. 3B). A high fraction of myofibers containing central nuclei is one of the most characteristic changes observed in DM1 muscle histology (1). MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) histology also showed myofiber atrophy, increased variation in myofiber size, evidence of degeneration marked by the presence of small myofibers with inflammatory infiltrate and pyknotic nuclear clumps (Fig. 3B and Supplementary Material, Fig. S1). Similar changes were seen after 4 weeks on dox diet, but by 8 weeks the histological abnormalities were milder, only occasionally showing central nucleated myofibers (Fig. 3C and data not shown).
Figure 3.
Induced MDAFrtTA/TRECUGBP1 mice exhibit DM1-like histological features. Light microscope analysis of gastrocnemius muscle stained with hematoxylin and eosin. (A) Uninduced control mice have normal myofiber size and peripherally located nuclei. Scale bar: 20 µm. (B and C) Induced MDAFrtTA/TRECUGBP1 mice have an abundance of central nuclei (arrows) and evidence of degeneration (small fibers with inflammatory infiltrate, double asterisks) by 2 and 4 weeks on dox diet. (D) Skeletal muscles from EpA960/HSA-Cre-ERT2 mice 4 weeks after the induction of DMPK-CUG960 RNA show similar changes as CUGBP1-overexpressing muscle, including hypotrophic myofibers with central nuclei (arrows) and degenerative changes (double asterisks). All panels are 40× magnification.
Type I fiber atrophy and type II predominance have been previously reported in patient biopsies in DM1 (1). Using immunolabelling on MDAFrtTA/TRECUGBP1 (+dox) gastrocnemius muscle to define fiber types, we did not detect any notable differences between type I and type II fibers in terms of myofiber size or abundance compared with uninduced control muscle (Supplementary Material, Fig. S2).
To make a direct histological comparison between adult muscle overexpressing CUGBP1 or DMPK-CUG960 RNA, gastrocnemius muscle from MDAFrtTA/TRECUGBP1 (+dox) mice was compared side by side with gastrocnemius muscle from tamoxifen-induced EpA960/HSA-Cre-ERT2 mice 4 weeks after the induction of DMPK-CUG960 RNA. Similar to CUGBP1 overexpressing mice, EpA960/HSA-Cre-ERT2 (+tam) mice showed myofiber atrophy, myofiber size variation, degenerative changes and an even greater presence of centrally nucleated myofibers [Fig. 3D and as described previously (21)]. The histopathological abnormalities present in both MDAFrtTA/TRECUGBP1 (+dox) muscle and EpA960/HSA-Cre-ERT2 (+tam) muscle are consistent with a dystrophic process and are very similar to those seen in patients with classical DM1. Some of these features, in particular the abundance of central nuclei, are observed much more frequently in DM1 than other muscular dystrophies and therefore serve as a valuable diagnostic attribute (1).
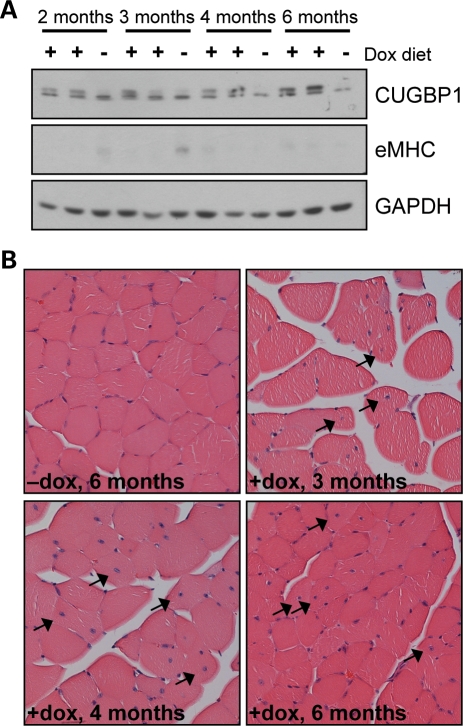
MDAFrtTA/TRECUGBP1 mice given food containing 2 g dox/kg food induce CUGBP1 expression 8-fold above endogenous levels but this level of expression was not sustained beyond 2 weeks. In DM1 skeletal muscle tissue, CUGBP1 levels are elevated 2–4-fold (8,9). We therefore tested whether a lower dosage of doxycycline would provide sustained elevation of CUGBP1 levels comparable to that observed in DM1 skeletal muscle. MDAFrtTA/TRECUGBP1 mice induced by a low dosage dox diet (0.05 g dox/kg food) had approximately a 2-fold increase in CUGBP1 levels in the gastrocnemius muscle with stable protein expression for up to 6 months (Fig. 4A). The muscle had no detectable expression of the regeneration marker, eMHC, consistent with an impaired regeneration response in DM1 patients (Fig. 4A) (29,30). This low induction of CUGBP1 was not enough to induce changes in alternative splicing (Supplementary Material, Fig. S3). However, MDAFrtTA/TRECUGBP1 mice with a 2-fold increase in CUGBP1 did have mild muscle histology with scattered myofibers containing centrally located nuclei, particularly by 4–6 months on dox diet, compared with uninduced control mice (Fig. 4B). There was little variability in myofiber size and few small regenerating myofibers were present (Fig. 4B).
Figure 4.
Long-term expression of CUGBP1 in MDAFrtTA/TRECUGBP1 mice induced with a low dosage dox diet (0.05 g dox/kg food). (A) Western blot of protein extracted from gastrocnemius muscle shows a 2-fold up-regulation of CUGBP1 in MDAFrtTA/TRECUGBP1 mice (two mice per time point) fed a low dosage dox diet compared with uninduced controls (one mouse per time point). The induction is stable over the 6-month time course. There is no detectable expression of the regeneration marker, eMHC. (B) Hematoxylin and eosin staining of gastrocnemius cross-sections from low dosage dox-induced MDAFrtTA/TRECUGBP1 mice shows an increase in myofibers containing centrally located nuclei (arrows) (20× magnification).
Overexpressing CUGBP1 in mouse skeletal muscle reproduces misregulation of alternative splicing that is characteristic of DM1
A distinctive molecular feature of DM1 skeletal muscle is the misregulation of alternative splicing, such that there is increased expression of embryonic splice variants in adult tissues. Detailed analysis of developmentally regulated alternative splicing events was previously conducted in mouse heart tissue overexpressing CUGBP1 or from MBNL1 knockout mice (11). Whereas some events were jointly regulated by both CUGBP1 and MBNL1, others responded only to an increase in CUGBP1 or loss of MBNL1, and are referred to as CUGBP1-responsive or MBNL1-responsive events, respectively.
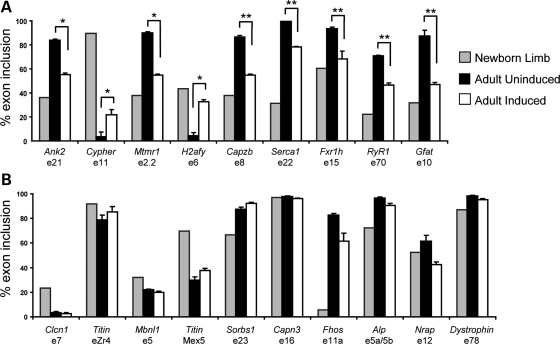
To determine whether MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) mouse muscle recapitulated the misregulation of alternative splicing observed in DM1, we analyzed 19 alternative splicing events previously demonstrated to be misregulated in DM1 patient skeletal muscle (20,21,24,31,32). Nine of the splicing events were significantly misregulated in skeletal muscle overexpressing CUGBP1, showing increased expression of the embryonic splice variant in adult tissues for all nine events (Fig. 5A). Splicing of H2afy exon 6 was regulated by both CUGBP1 and MBNL1 in mouse heart tissue, whereas three of the events (Ank2 exon 21, Capzb exon 8 and Fxr1h exon 15) were previously demonstrated to be CUGBP1-responsive events in mouse heart (11). Additionally, we showed that these three CUGBP1-responsive events are misregulated in induced EpA960/HSA-Cre-ERT2 mouse muscle which has increased expression of CUGBP1, but not in HSALR or MBNL1ΔE3/ΔE3 muscle without CUGBP1 up-regulation (21). Since these three alternative splicing events change in response to CUGBP1 overexpression but not MBNL1 depletion, up-regulation of CUGBP1 is essential for at least some molecular features of DM1. Furthermore, these data show that overexpression of CUGBP1 is sufficient to reproduce some of the embryonic splicing patterns that are observed in DM1 skeletal muscle.
Figure 5.
Induced MDAFrtTA/TRECUGBP1 mice reproduce the characteristic alternative splicing misregulation observed in DM1 patients. (A) MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) mice have increased expression of the embryonic splice variants in adult gastrocnemius muscle for the indicated alternative splicing events, *P < 0.01 and **P < 0.001. t-Test for statistical significance was performed between adult uninduced and induced mice. (B) Alternative splicing events that are not significantly misregulated in induced MDAFrtTA/TRECUGBP1 gastrocnemius muscle. Newborn limb (n = 1, pooled limbs from 12 mice), adult uninduced (n = 4, individual) and adult induced (n = 4, individual).
The remaining 10 splicing events tested showed little or no misregulation in MDAFrtTA/TRECUGBP1 (+dox) mouse muscle (Fig. 5B). For four events (Titin exon Zr4, Capn3 exon 16, Nrap exon 12 and Dystrophin exon 78), there was only a weak developmental regulation between newborn limb and adult control in the mouse. One event (Sorbs1 exon 23) was previously characterized as a MBNL1-responsive event in mouse heart (11) and was therefore not expected to change in muscle overexpressing CUGBP1. Splicing of Clcn1 exon 7a is well characterized in other DM1 mouse models and its splicing misregulation is responsible for myotonia (15,16,18). Clcn1 exon 7a is not misregulated in MDAFrtTA/TRECUGBP1 (+dox) muscle, and in agreement with this result, six out of six mice tested by electromyography did not show evidence of myotonia (data not shown). Alternative splicing events that are misregulated in DM1 patients, but not in MDAFrtTA/TRECUGBP1 (+dox) mice are likely MBNL1-responsive events or are effects secondary to other disease processes.
DISCUSSION
The expression of CUG repeat RNA in individuals with DM1 induces alterations in two RNA binding proteins; namely, the depletion of MBNL1 and up-regulation of CUGBP1. The role of MBNL1 in disease pathogenesis has been well established through the generation and characterization of MBNL1 knockout mice (23). Although MBNL1 knockout is sufficient for the development of myotonia and dystrophic muscle histology, MBNL1 knockout mice do not possess a strong muscle wasting phenotype (23). More recently, transcriptome profiling in MBNL1 knockout mice has revealed widespread changes in transcription, RNA processing and mRNA decay which largely overlap with changes in transgenic mice generated by the expression of CUG250 RNA in the context of the human skeletal actin gene (HSA-CUG250 RNA) (13,14). Neither MBNL1 knockout mice nor transgenic mice expressing HSA-CUG250 RNA have increased CUGBP1 levels. These studies indicate that MBNL1 can reproduce some features of DM1 independent of alterations in CUGBP1 expression.
So what is the role of CUGBP1, if any, in DM1 pathogenesis? CUGBP1 protein levels are increased in DM1 heart and skeletal muscle (8,9,33), but whether this increase plays a primary role in muscle degeneration or is simply an effect secondary to muscle damage is not known. Several studies using transgenic mice support a pathogenic function of CUGBP1. First, mice expressing DMPK-CUG5 RNA in skeletal muscle had elevated levels of CUGBP1 with no change in MBNL expression or sequestration and showed characteristic DM1 histopathology, myotonia and splicing abnormalities (34). Second, induction of DMPK-CUG960 RNA in mouse skeletal muscle resulted in a severe muscle wasting phenotype that correlated with an increase in CUGBP1, which has not been mimicked by MBNL1 depletion alone (21). Third, within 6h following induction of DMPK-CUG960 RNA in mouse cardiac muscle, CUGBP1 levels were increased and pharmacological inhibition of CUGBP1 up-regulation prevented the DM1-like heart phenotype (35,36). Fourth, we recently showed that 4-fold overexpression of exogenous CUGBP1 in mouse cardiac tissue reproduced molecular, histopathological and functional changes observed in DM1 mouse models and individuals with DM1 (27).
In this paper, we tested the consequence of transgenic expression of exogenous CUGBP1 in adult mouse skeletal muscle. We showed that an 8-fold increase in CUGBP1 resulted in decreased muscle weight and impaired muscle function. By histology, the muscle was reminiscent of DM1 muscle biopsies with a large fraction of myofibers containing centrally located nuclei, pyknotic nuclear clumps and evidence of muscle degeneration. We estimate that less than 10% of the gastrocnemius muscle exhibits regeneration consistent with the low expression of eMHC. In contrast, some of the misregulated splicing changes assayed from gastrocnemius tissue show nearly full reversion to the embryonic patterns. Therefore, we conclude that the large extent of the molecular and phenotypic changes cannot be explained solely based on non-specific responses to muscle regeneration rather than direct effects of CUGBP1 expression.
CUGBP2 and MBNL1 were also increased in MDAFrtTA/TRECUGBP1 mice with an 8-fold increase in CUGBP1. Although it is possible that elevated MBNL1 may have some consequences on the skeletal muscle phenotype in these mice, MDAFrtTA/TRECUGBP1 mice fed a low dose dox diet with only a 2-fold increase in CUGBP1 did not exhibit elevated MBNL1 and yet show histological abnormalities.
A number of alternative splicing events misregulated in DM1 skeletal muscle were shown to be regulated by CUGBP1 and not MBNL1 in transgenic and knockout mice (11). It is likely that misregulation of these CUGBP1-responsive events in DM1 tissues are a molecular signature of elevated CUGBP1, independent of MBNL1 depletion. In this study, we showed that nine misregulated alternative splicing events identified in DM1 skeletal muscle are also misregulated in the MDAFrtTA/TRECUGBP1 (+dox, 2 weeks) mice with the expression of the embryonic splice variants in adult muscle for all events. Many of the affected transcripts have important functions in skeletal muscle, and their aberrant splicing may have adverse consequences on muscle integrity and function. It has already been reported that alternative splicing of RyR1 exon 70 alters excitation–contraction coupling (19) and this exon is misregulated in CUGBP1 overexpressing muscle. These data suggest that down-regulation of CUGBP1 in DM1 mouse models expressing DMPK-CUG repeat RNA even in the presence of MBNL1 depletion may ameliorate the disease phenotype.
A progressive loss of inducibility occurred in the MDAFrtTA/TRECUGBP1 mice fed 2 g dox/kg food for 8 weeks, possibly due to the induction of post-transcriptional down-regulation by high CUGBP1 expression or increased doxycycline metabolism by the liver. The inability to maintain long-term induction of CUGBP1 was overcome using a lower doxycycline dosage (0.05 g dox/kg food), but we only achieved a 2-fold up-regulation of CUGBP1 by this method. Although there were some mild histological features in the MDAFrtTA/TRECUGBP1 mice induced with the lower doxycycline dosage diet, there were not other features of DM1 including misregulated alternative splicing. In DM1 tissue and cell cultures, CUGBP1 is stabilized by hyperphosphorylation (10). It may be possible that in addition to increasing CUGBP1 steady-state levels, hyperphosphorylation alters the protein activity or function and this may be necessary for mediating the effect of CUGBP1 on muscle wasting at lower levels of the protein. Although this very low, stable induction of CUGBP1 over a long time course did not produce a severe muscle wasting phenotype, it may more accurately represents the progressive nature of muscle atrophy in DM1.
The results presented here demonstrate that aberrant expression of CUGBP1 in adult skeletal muscle is pathogenic and produces a phenotype reminiscent of DM1 skeletal muscle. In the future, it will be important to determine the alterations that occur downstream of CUGBP1 up-regulation and which of those events are responsible for the specific phenotypes observed in CUGBP1 overexpressing muscle and individuals with DM1. Although some alternative splicing and translational targets of CUGBP1 have already been identified, global transcriptome profiling will undoubtedly identify new CUGBP1 targets with vital roles in skeletal muscle tissue.
MATERIALS AND METHODS
Transgenic mice
TRECUGBP1 (line 3413) transgenic mice were generated as previously described and maintained on an FVB background (27). MDAFrtTA transgenic mice express a modified rtTA from the myosin light chain 1/3 promoter/enhancer and were maintained on a mixed C57BL/6× DBA background (28). All mice used in these studies were F1 progeny from TRECUGBP1 × MDAFrtTA matings. At 2–3 months of age, bitransgenic MDAFrtTA/TRECUGBP1 mice were given free access to food containing 0.05 g dox/kg food (Teklad Harlan Inc., Madison, WI, USA) or 2 g dox/kg food (Bio-Serv, Frenchtown, NJ, USA). Whenever possible, littermate TRECUGBP1 or MDAFrtTA (+dox) or MDAFrtTA/TRECUGBP1 (–dox) controls were used.
Treadmill assay
Mice were placed in an AccuPacer treadmill with an electric shock stimulus grid (AccuScan Instruments, Inc.) and muscle function was assessed by a graded treadmill protocol. The starting treadmill speed was 10 m/min and the pace was increased by 2 m/min every 2min until a maximum speed of 30 m/min was reached. Exhaustion was declared the third time a mouse spent 3–5 s on the shock grid without returning to the treadmill. The experiment was stopped immediately at the time of exhaustion or 30 min, whichever came first.
Histology
Tissues were fixed overnight in 10% formalin and then stored in 70% ethanol until sectioning. Tissues were then paraffin-embedded and cut in cross-section at 10 µm. Hematoxylin and eosin staining was done according to the standard procedures. Immunohistochemistry for slow twitch and fast twitch myosin heavy chain isoforms was performed with monoclonal anti-myosin slow clone NOQ7.5.4D (Sigma) and monoclonal anti-myosin fast clone MY-32 (Sigma) antibodies.
RNA isolation and RT–PCR
Total RNA was isolated from skeletal muscle using TRIzol reagent (Invitrogen) with the Omni TH homogenizer (Omni International). RNA integrity was assessed on a formaldehyde gel. cDNA was generated from 4 µg of RNA using oligo(dT) and AMV reverse transcriptase (Life Sciences, Inc.). The PCR program for all transcripts was 95°C for 1 min 15 s (95°C for 45 s, 57°C for 45 s, 72°C for 1 min), 25 cycles, 72°C for 10 min. For the Serca1 and Cypher transcripts, the program was modified to include only 20 cycles, and for the Clcn1 transcript, 26 cycles was used. All primer sequences were designed to flank the alternative exon and are provided in Supplementary Material, Table S1. After separation on a 5% non-denaturaing acrylamide gel, band products were quantified using the Kodak Gel Logic 2200 (Rochester, NY, USA) and Molecular Imaging software. The percentage of exon inclusion was calculated as [exon inclusion band/ (exon inclusion band + exon exclusion band)] × 100 with a correction factor for the amount of ethidium bromide bound per base pair of DNA.
Protein isolation and western blot
Skeletal muscle was subject to dounce homogenization in lysis buffer [10 mm HEPES pH 7.5, 0.32 m sucrose, 5 µm MG132, 5 mm EDTA, protease inhibitor cocktail tablet (Roche) and 1% SDS]. Protein concentration from the supernatant was determined with the BCA protein assay kit (Thermo Scientific). Protein samples (50 µg each) were separated on a 10% SDS–PAGE gel and transferred to Immobilon-P membrane (Millipore), after which the membranes were blocked and probed with one of the following antibodies: CUGBP1 clone 3B1 (M. Swanson, 1:1000) and CUGBP2 clone IH2 (M. Swanson, 1:500) followed by goat anti-mouse light chain (Jackson ImmunoResearch, 1:10 000); MBNL1 (C. Thornton, 1:1000) followed by goat anti-rabbit (Calbiochem, 1:10 000); eMHC F1.652 (Developmental Studies Hybridoma Bank, 1:200) and GAPDH (Abcam, 1:5000) followed by sheep anti-mouse (Jackson ImmunoResearch 1:10 000).
Statistical analysis
All statistical analysis was performed with GraphPad InStat software. When two groups were compared, a two-tailed Student's t-test with equal or unequal variance was performed as appropriate, and the significance level was set at P-values less than 0.05 for all statistical analyses. All data are expressed as mean ± standard error of the mean.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health predoctoral NRSA fellowship (F31NS067740 to A.J.W.), National Institutes of Health (R01AR45653, R01GM076493 to T.A.C.), the Muscular Dystrophy Association (T.A.C.), and set-up funds from Texas A&M Health Science Center College of Medicine (M.R.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Donnie Bundman for technical assistance in generating the TRECUGBP1 transgenic line and Xander Wehrens (Baylor College of Medicine, Houston, TX, USA) for generous access to the treadmill apparatus. All histology was performed by the Center for Comparative Medicine pathology core facility at Baylor College of Medicine, with a special thanks to Bilqees Bhatti for her expertise.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harper P.S., Brook J.D., Newman E. Myotonic dystrophy. London, New York: W. B. Saunders; 2001. [Google Scholar]
- 2.Moxley R.T., III, Griggs R.C., Goldblatt D., VanGelder V., Herr B.E., Thiel R. Decreased insulin sensitivity of forearm muscle in myotonic dystrophy. J. Clin. Invest. 1978;62:857–867. doi: 10.1172/JCI109198. doi:10.1172/JCI109198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groh W.J., Groh M.R., Saha C., Kincaid J.C., Simmons Z., Ciafaloni E., Pourmand R., Otten R.F., Bhakta D., Nair G.V., et al. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N. Engl. J. Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. doi:10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- 4.Mathieu J., Allard P., Potvin L., Prevost C., Begin P. A 10-year study of mortality in a cohort of patients with myotonic dystrophy. Neurology. 1999;52:1658–1662. doi: 10.1212/wnl.52.8.1658. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O'Hoy K., et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. doi:10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 6.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. doi:10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. doi:10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. doi:10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 9.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. doi:10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 10.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. doi:10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. doi:10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlasova I.A., Tahoe N.M., Fan D., Larsson O., Rattenbacher B., Sternjohn J.R., Vasdewani J., Karypis G., Reilly C.S., Bitterman P.B., et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. doi:10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du H., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A., et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. doi:10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborne R.J., Lin X., Welle S., Sobczak K., O'Rourke J.R., Swanson M.S., Thornton C.A. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. doi:10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlet B.N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 16.Lueck J.D., Lungu C., Mankodi A., Osborne R.J., Welle S.L., Dirksen R.T., Thornton C.A. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am. J. Physiol. Cell Physiol. 2007;292:C1291–C1297. doi: 10.1152/ajpcell.00336.2006. doi:10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 17.Mankodi A., Takahashi M.P., Jiang H., Beck C.L., Bowers W.J., Moxley R.T., Cannon S.C., Thornton C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. doi:10.1016/S1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura T., Lueck J.D., Harvey P.J., Pace S.M., Ikemoto N., Casarotto M.G., Dirksen R.T., Dulhunty A.F. Alternative splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell Calcium. 2009;45:264–274. doi: 10.1016/j.ceca.2008.11.005. doi:10.1016/j.ceca.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura T., Nakamori M., Lueck J.D., Pouliquin P., Aoike F., Fujimura H., Dirksen R.T., Takahashi M.P., Dulhunty A.F., Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. doi:10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 21.Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J., Cooper T.A. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. doi:10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. doi:10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 23.Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. doi:10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 24.Ho T.H., Bundman D., Armstrong D.L., Cooper T.A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. doi:10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 25.Timchenko N.A., Patel R., Iakova P., Cai Z.J., Quan L., Timchenko L.T. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. doi:10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 26.de Haro M., Al-Ramahi I., De Gouyon B., Ukani L., Rosa A., Faustino N.A., Ashizawa T., Cooper T.A., Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. doi:10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 27.Koshelev M., Sarma S., Price R.E., Wehrens X.H., Cooper T.A. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. doi:10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponomareva O.N., Ma H., Vock V.M., Ellerton E.L., Moody S.E., Dakour R., Chodosh L.A., Rimer M. Defective neuromuscular synaptogenesis in mice expressing constitutively active ErbB2 in skeletal muscle fibers. Mol. Cell Neurosci. 2006;31:334–345. doi: 10.1016/j.mcn.2005.10.004. doi:10.1016/j.mcn.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Furling D., Coiffier L., Mouly V., Barbet J.P., St Guily J.L., Taneja K., Gourdon G., Junien C., Butler-Browne G.S. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001;10:2079–2087. doi: 10.1093/hmg/10.19.2079. doi:10.1093/hmg/10.19.2079. [DOI] [PubMed] [Google Scholar]
- 30.Thornell L.E., Lindstom M., Renault V., Klein A., Mouly V., Ansved T., Butler-Browne G., Furling D. Satellite cell dysfunction contributes to the progressive muscle atrophy in myotonic dystrophy type 1. Neuropathol. Appl. Neurobiol. 2009;35:603–613. doi: 10.1111/j.1365-2990.2009.01014.x. doi:10.1111/j.1365-2990.2009.01014.x. [DOI] [PubMed] [Google Scholar]
- 31.Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. doi:10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 32.Nakamori M., Kimura T., Fujimura H., Takahashi M.P., Sakoda S. Altered mRNA splicing of dystrophin in type 1 myotonic dystrophy. Muscle Nerve. 2007;36:251–257. doi: 10.1002/mus.20809. doi:10.1002/mus.20809. [DOI] [PubMed] [Google Scholar]
- 33.Timchenko N.A., Cai Z.J., Welm A.L., Reddy S., Ashizawa T., Timchenko L.T. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. doi:10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 34.Mahadevan M.S., Yadava R.S., Yu Q., Balijepalli S., Frenzel-McCardell C.D., Bourne T.D., Phillips L.H. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. doi:10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. doi:10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G.S., Kuyumcu-Martinez M.N., Sarma S., Mathur N., Wehrens X.H., Cooper T.A. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 2009;119:3797–3806. doi: 10.1172/JCI37976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.