Abstract
Botulinum neurotoxins (BoNTs) typically bind the neuronal cell surface via dual interactions with both protein receptors and gangliosides. We present here the 1.9 Å X-ray structure of the BoNT serotype G (BoNT/G) receptor binding domain (residues 868-1297) and a detailed view of the protein receptor and ganglioside binding regions. The ganglioside binding motif (SxWY) has a conserved structure compared to the corresponding regions in BoNT/A and BoNT/B, but several features of the interactions with the hydrophilic face of the ganglioside are absent from the opposite side of the motif in the BoNT/G ganglioside binding cleft. This may significantly reduce the affinity between BoNT/G and gangliosides. BoNT/G and BoNT/B share the protein receptor, synaptotagmin I/II (Syt-I/II). The Syt binding site has a conserved hydrophobic plateau located centrally in the proposed protein receptor binding interface (Tyr1189, Phe1202, Ala1204, Pro1205 and Phe1212). Interestingly, only 5 of 14 residues that are important for the binding between Syt-II and BoNT/B are conserved in BoNT/G, suggesting that the means by which BoNT/G and BoNT/B bind Syt diverges more than previously appreciated. Indeed, substitution of Syt-II Phe47 and Phe55 with alanine residues had little effect on the binding of BoNT/G, but strongly reduced binding of BoNT/B. Furthermore, an extended solvent exposed hydrophobic loop, located between the Syt binding site and the ganglioside binding cleft, may serve as a third membrane association and binding element to contribute to the high affinity binding to the neuronal membrane. While BoNT/G and BoNT/B are homologous to one-another and both utilize Syt-I/II as their protein receptor, the precise means by which these two toxin serotypes bind to Syt appears surprisingly divergent.
Keywords: botulinum, neurotoxin, Synaptotagmin, ganglioside, toxin
Introduction
Botulinum neurotoxins (BoNTs) are the most toxic biological substances known with LD50s as low as 1 ng/kg1. The toxin is a zinc metalloprotease that targets the SNARE (soluble N-ethylmaleimide-sensitive-factor attachment protein receptor) proteins that are critical for the release of acetylcholine in neuromuscular junctions 2; 3. A multitude of medical applications exist where BoNTs are used to paralyze specific muscles and new applications are regularly being discovered 3; 4; 5. There are seven known serotypes of BoNTs, BoNT/A-G that have different receptor binding and SNARE-cleavage site specificities, although they share several structural and functional features. BoNT is produced mainly by Clostridium botulinum as a 150 kDa protein consisting of a 50 kDa light chain (LC) and a 100 kDa heavy chain. The LC has proteolytic activity, and crystal structures of the LCs from all seven BoNT serotypes (A-G) have been solved, including the BoNT/G LC structure 6.
The heavy chain consists of the N-terminal translocation domain (Hn) that moves the LC across the endosome membrane and into the cytosol, and the C-terminal domain (Hc) that is responsible for the high affinity and specificity of the toxin's binding to neuronal membranes. Recognition and binding of BoNTs to neuronal membranes was originally proposed to occur via a “dual receptor” model, where BoNTs bind the neuronal cell surface via interactions with both protein components and gangliosides. This model was first proposed more than 20 years ago7 and now has extensive experimental support 8; 9; 10; 11; 12.
After binding, BoNT enters the cell by endocytosis. The low pH of the endocytosed vesicle induces conformational changes in the heavy chain Hn domain, which somehow forms a channel that translocates the LC across the membrane into the cytosol of the neuron, thus allowing access to its substrates,the SNARE proteins1; 13. Cleavage site specificity is dependent on the BoNT serotype. BoNT serotype G cleaves synaptobrevin 14 (also referred to as VAMP), BoNT serotypes B, D and F also cleave synaptobrevin 2, BoNT serotypes A and E cleave SNAP-25, and BoNT serotype C can cleave both syntaxin and SNAP-25 2.
Different toxin serotypes may recognize different cell surface receptors. All BoNTs, except D, are known to interact with gangliosides. A ganglioside binding motif SxWY has been proposed that can be found in six of the seven toxins except BoNT/D 15; 16. The protein receptors have been established for serotypes A, B, E and G. BoNT/A uses SV2A, SV2B and SV2C 17; 18. BoNT/E binds SV2A and SV2B 19 while BoNT/B and BoNT/G both bind synaptotagmin (Syt) I/II 20; 21; 22. It was recently reported that BoNT/F uses SV2 as its receptor 16; 23. Both SV2 and Syt are synaptic vesicle membrane proteins.
BoNT/B and BoNT/G are the only serotypes that are known to utilize Syt proteins as receptors 21. The structures of the binding domains of Hc/A, Hc/B, Hc/E, Hc/F have previously been reported 23; 24; 25; 26 as has the structure of the binding domain of the related tetanus toxin 27. The only structure of a BoNT in a complex with its receptor protein is BoNT/B bound to Syt-II 8; 9. BoNT/G was reported to recognize the same region of Syt-II as BoNT/B 12; 21. A comparative study between these two toxins will shed light onto the molecular details about how closely related toxins achieve specific recognition of the same protein receptors. In order to understand the differences in binding properties for two different BoNT serotypes that recognize the same cell surface receptor, we also determined the structure of the receptor binding domain of BoNT/G (Hc/G). The Hc/G structure presented here expands our structural knowledge of this domain and facilitates modeling of its interactions with gangliosides and protein receptors. A detailed understanding of BoNT-receptor interfaces is crucial not only for rational redesign of the neurotoxins for medical purposes, but also for the development of inhibitors targeting dual receptor binding.
Results and Discussion
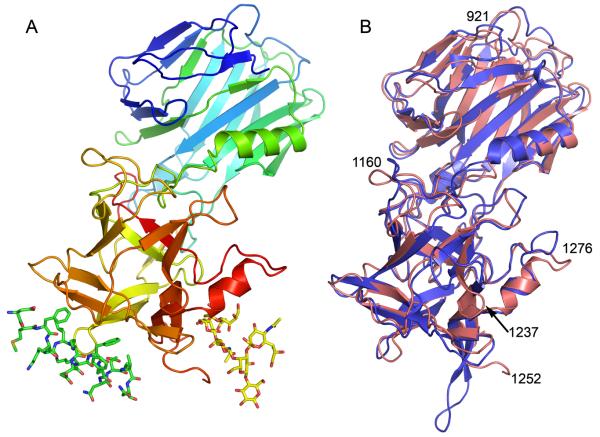
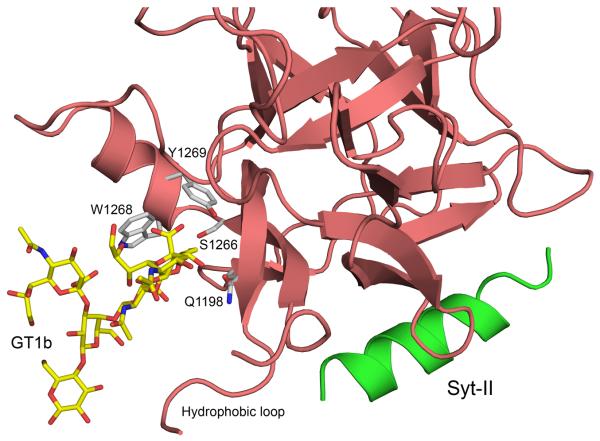
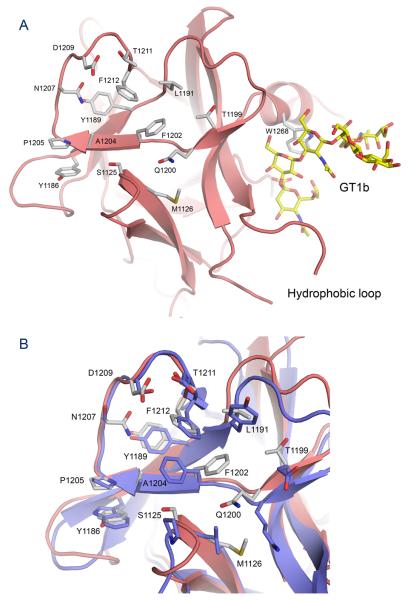
The structure of the BoNT/G binding domain (Hc/G) residues 868-1297 has been determined at 1.9 Å resolution using x-ray crystallography. The structure can be divided into two subdomains, the N-terminal subdomain (Hcn) composed of residues 868-1073 and the C-terminal subdomain (Hcc), composed of residues 1089-1297. Hcn is all β-sheet organized into a jelly roll barrel fold, and has unknown function. Hcc has a β-trefoil motif and harbors the ganglioside binding site and the protein receptor binding site. The protein receptor binding site is located in a distinct crevice separate from the ganglioside binding site (Figure 1a). Three residues in Hcc (Gly1253, Tyr1254 and Val1255) located in a flexible surface loop were disordered with minimal electron density and were not modeled.1
Figure 1. Overall structure of Hc/G and comparison with Hc/B.
(A) Ribbon representation of the BoNT/G binding domain (Hc/G) structure, in rainbow color representation, from the N-terminus (blue) to the C-terminus (red). The position of GT1b has been modeled from the Hc/A GT1b complex 11 and GT1b is shown as yellow sticks. The position of the Syt-II receptor has been modeled from the BoNT/B – Syt-II complex 8, and Syt-II is shown as green sticks. The exposed hydrophobic loop of Hc/G can be seen between GT1b and Syt-II. (B) Overlay of the Hc/G structure shown as light red ribbon and the Hc/B structure (2NP0) shown as blue ribbon. Residue numbers (BoNT/G) for some of the important areas are indicated.
The most similar structure to Hc/G in the Protein Data Bank is BoNT/B binding domain (Hc/B; PDB ID code 2NM1) with a root mean square deviation (RMSD) of 1.2 Å over 393 amino acids, (Figure 1b). A comparison with Hc/A (2VU9) shows a RMSD of 1.4 Å over 388 residues. The sequence identity between Hc/G and Hc/B is 49%, while the overall identity between the full length holotoxins is 58% (Figure 2). There is a lower degree of conservation for the Hc domain than the rest of the neurotoxin.
Figure 2. Structure-based sequence alignment.
Structure-based sequence alignment including the binding domains of BoNT/G (Hc/G; this work), BoNT/B (Hc/B; PDB: 2NM1), BoNT/A (Hc/A; PDB: 2VU9), and TeNT (Hc/T; PDB: 1AF9). The key residues for Syt binding are indicated with filled circles. These residues have been shown by mutagenesis to be important for Syt binding or correspond to the residues that directly interact with Syt-II in the complex between Hc/B and Syt-II 8; 9. The key residues in ganglioside binding from the Hc/A-GT1b complex are indicated with filled triangles. The three residues that were disordered in the Hc/G structure are indicated with stars, these residues also constitute the center of the hydrophobic loop that might be involved in membrane binding. The numbering corresponds to BoNT/G and secondary structure elements from Hc/G are indicated above the sequence.
The overall Hc/G structure fold is conserved among the Hc domains of known structure, with the exception of a few specific areas. One of these areas is Hc/G 921-932, located in the interface between Hc and the translocation domain, which has a very different conformation compared to the corresponding region in the BoNT/B Hc domain in both the holotoxin 25 and the isolated domain 28 structures. In both BoNT/A and BoNT/B, the detachment of the Hc domain from the rest of the toxin leads to a drastic rearrangement of this region 11; 28. Interestingly, the structure of the 921-932 region in Hc/G is more similar to the corresponding region in the isolated Hc/A structure 11 than the isolated Hc/B structure. This was unexpected since BoNT/G has a higher degree of overall sequence identity to BoNT/B. However, a closer examination of the Hc/G 921-932 region reveals that only 2 of 12 residues are homologous to Hc/B while 5 of 12 are homologous to Hc/A. The rearrangement of this region has now been observed for Hc/A, Hc/B and is likely to have occurred also for Hc/G.
The largest structural differences between Hc/G and Hc/B are in loop regions (1160, 1195, 1237, 1255 and 1276) (Figure 1). The Tyr1252-Trp1256 loop is located between the binding sites for the ganglioside and the protein receptor (Figure 3) and this hydrophobic loop is likely involved in membrane association. The 1276 loop is close to the ganglioside-binding site and has a different configuration than in the Hc/B structure (Figure 1). Lastly, the Leu1191-Gln1200 loop, which has a critical role in receptor binding since it is likely to be in direct contact with both the ganglioside and the protein receptor 8; 9; 11, also has a different conformation in Hc/G compared to Hc/B (Figure 4). A well-defined glycerol molecule in the Hc/G structure interacts with the backside of this loop (Leu1191-Gln1200), indicating that this area may also be involved in direct interactions with either the membrane or a receptor.
Figure 3. Ganglioside binding grove.
The ganglioside binding grove of BoNT/G with a modeled GT1b ganglioside (from the Hc/A-GT1b complex). The position of GT1b has been modeled from the Hc/A - GT1b complex, and is shown as yellow sticks 11. The key residues in ganglioside binding are shown as grey sticks. The position of the Syt-II receptor has been modeled from the Hc/B – Syt-II complex. Syt-II is shown as green ribbon in a neighboring binding grove 8. The exposed hydrophobic loop of Hc/G can be seen between GT1b and Syt-II.
Figure 4. Synaptotagmin binding site.
(A) The Syt binding site of the Hc/G structure, looking down from the Syt receptor, Hc/G is shown as a light red ribbon. Key residues in Syt binding are shown as grey sticks. These residues have been shown by mutagenesis to be important for binding, or correspond to the residues that directly interact with Syt-II in the complex between Hc/B and Syt-II. Trp1268 (grey sticks) and a modeled GT1b (yellow sticks) are also shown to indicate the position of the ganglioside binding site. (B) An overlay of the Syt binding sites of the Hc/G structure (light red ribbon) and the Hc/B structure (blue ribbon) 8. Numbering corresponds to BoNT/G and key residues are shown as grey sticks; the corresponding residues in Hc/B are shown as blue sticks. The hydrophobic center of the binding site (5 of 14 residues) is conserved while large differences are present for the other key residues in binding.
It is interesting to note that most of the structural differences between Hc/G and Hc/B are localized to the Hcc domain, the domain that harbors the binding sites for the receptors and that interacts with the membrane. Apart from the interaction area between the Hcn domain and the translocation domain, there are few major differences in the backbone structure of the Hcn domain between Hcn/G and Hcn/B. One could argue that there should be fewer changes tolerated in the Hcc domain than the Hcn domain because of the need for structural conservation of the receptor interactions that are located in the Hcc subdomain, but the opposite appears to be the case. Our data suggest that there appears to be a higher need or tolerance for structural flexibility in the Hcc domain.
The ganglioside binding site
The structure of the complex between BoNT/A and the ganglioside GT1b has recently been determined11. This structure makes it possible to model the interaction between GT1b and Hc/G based on an overlay of the Hc/A and Hc/G structures. The overall structure of the ganglioside binding cleft and the orientation and position of the SxWY motif required for interactions are conserved in the Hc/G structure (Ser1266, Trp1268 and Tyr1269) (Figure 2 and 3). The position of these three residues is almost identical to the corresponding residues in BoNT/A and B. However, there are considerable differences located on the side opposite of the SxWY motif in the ganglioside binding cleft and the region that interacts with the hydrophilic face of the ganglioside.
There appears to be a cumulative effect of the interaction with the Syt receptor and the ganglioside and serotype B. The dissociation constant between BoNT/B and Syt-I is 2.3 nM, while it is 0.23 nM for the interaction with Syt-II 29, showing a clear preference for the binding to Syt-II. BoNT/G does not appear to have the same preference for Syt-II over Syt-I as BoNT/B 10. We have studied the apparent affinity of Syt-I and Syt-II for BoNT/G-HCR in the presence and absence of gangliosides using a pull down assay (Supplementary Figure S1). The apparent dissociation constants were estimated from fitted curves as: 6.1±0.4 nM (Syt-I plus gangliosides); 3.7±0.4 nM (Syt-II plus gangliosides); 10.8±1.9 nM (Syt-I); 8.1±0.9 nM (Syt-II). These data suggest that BoNT/G-HCR has similar affinity for Syt-I and Syt-II, and the presence of ganglioside can modestly enhance the apparent affinity.
Gangliosides bind BoNT/G using the same binding pocket as BoNT/A and B 10, but there are several lines of evidence that indicate that BoNT/G is a weaker binding partner for gangliosides 12. First, the ganglioside binding pocket in Hc/G appears to lack some of the interactions that are important for the interaction between GT1b and BoNT/A. The most obvious difference is that in BoNT/A, His1253 makes an important hydrogen bond to GT1b, but in BoNT/G, this interaction is missing as BoNT/G has a glycine (Gly1246) in the corresponding position11; 15. The role of His1253 in BoNT/A and the corresponding residue in BoNT/B (His1240) have previously been investigated by mutagenesis and alterations of this residue are shown to lower binding significantly. Interestingly, BoNT/B is considerably less sensitive to mutation of this residue than BoNT/A, retaining 50% of its binding to GT1b 15. The differential sensitivity to mutation at this site on GT1b-binding is consistent with the finding that BoNT/G, which has greater overall homology to BoNT/B than BoNT/A, still binds gangliosides even though it has a glycine in the corresponding position.
The second line of evidence that that BoNT/G is a weaker binding partner for gangliosides is Gln1198 in BoNT/G. Glu1203, which makes two hydrogen bonds to GT1b in BoNT/A, corresponds to Gln1198 that still can make hydrogen bonds to GT1b in BoNT/G; however, the interaction is weaker due to the absence of the negative charge of the glutamic acid.
A third line of evidence that BoNT/G is a weaker binding partner for gangliosides only applies to gangliosides that have a monosaccharide in a position corresponding to Sia5 in GT1b. In the BoNT/A GT1b complex Tyr1117 interacts with Sia5, this interaction is absent in BoNT/G were the corresponding residue is Gly1112. The backbone of this residue is located in the same position when comparing Hc/A to Hc/G, but the absence of the tyrosine side chain opens up the binding site and removes the possibility of hydrogen bonding.
Taken together, these three differences between Hc/G and Hc/A structures are likely to contribute to a lower affinity between Hc/G and GT1b than for the Hc/A – GT1b interaction. However, we also note that when GT1b is modeled into Hc/G, Arg1271 is located in a position where it could interact with the negatively charged carboxyl group of Sia6 on GT1b and could have stabilizing influence on the interaction.
The Syt binding site
The general protein architecture of the Syt binding site in BoNT/G and BoNT/B are primarily conserved, with a few key differences (Figure 4B). Of the fourteen amino acids that make up the binding pocket for Syt-II in BoNT/B, only 5 of these are conserved in BoNT/G. Mutational studies presented below, together with several other studies, show the Syt binding site to be located in similar positions in BoNT/G and BoNT/B. The binding of Syt as well as gangliosides could induce conformational changes in the binding sites of BoNT/G, however these changes have been small for the complexes of BoNT/A with ganglioside and BoNT/B with Syt-II 8; 9; 11. BoNT/G has previously been shown to bind Syt-I in a ganglioside-independent manner, while BoNT/B binds Syt-I when gangliosides are present simultaneously within the same micelle 21; 22. Our data indicate a small increase in the apparent affinity of BoNT/G for Syt-I and Syt-II in the presence of gangliosides (Supplementary Figure S1).
The residues in the hydrophobic plateau located in the center of the binding area are conserved between the two BoNT serotypes. In the Hc/G structure this hydrophobic plateau is composed of Tyr1189, Phe1202, Ala1204, Pro1205 and Phe1212, (Tyr1180, Phe1193, Ala1195, Pro1196 and Phe1203 in BoNT/B) (Figure 4). These five residues have similar conformations to their corresponding residues in Hc/B except for Phe1202 from Hc/G which has adopted an alternative conformation induced by a crystal packing contact (Figure 4B).
Overall, the Syt binding pocket in Hc/G is more structurally similar to the Syt binding pocket in Hc/B in the region where the N-terminus of the Syt-II peptide is bound, than in the region where the C-terminus of the Syt-II peptide is bound. When bound, the C-terminal part of Syt (Ile58 and Asn59 of Syt-II) is close to the membrane surface and the ganglioside binding cleft. Thr1199, Gln1200 and Leu1191 in the Hc/G structure (Figure 4) are residues in one of the loops discussed above that is structurally different in Hc/B compared to Hc/G. These residues contribute to ganglioside binding, bind a well-defined glycerol molecule, and are located in an area that could interact with Ile58 and Asn59 of Syt-II 8; 9. This area is likely to contribute to most of the differences in Syt-I binding between Hc/G and Hc/B. It is also possible that the Syt receptor binds differently to BoNT/G than to BoNT/B 10, especially in the region interacting with the C-terminal end of the Syt receptor where the two BoNTs are less structurally similar.
In the complex between Syt-II and BoNT/B, Glu1202 interacts with Lys51 from Syt-II 8; 9. The negative charge of Asp1209 in the Hc/G structure is positioned so that it can interact with Lys51 of Syt-II or the equivalent Lys43 of Syt-I. Asp1209 in BoNT/G and Glu1202 in BoNT/B are likely to fulfill the same function in the interaction. However, the negative charge from these two amino acids is coming from different positions in the two structures (Figure 4B). Thus, the surface properties of the Syt binding site have been conserved while the sequence has been drastically changed and the negative charge is being provided from different positions in the BoNT/G and the BoNT/B structures.
The binding of synaptotagmin point mutations to BoNT/G and BoNT/B
Using a pull down assay based on glutathione s-transferase (gst) fused to different wild type and mutant fragments of Syt, we find that Hc/G binds Syt-I 1-265 and 32-265, and Syt-II 1-267 and 47-267, both in the presence and absence of gangliosides under our assay conditions (Figure 5A). The Syt-II 56-267 construct lacks the key interaction area and does not bind Hc/G (Figure 5A). This mapping points to a region in Syt-II from 47-55 as being critical for binding of BoNT/G. All Syt constructs used in this study contain the transmembrane segment immediately following the key recognition areas. The transmembrane segments start at position 57 for Syt-I and 65 for Syt-II.
Figure 5. Synaptotagmin binding analysis.
(A) Truncation mutants of Syt-I and Syt-II were purified as GST-fusion proteins, immobilized on beads, and incubated with the receptor binding domain of BoNT/G (BoNT/G-HCR, 30 nM, with a 3xFLAG tag at the N-terminus), either with (+; 25 μg/ml) or without (−) gangliosides in 100 μl of TBS. Bound materials were analyzed by immunoblotting assay using an anti-FLAG tag antibody. (B) Binding assays were carried out as in panel A using Syt-II fragments containing indicated point mutations. (C) Mutants of Syt-II that were tested for BoNT/B (50 nM) binding. Bound materials were analyzed by immunoblotting assay using a polyclonal anti-BoNT/B antibody. GST = glutathione S-transferase.
Mutations at residue Phe55 in Syt-II is tolerated for the interaction between Syt-II and Hc/G, the binding is unaffected by mutation of this residue to Ala (Figure 5B). However, the Phe55Ala mutation of Syt-II has been reported to diminish binding of Syt-II to BoNT/B 8; 9, although binding can be rescued to some extent by the addition of gangliosides (Supplementary Figure S2). This indicates that the lack of the Syt-II-Phe55 interaction with BoNT/G in the Phe55Ala mutant is compensated for by other Syt-II – BoNT/G contacts. Based on the BoNT/B – Syt-II complex, we predict that Phe55 of Syt-II interacts with an area of BoNT/G with low structural conservation. The corresponding residue to Syt-II Phe55 in Syt-I is Met47, the differences in the binding site for these residues could be part of the explanation for the high affinity of Hc/G for Syt-I. The area of Hc/G interacting with this Syt residue could have the capacity to accommodate either phenylalanine (Syt-II) or methionine (Syt-I).
Similarly, the mutation of Syt-II Phe47 to alanine diminished the binding of Syt-II to BoNT/B, which can be restored by adding gangliosides (Figure 5C). This mutant did not affect the binding of BoNT/G to Syt-II (Figure B). This is because two residues in the Syt-II Phe47 binding pocket are larger in Hc/G than in Hc/B; Asn1207 and Asp1209 in Hc/G (Figure 4) correspond to Ser1198 and Ser1200 in BoNT/B, respectively, and both of these residues contribute to a more hydrophilic environment for this area of the binding pocket in Hc/G. The Syt-II Phe47 interaction with Hc/G does not appear to be critical for binding, and the absence of the Syt-II Phe47 interaction is compensated for by other interactions between Syt-II and Hc/G.
The opposite appears to be the case for the Syt-II Phe54Met mutation reported here. Hc/G loses almost all of its binding to this Syt-II mutant both in the presence and absence of gangliosides (Figure 5B), while BoNT/B is able to bind the Syt-II Phe54Met mutant in the presence of gangliosides (Figure 5C). The Syt-II Phe54Ala mutant also does not interact with Hc/G (Figure 5B) and, in contrast to the Syt-II Phe54Met mutant, the Syt-II Phe54Ala mutant does not interact with BoNT/B (Supplementary Figure S2). There are rather large differences between Hc/G and Hc/B in the Syt-II Phe54 binding area that could be the reason for these differences. They map, once again, to the loop that includes Leu1191, Thr1199 and Gln1200 in Hc/G (corresponding residues in Hc/B are Tyr1182, Glu1190 and Lys1191, Figure 4B).
Mutational studies of Syt-II have previously shown that Syt-II Glu57 of the receptor does not contribute to the interaction with BoNT/G 10. In BoNT/B Syt-II Glu57 interacts with Lys1112 and Lys1191. The Syt-II Glu57Lys mutation leads to a strong charge repulsion with BoNT/B, whereas the same Syt-II mutation does not alter the binding to BoNT/G. In the Hc/G structure, the amino acids in the positions corresponding to the BoNT/B lysines (1112 and 1191) are Phe1121 and Gln1200, and these residues do not repel the positive charge that the Syt-II Glu57Lys mutation introduces 10. Unexpectedly, the Gln1200Glu mutation that introduces a negative charge in the BoNT/G site (which would be expected to stabilize the positive charge introduced by the Syt-II Gly57Lys mutation) leads to a 70% loss of activity, while the corresponding Lys1191Glu mutation in BoNT/B leads to a near complete loss of toxicity 10. The Gln1200Lys mutation of BoNT/G changes this position to its counterpart in BoNT/B, where it is a lysine, and lowers the binding by 95% 10. This very strong negative effect, together with the positive effect of the Tyr1262Phe mutation that is very close to Gln1200 in the Hc/G structure, could be explained by differences in the Syt binding of Hc/G in this region of the binding site. The structure of the complex between Syt-I or Syt-II and BoNT/G is currently not known, but would give critical information about the nature of this interaction.
Residues Gln1198 and Gln1200 in Hc/G are very close in sequence and structure, and are located in an N-terminal part of a loop that has adopted a different orientation in the known BoNT structures of different serotypes. Gln1198 in the Hc/G structure corresponds to Glu1203 in Hc/A that coordinates with GT1b. Gln1200 of Hc/G corresponds to Lys1191 in Hc/B and mutating this residue in either serotype has a very strong effect on the binding of Syt 10. Glu1190 located between these two key residues in BoNT/B is important for Syt specificity; mutating it to a leucine in BoNT/B leads to a higher affinity for Syt-I 10.
Mutations of Syt-II Ile58 to leucine (or Leu50Ile in Syt-I) highlights the importance of this residue in the affinity and specificity of interactions with BoNT/B.8;9. The interaction area of this residue is not conserved in either structure or sequence between Hc/G and Hc/B. Therefore, it is difficult to predict the exact nature of this interaction. Ile58 of Syt-II interacts with Glu1190 of BoNT/B in the complex structure 8; 9. Ile58 is in close contact with a negatively charged residue and this interaction is negative for binding. Removal of the negative charge by Glu1190Leu mutation in BoNT/B enhances the interaction to Syt-I 10. In the BoNT/G structure, an uncharged residue, Thr1199, occupies the position of Glu1190 in BoNT/B, and may contribute to the strong interaction between Syt-I and BoNT/G. The Syt-II double mutant Ile58Leu, Asn59His turns these to positions into their Syt-I counterparts, whose binding to BoNT/B is similar to Syt-I binding 12. The reverse two Syt-I mutations (Leu50Ile, His51Asn), change these positions into the Syt-II counterparts and results in a Syt-I that binds to BoNT/B in a largely ganglioside independent-manner 12. The Ile58Leu mutation of Syt-II introduces a steric clash that is suggested to lower the interaction between Syt-I and BoNT/B 8. In the Hc/G structure, the binding pocket for Ile58 is bigger and the predicted steric clash between Leu50 of Syt-I and BoNT/B Glu1190 is absent. Instead, Thr1199 is present in the Hc/G structure.
The hydrophobic loop
One region of particular interest is a hydrophobic and solvent exposed loop region (Tyr1252, Gly1253, Tyr1254, Val1255 and Trp1256) located between the Syt binding site and the ganglioside-binding site (Figure 3). This hydrophobic loop is present in both BoNT/G and BoNT/B which have the same protein receptors, but is not present in other BoNT serotypes which utilize different protein receptors. Residues Gly1253-Val1255 of this loop could not be modeled because of the lack of electron density, further highlighting their dynamic properties. Ile1247, Val1248 and Phe1249 are the hydrophobic residues located on the tip of the somewhat shorter loop in BoNT/B. Tyrosine residues located at both ends of this loop have been suggested to be involved in membrane binding 8. This hydrophobic loop could play an important role in the binding of BoNT/G and BoNT/B to the presynaptic membrane. It is energetically costly to have extended hydrophobic residues exposed to the surrounding aqueous environment and this, together with the highly strategic position of the loop, suggests that this loop could be involved in a nonspecific association of the toxin to the membrane, increasing the probability of encountering the gangliosides and protein receptors located on the neuronal surface. The hydrophobic loop could also function in a similar manner to the gangliosides, localizing the toxin to the membrane surface in order to increase the chance of locating the scarcely dispersed protein receptors on the membrane. It is also possible that this loop plays a more critical role in BoNT/G where ganglioside binding appears to be less important and the hydrophobic loop is longer. Finally, it is also possible that this loop contributes to the correct positioning of the translocation domain of the toxin during insertion into the membrane during translocation of the toxin light chain. Further experiments are necessary to clarify the role of this loop.
Based on the complex between GT1b and BoNT/A 11, the position of the Sia5 monosaccharide of GT1b would be located in the vicinity of this hydrophobic loop. The carbonyl oxygen of Ile1245 in Hc/G is equivalent to the backbone position of Phe1252 that directly coordinates to GT1b in the Hc/A GT1b complex. This residue constitutes the start of the hydrophobic loop in BoNT/B and BoNT/G. The stretch Ile1245-Val1249 of the hydrophobic loop in BoNT/G is located in a position where it could directly interact with a bound ganglioside.
In contrast to the dual receptor model previously discussed, there are three possible membrane interaction points for BoNT/G and BoNT/B that would be located very closely together in the area that interacts with the presynaptic membrane. The transmembrane helix of the Syt receptor, the lipid part of the ganglioside, and the hydrophobic loop could all contribute to the binding of the neurotoxin. The hydrophobic loop discussed above could enhance binding of the serotype B and G neurotoxins, further adding to the total affinity previously described by the “dual receptor model” 7; 10. It is intriguing to speculate on how these three parts may work in concert together, and will require additional structural and biochemical studies in order to complete the picture of toxin recognition and binding.
Material and Methods
Protein expression and purification
Amino acid 860-1297 of BoNT/G was expressed from a pET28a (Novagen) expression vector 30. The protein was expressed in Escherichia coli (BL21 ai) grown in LB with 50 μg/ml kanamycin and incubated at 37°C until the OD600 nm = 0.8. The temperature was lowered to 18°C and expression was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 0.2% arabinose. After incubation overnight, 23g of cells were harvested, resuspended in 35 ml of 500 mM NaCl, 50 mM Tris pH 8, 5 mM Imidazole, 0.7 mg/ml lysozyme, 0.7 mM TCEP and disrupted by sonication. The sample was centrifuged at 40 k for 15 min, passed through a 0.22 μm filer and loaded on a 1 ml HiTrap Chelating HP column charged with NiSO4 (GE healthcare). The column was washed with 5 ml of 500 mM NaCl, 50 mM Tris pH 8 and 5 mM Imidazole, then 30 ml of the same buffer with 28 mM Imidazole; the elution buffer contained 500 mM Imidazole and 0.5 mM TCEP. The eluted sample was used in size-exclusion chromatography using a Sephacryl S-200 HR column (GE Healthcare) pre-equilibrated with 10% Glycerol, 300 mM NaCl, 20 mM Tris pH 8 and 0.5 mM TCEP. The highly pure protein was concentrated to 10.4 mg/ml using a 10 kDa cut off Amicon Ultra. The protein concentration was determined by OD280 using a calculated extinction coefficient of 104 830 M−1 cm−1.
Crystallization and structure determination
Spherulites from condition 13 in the JCSG+ screen (QIAGEN) was optimized to yield the final crystallization condition. 200 nl of protein solution was mixed with 100 nl of well solution in a sitting drop crystallization plate. The well solution consisted of 1.2 M ammonium sulphate, 85 mM Citrate pH 4.0 and 8% glycerol. Small crystals with the dimensions of 10×10×100 μm grew from heavy precipitate after several weeks. The crystals were flash frozen in liquid nitrogen and the diffraction data was collected at the Advanced Photon Source on beamline GM/CA CAT BL23 ID-B. The data were processed using XDS and scaled using XSCALE 31. Molecular replacement was performed using MOLREP 32 with the search model 2NP0. The structure was built using Arp/warp 33 and Coot 34; refinement was carried out with Refmac5 35 using TLS 36. Data processing and refinement statistics are presented in Table 1. Coordinates for the BoNT/G binding domain structure presented here have been deposited in the PDB with the accession code 2VXR. ESPript 37 and SSM 38 were used to generate the structure-based alignment in Figure 2. A Fo-Fc omit map of residues Gln1200 – Pro1205 in the Syt binding site is available as Supplementary Figure S3.
Table 1.
Data processing and refinement statistics
| BoNT/G binding domain | |
|---|---|
| Space group | P212121 |
| Unit cell (a,b,c in Å) | 41.5, 93.4, 131.4 |
| Resolution (Å) | 20-1.9 (2.0-1.9) |
| Rsym (%) | 10.4 (44.5) |
| <I/σI> | 11.66 (3.1) |
| Completeness (%) | 98.6 (99.8) |
| Redundancy | 4.2 (4.2) |
| Rcryst (%) | 15.9 |
| Rfree (%) | 19.6 |
| r.m.s. deviation bond length (Å) | 0.014 |
| r.m.s. deviation bond angle (°) | 1.4 |
| Average B factors (Å2) | |
| Protein | 13.4 |
| Solvent | 30.8 |
| Glycerols | 31.6 |
| Sulphates | 30.0 |
| Ramachandran plot | |
| Most favored (%) | 86.6 |
| Additional allowed (%) | 13.4 |
| Generously allowed (%) | 0.0 |
| Disallowed (%) | 0.0 |
Co-purification assays
Constructs of rat Syt-I or mouse Syt-II fragments harboring point-mutations described in Figure 5 were generated by the overlapping PCR method and subcloned into pGEX-2T. These constructs were expressed and purified as GST fusion proteins and immobilized on glutathione-Sepharose beads. Triton X-100 (1%) was used to solubilize the recombinant Syt proteins. BoNT/G-HCR (residues 863-1297 of BoNT/G) recombinant protein was generously provided by J. Barbieri (Milwaukee, WI). This fragment contains a His6 tag and 3xFLAG tag at the N-terminus. Vector construction and protein purification were described previously 39.
Briefly, 8 μg of immobilized protein was mixed with the BoNT/G-HCR (30 nM) or BoNT/B (50 nM), either with (+; 25 μg/ml) or without (−) gangliosides (mixed bovine brain gangliosides, Matreya LLC) in 100 μl of Tris buffered saline (TBS; 20 mM Tris, 150 mM NaCl, pH 7.4) plus 0.5% Triton X-100 for 1 hour at 4°C. Beads were washed three times, bound proteins were subjected to SDS-PAGE and immunoblot analysis using an anti-FLAG tag antibody (M2, Sigma Aldrich, St. Louis, MO), or a polyclonal anti-BoNT/B antibody described previously 22.
Supplementary Material
Acknowledgments
This work was supported by the Pacific Southwest Research Center for Excellence (PSWRCE; NIH/NIAID award U54 AI065359 to R.C.S.) and a grant from the NIH (NIAID R01 AI057744) to E.R.C. E.R.C. acknowledges membership and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases (GLRCE; NIH/NIAID award U54 AI057153). E.R.C. is an Investigator of the Howard Hughes Medical Institute. P.S. was supported by a fellowship from the Wenner-Gren Foundations and with funds from the Center for Biomembrane Research and the Swedish Foundation for Strategic Research during the preparation of this manuscript. We thank Angela Walker for assistance with manuscript preparation. The GM/CA CAT beamline (23 ID-B) is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104).
Abbreviations
- BoNT
botulinum neurotoxin
- gst
glutathione s-transferase
- Hc
C-terminal binding domain
- Hcc
C-terminal subdomain
- Hcn
N-terminal subdomain
- Hn
N-terminal translocation domain
- LC
light chain
- RMSD
root mean square deviation
- SNARE
soluble N-ethylmaleimide-sensitive-factor attachment protein receptor
- Syt
Synaptotagmin
- TBS
Tris buffered saline
Footnotes
The numbering of BoNT/B residues differ in varying reports on BoNT/B structure, for clarity, we have used the numbering from the structure 2NP08 in our discussion; there is a −1 difference to the other reports 9,10.
Accession Number
Coordinates for the BoNT/G binding domain structure presented here have been deposited in the PDB with the accession code 2VXR.
References
- 1.Willis B, Eubanks LM, Dickerson TJ, Janda KD. The Strange Case of the Botulinum Neurotoxin: Using Chemistry and Biology to Modulate the Most Deadly Poison. Angew Chem Int Ed Engl. 2008;47:8360–8379. doi: 10.1002/anie.200705531. [DOI] [PubMed] [Google Scholar]
- 2.Breidenbach MA, Brunger AT. New insights into clostridial neurotoxin-SNARE interactions. Trends Mol Med. 2005;11:377–381. doi: 10.1016/j.molmed.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Turton K, Chaddock JA, Acharya KR. Botulinum and tetanus neurotoxins: structure, function and therapeutic utility. Trends Biochem Sci. 2002;27:552–558. doi: 10.1016/s0968-0004(02)02177-1. [DOI] [PubMed] [Google Scholar]
- 4.Bhidayasiri R, Truong DD. Expanding use of botulinum toxin. J Neurol Sci. 2005;235:1–9. doi: 10.1016/j.jns.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Shukla HD, Sharma SK. Clostridium botulinum: A bug with beauty and weapon. Crit Rev Microbiol. 2005;31:11–18. doi: 10.1080/10408410590912952. [DOI] [PubMed] [Google Scholar]
- 6.Arndt JW, Yu W, Bi F, Stevens RC. Crystal structure of botulinum neurotoxin type G light chain: serotype divergence in substrate recognition. Biochemistry. 2005;44:9574–80. doi: 10.1021/bi0505924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montecucco C. How Do Tetanus and Botulinum Toxins Bind to Neuronal Membranes. Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- 8.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 9.Jin RS, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 10.Rummel A, Eichner T, Weil T, Karnath T, Gutcaits A, Mahrhold S, Sandhoff K, Proia RL, Acharya KR, Bigalke H, Binz T. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc Natl Acad Sci U S A. 2007;104:359–64. doi: 10.1073/pnas.0609713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong M, Tepp WH, Liu H, Johnson EA, Chapman ER. Mechanism of botulinum neurotoxin B and G entry into hippocampal neurons. J Cell Biol. 2007;179:1511–22. doi: 10.1083/jcb.200707184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunger AT, Jin R, Breidenbach MA. Highly specific interactions between botulinum neurotoxins and synaptic vesicle proteins. Cell Mol Life Sci. 2008;65:2296–306. doi: 10.1007/s00018-008-8088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiavo G, Malizio C, Trimble WS, Polverino de Laureto P, Milan G, Sugiyama H, Johnson EA, Montecucco C. Botulinum G neurotoxin cleaves VAMP/synaptobrevin at a single Ala-Ala peptide bond. J Biol Chem. 1994;269:20213–6. [PubMed] [Google Scholar]
- 15.Rummel A, Mahrhold S, Bigalke H, Binz T. The H-CC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol Microbiol. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 16.Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–54. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- 17.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 18.Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–4. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 19.Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B Mediate the Entry of Botulinum Neurotoxin E into Neurons. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiki T, Kamata Y, Nemoto Y, Omori A, Ito T, Takahashi M, Kozaki S. Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. J Biol Chem. 1994;269:10498–503. [PubMed] [Google Scholar]
- 21.Rummel A, Karnath T, Henke T, Bigalke H, Binz T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J Biol Chem. 2004;279:30865–70. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- 22.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Z, Chen C, Barbieri JT, Kim JJ, Baldwin MR. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry. 2009;48:5631–41. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neuro-toxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 25.Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol. 2000;7:693–9. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- 26.Kumaran D, Eswaramoorthy S, Furey W, Navaza J, Sax M, Swaminathan S. Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation. J Mol Biol. 2009;386:233–45. doi: 10.1016/j.jmb.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 27.Umland TC, Wingert LM, Swaminathan S, Furey WF, Schmidt JJ, Sax M. Structure of the receptor binding fragment HC of tetanus neurotoxin. Nat Struct Biol. 1997;4:788–92. doi: 10.1038/nsb1097-788. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman S, Eswaramoorthy S, Ahmed SA, Smith LA, Swaminathan S. N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B. Biochem Biophys Res Commun. 2005;330:97–103. doi: 10.1016/j.bbrc.2005.02.123. [DOI] [PubMed] [Google Scholar]
- 29.Nishiki T, Tokuyama Y, Kamata Y, Nemoto Y, Yoshida A, Sato K, Sekiguchi M, Takahashi M, Kozaki S. The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 1996;378:253–7. doi: 10.1016/0014-5793(95)01471-3. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin MR, Tepp WH, Przedpelski A, Pier CL, Bradshaw M, Johnson EA, Barbieri JT. Subunit vaccine against the seven serotypes of botulism. Infect Immun. 2008;76:1314–8. doi: 10.1128/IAI.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 32.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 33.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D-Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D-Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D-Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 37.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D-Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin MR, Barbieri JT. Association of botulinum neurotoxin serotypes A and B with synaptic vesicle protein complexes. Biochemistry. 2007;46:3200–10. doi: 10.1021/bi602396x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.