Abstract
In neurons, SNAREs, synaptotagmin, and other factors catalyze Ca2+-triggered fusion of vesicles with the plasma membrane. The molecular mechanism of this process remains an enigma, especially regarding the interaction between synaptotagmin and SNAREs. Here we characterized this interaction by single-molecule fluorescence microscopy and crystallography. The two rigid Ca2+-binding domains of synaptotagmin 3 undergo large relative motions in solution. Interaction with SNARE complex amplifies a particular state of the two domains that is further enhanced by Ca2+. This state is represented by the first SNARE-induced Ca2+-bound crystal structure of a synaptotagmin fragment containing both domains. The arrangement of the Ca2+-binding loops of this structure of synaptotagmin 3 matches that of SNARE-bound synaptotagmin 1, suggesting a conserved feature of synaptotagmins. The loops resemble the membrane-interacting loops of certain viral fusion proteins in the postfusion state, suggesting unexpected similarities between both fusion systems.
Keywords: neurotransmitter release, synaptic vesicle, SNARE-induced fusion, virus-induced fusion, Ca2+ sensor, single molecule fluorescence microscopy
Regulated fusion of biological membranes is triggered by signals such as an increase in Ca2+ concentration in the case of neurotransmitter release or by a pH change in the case of enveloped virus fusion1,2. In neurons, SNARE (SNAP receptor, where SNAP is defined as soluble NSF attachment protein and NSF as N-ethylmaleimide-sensitive factor) complexes consisting of syntaxin, SNAP-25, and synaptobrevin, in conjunction with other factors, catalyze Ca2+-triggered fusion of synaptic vesicles within 1 msec after Ca2+ influx1,3. Zippering of the SNARE complex brings membranes together and provides energy for fusion of membrane compartments in eukaryotic cells1. In neurons, this process is tightly regulated by Ca2+ concentration, yet SNARE complex assembly does not exhibit any Ca2+ sensitivity. Thus, other factors are responsible for Ca2+ sensing, coordination of SNARE complex assembly, and triggering of membrane fusion. In particular, synaptotagmin 1 (Syt1) is the primary Ca2+ sensor for fast synchronous neurotransmitter release4. Synaptotagmins are comprised of a short intralumenal/extracellular sequence, a single transmembrane helix, linker, and two Ca2+-binding domains, termed C2A and C2B. Interactions of Syt1 with both lipid membrane and the SNARE complex are physiologically important4,5, yet the molecular mechanism by which they link Ca2+ influx to synaptic vesicle fusion is unknown. In vitro, SNAREs alone promote slow, Ca2+-independent, lipid mixing/fusion6 while synaptogmins alone, in the presence of Ca2+, bind anionic lipids7-9, modify membrane curvature10, and promote membrane juxtaposition without fusion11. When combined, SNARE proteins and synaptotagmin enhance lipid mixing12,13, but the large physiological sensitivity of the system to Ca2+ concentration has not yet been reproduced in vitro.
The synaptotagmin family has at least 16 isoforms with different regional and cellular expression in the brain14 and throughout the body15. Except for Syt1, the functions of most other isoforms have not yet been identified. Some of them are also likely Ca2+ sensors for SNARE-dependent fusion processes based on their phospholipid- and SNARE-binding properties10,13,15. Among these putative Ca2+-sensors are synaptotagmins 3,5,6, and 10, since cultured olfactory neurons lacking them exhibit decreased inhibitory postsynaptic current amplitudes (P. Cao, T. C. Sudhof, personal communication). As a particular case in point, synaptotagmin 3 (Syt3) exhibits biochemical properties similar to Syt1, yet Syt3 is not involved in fast neurotransmitter release16. Syt1 and Syt3 both bind SNARE complex17 (Supplementary Fig. 1), syntaxin–SNAP-25 complex13, anionic lipids7,13 (Supplementary Fig. 2a), modify membrane curvature10, and enhance SNARE-catalyzed lipid mixing13 in a Ca2+-dependent manner. Our results presented here and elsewhere18 now show further remarkable similarities.
Several structures of individual synaptotagmin C2 domains revealed that Ca2+ has little effect on the individual C2 domains, suggesting a mechanistic role of the relative domain orientation. However, the two available structures containing fragments with both C2 domains (termed C2AB)19,20 were solved in the absence of Ca2+, thus providing only limited information. Elucidation of the Ca2+-triggering mechanism requires a spatial and dynamic understanding of synaptotagmin conformations in isolation, upon Ca2+, and SNARE complex binding.
Here we characterized the interaction between SNARE complex and Syt3 by a combination of single-molecule fluorescence microscopy and X-ray crystallography. The two rigid Ca2+-binding domains, C2A and C2B, of Syt3 undergo large relative motions in solution. The interaction with SNARE complex greatly enhances a particular conformation of Syt3. This conformation matches that of the first SNARE-induced Ca2+-bound structure of a synaptotagmin containing both C2 domains that we obtained by X-ray crystallography. The induction of this Ca2+-bound conformation of Syt3 by the SNARE complex already occurs independent of Ca2+, but it is enhanced in the presence of Ca2+. We found similar effects also for Syt118, suggesting a conserved molecular mechanism among synaptotagmins. Remarkably, the spatial arrangement of the Ca2+-binding loops in the Ca2+-bound conformation of Syt3 resembles that of membrane-interacting loops of certain viral fusion proteins in their postfusion state21,22. Thus, both fusion systems have two similar membrane interacting elements: transmembrane helical bundles and membrane interacting loops. Based on the similarity to viral fusion systems and the conservation of the SNARE-induced changes in synaptotagmin dynamics and conformation, we propose a general model for the SNARE–synaptotagmin fusion “machine”.
RESULTS
Structure of the SNARE-induced Ca2+-bound synaptotagmin 3
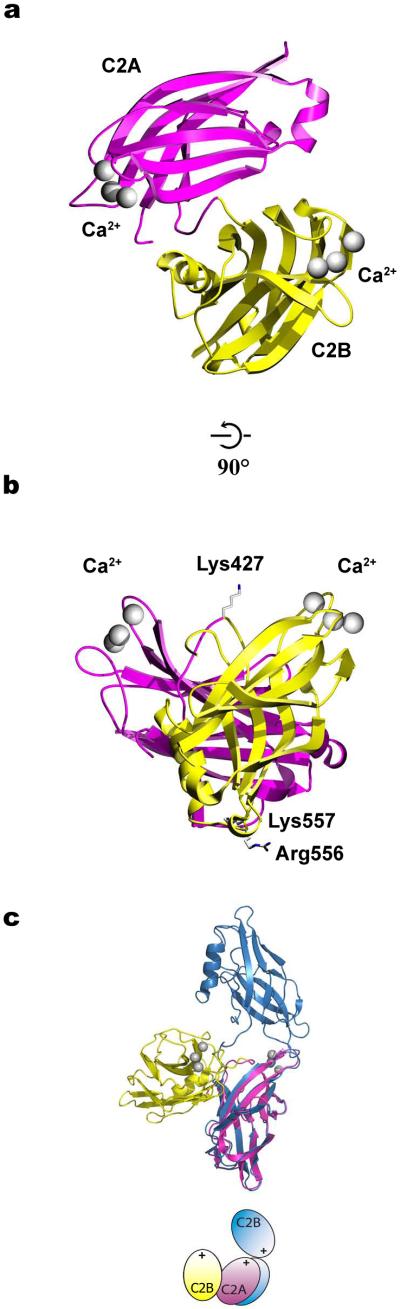
We determined the first crystal structure of a SNARE-induced Ca2+-bound conformation of any synaptotagmin containing both Ca2+-binding C2 domains (C2AB of Syt3, Fig. 1a and Table 1). Crystallization of this conformation required the presence of SNARE complex. Although there is no well-defined electron density for SNAREs in the crystal structure, the presence of the SNARE complex may have catalyzed this particular conformation, a notion that is supported by single molecule experiments discussed below. The structure reveals a previously unseen arrangement of the C2A and C2B domains. The short linker between the C2A and C2B domains adopts a hairpin conformation allowing the two domains to position the Ca2+-binding loops on the same face of the molecule (Fig. 1a,b and Supplementary Fig. 3a). The Ca2+-binding loops are both surface exposed, protrude from the two C2 domains, and can therefore insert simultaneously into the same membrane. The linker between the C2 domains is folded such that a positively charged lysine residue (Fig. 1b) could also interact with anionic lipid headgroups of the same membrane as the Ca2+-binding loops. In the C2A domain, three Ca2+ ions are bound to the Ca1, Ca2, and Ca3 sites, while the Ca4 site is unoccupied (Supplementary Fig. 3c,d and Supplementary Methods). The Syt3 C2B domain contains two Ca2+ ions in the Ca1 and Ca2 positions as well as a third Ca2+ ion at lower occupancy in the Ca4 position (Supplementary Fig. 3c,d and Supplementary Methods). Thus, depending on Ca2+ concentration or the presence of coordinating lipids, Syt3 can bind 5 or 6 Ca2+ ions. The positively charged patch identified in Syt1 (Arg398 and Arg399)11 (equivalent residues in Syt3: Arg556 and Lys557) is surface exposed and available for other interactions (Fig. 1b).
Figure 1.
Crystal structure of SNARE-induced Ca2+-bound Syt3. (a) Showing a ribbon diagram of the C2AB fragment of Syt3 including the C2A (magenta) and C2B (yellow) domains, and bound Ca2+ ions (gray spheres). (b) Side view illustrating that the Ca2+-binding loops of both C2 domains emerge from the same side of the molecule. Residues Lys427, Lys557, and Arg556 are shown as sticks. (c) Superposition of the crystal structures of SNARE-induced Ca2−-bound (magenta and yellow) and Ca2+-free (blue, PDB ID 1DQV20) Syt3 C2AB fragments. We aligned structures by superposition of the Cα positions of their respective C2A domains (see cartoon where the “+” sign indicates the Ca2+-binding regions).
Table 1.
Data collection and refinement statistics (molecular replacement)
| Syt3 C2AB | |
|---|---|
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 205.7, 205.7, 143.1 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50-3.5 (3.63-3.5)* |
| R merge | 16.7 (57.1) |
| I / σI | 10.4 (3.5) |
| Completeness (%) | 99.7 (100.0) |
| Redundancy | 5.8 (5.9) |
| Refinement | |
| Resolution (Å) | 50-3.5 |
| No. reflections | 39136 |
| Rwork / Rfree | 22.9 / 25.5 |
| No. atoms | |
| Protein | 6651 |
| Ligand/ion | 21 |
| Water | 0 |
| B-factors | |
| Protein | 84 |
| Ligand/ion | 67.3 |
| Water | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.269 |
One crystal was used for the structure of Syt3 C2AB.
Values in parentheses are for the highest-resolution shell.
The interface between the C2 domains of the SNARE-induced Ca2+-bound structure of Syt3 is extensive. The size of the buried surface area at the C2A–C2B interface (775 Å2 per monomer) is similar to certain antibody–antigen complexes or to other protein complexes of similar size (Supplementary Table 1). The interface also has a shape complementarity and number of hydrogen bonds similar to other protein complexes. In addition, the asymmetric unit contains three independent molecules of Syt3 C2AB domains, with different crystal packing contacts, yet they all adopt the same conformation. The three independent C2AB molecules form relatively small crystal contacts with themselves and other molecules in the crystal, producing large solvent channels with an unusually high solvent content of 84% (Supplementary Fig. 4). Taken together, the observed conformation of the C2 domains of SNARE-induced Ca2+-bound Syt3 is unlikely affected by crystal packing.
Superposition of the SNARE-induced Ca2+-bound structure of Syt3 with the previously determined Ca2+-free structure20 reveals large rigid body displacements of the C2A and C2B domains and a consequent change in the position of the Ca2+-binding loops (Fig. 1c). Relative to the Ca2+-free conformation, the SNARE-induced Ca2+-bound conformation has an interface between C2 domains that is more than twice as large (Supplementary Table 1). Interestingly, the highly conserved WHXL motif, proposed to regulate synaptotagmin endocytosis23, is buried at this interface, while it is exposed in the Ca2+-free Syt3 structure.
Similarity to viral fusion proteins
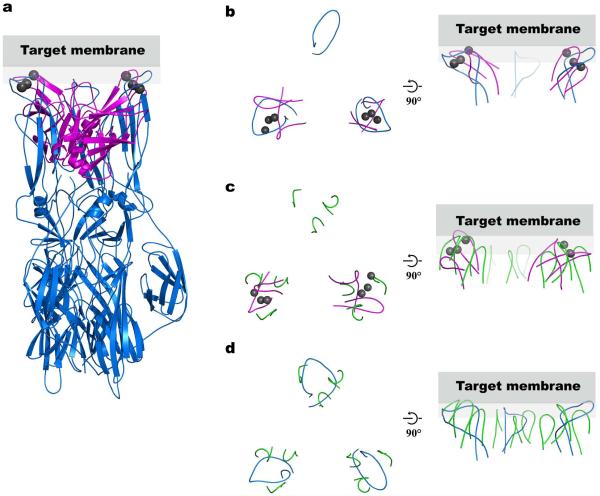
We noticed a striking analogy in position and structural arrangement of the Ca2+-binding loops in the SNARE-induced Ca2+-bound Syt3 structure and fusion loops of the glycoprotein E1 of Semliki forest virus in their low pH/post-fusion state21 (Fig. 2a,b). The separation between Ca2+-binding loops in the SNARE-induced Ca2+-bound Syt3 structure (~35–40 Å) is similar to that of two of the three viral fusion loops. A similar separation is also observed for the membrane interacting loops of the VP5 domain of spike protein VP4 of the non-enveloped rotavirus22 (Fig. 2c). Structural similarities of the fusion loops from the Semliki forest virus glycoprotein E1 and membrane-interacting loops of rotavirus VP5 domain in their activated states are shown in Fig. 2d. Analogous to the Ca2+-binding loops of synaptotagmins, viral fusion loops are conserved within their respective families, and mutations in the loops shift the threshold of the fusion trigger24 or affect membrane permeability25. Viral fusion loops are thought to act in concert with transmembrane helices to trigger fusion upon activation (enveloped virus, low pH; rotavirus, trypsin cleavage; see Fig. 5 of ref. 21). Based on this analogy we hypothesize that the Ca2+–SNARE–synaptotagmin complex couples the action of the SNARE transmembrane helices and the synaptotagmin Ca2+-binding loops to catalyze Ca2+-triggered exocytosis.
Figure 2.
Comparison of the structural arrangement of the Syt3 Ca2+-binding loops with Semliki Forest virus E1 fusion loops and Rhesus Rotavirus VP5 membrane-interacting loops. (a) We superimposed two of the three E1 loops of the Semliki Forest virus E1 protein (blue, PDB ID 1RER21) on the Ca2+-binding loops of Syt3 (magenta, Ca2+ ions are in gray). (b–d) A top view looking from the target membrane onto the proteins is shown in the left panels, and a side view in the right panels. Gray rectangles represent the target membrane. Only the fusion membrane-interacting loops are shown in all panels for clarity. (b) Top view of panel (a) showing only the superimposed loops. (c) We superimposed two of the three membrane-interacting loop regions of the Rhesus Rotavirus VP5 protein trimer (green, PDB ID 1SLQ22) on the Ca2+-binding loops of Syt3 (magenta, Ca2+ ions are in gray). (d) Superposition of the Semliki Forest virus E1 protein (blue) and the membrane-interacting loops of Rhesus Rotavirus VP5 protein (green) using all three loop regions. Both E1 of Semliki Forest virus and VP5 of Rhesus Rotavirus are trimers in their post-fusion state. In addition to structural similarities of the Semliki Forest virus fusion loops and the membrane interacting loops of VP5 Rhesus Rotavirus, these loops also share sequence homology22. The fusion/cell entry mechanism of non-enveloped rotavirus is not fully understood, however it is able to mediate cell-cell fusion in vitro47.
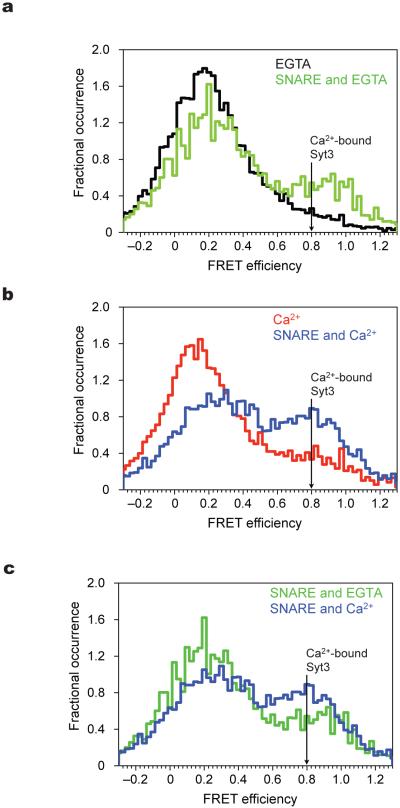
Figure 5.
Syt3 C2AB conformational dynamics in the presence of SNARE complex. Experimental conditions as in Fig. 3. (a) smFRET efficiency histograms of Syt3 in the absence of Ca2+ and absence (black) and presence (green) of SNARE complex. (b) smFRET efficiency histograms of Syt3 in the presence of Ca2+ and absence (red) and presence (blue) of SNARE complex Ca2+. Position of FRET efficiency expected for SNARE induced Ca2+-bound Syt3 crystal structure is shown (black arrow). The distance between ‘ticks’ on the X-axis corresponds to histograms’ bin width. (c) For better comparison, shown is an overlay of the same smFRET efficiency histograms from panels a and b in the presence of SNARE complex and presence (blue) or absence (green) of Ca2+. In order to quantify the increase in the high FRET population in panels a–c, we have fitted the FRET efficiency histograms to the sum of two Gaussian distributions. This fitting method is based on the observation that a smFRET distribution arising from a single state can be approximated by a Gaussian distribution. We chose the minimum number of states (two) that are required to fit the observed smFRET distributions. In the absence of Ca2+ and SNARE complex (panel (a), black line), the area of the high FRET population is 12%. The increases over this background of the high FRET state are: 10% for Ca2+ only (panel (b), red line), 14% for SNARE complex only (panel (a), green line), and 36% for SNARE complex plus Ca2+ (panel (b), blue line).
Conformational dynamics of Syt3
Syt3–SNARE complex interactions are affected by Ca2+ and ionic strength (Supplementary Fig. 1) in a manner analogous to that of Syt1–SNARE interactions17. To test the hypothesis that coupling between SNARE complex and synaptotagmin goes beyond a simple rigid body interaction, we wanted to probe if SNARE complex binding has an effect on synaptotagmin dynamics. Conventional ensemble methods are unsuited for this purpose since synaptotagmin exhibits significant conformational variability and observed averages could mask important effects. By contrast, recently developed single molecule methods enable study of individual complexes, bypassing the need for a homogenous population of identical complexes26. We measured single-molecule fluorescence resonance energy transfer (smFRET) to characterize the conformation of doubly-fluorophore labeled Syt3 C2 domains in solution, in the presence of Ca2+, and/or SNARE core complex. The SNARE core complex represents the cis (post-fusion) state, which is being used for all biophysical and biochemical studies to date. Substantial technological advances will be needed to study interactions with the trans (pre-fusion) state. Still, interactions with the cis state are likely related to interactions with the trans state, possibly with somewhat different affinities and kinetics.
Individual, doubly-fluorophore labeled Syt3 molecules were tethered to a surface (Fig. 3a and Supplementary Fig. 5), and their smFRET efficiencies were measured with and without Ca2+ (Fig. 4). The corresponding smFRET efficiency distributions are very similar and exhibit a broad peak with the maximum at 0.13–0.17 and a tail towards higher FRET states (Fig. 3b). The broad character of the distributions suggests multiple conformations, most of which are different from those observed in the available crystal structures of Syt3 (see arrows in Fig. 3b). In order to estimate the range of conformations consistent with the observed smFRET efficiency distribution, we performed a reduced-variable molecular dynamics simulation of the isolated C2A–C2B fragment. The calculated FRET efficiency distribution is also broad with a pronounced maximum at 0.1 (Fig. 3c). Considering that the calculated distribution is noise-free, the two distributions share the properties of a single major maximum at low FRET efficiency and a long tail towards higher FRET efficiency states. However, the lower value of the FRET efficiency maximum and the presence of a small high FRET peak in the simulation suggest that, in solution, Syt3 adopts fewer “extended” and “compact” conformations (cf. Figs. 1c and 3a). We note that, due to the time resolution of the smFRET experiment (100 msec), the observed smFRET efficiency states of the individual molecules could themselves represent fast motional averages. Most of the observed states are stable on the second timescale (Fig. 4), while some molecules exhibit very dynamic behavior (Fig. 4b, right column). Thus, the Syt3 C2 domains undergo large fluctuations in solution with a complex dynamic behavior. Uncomplexed Syt1 exhibits a very similar conformational dynamics18,27.
Figure 3.
Uncomplexed Syt3 conformational dynamics in solution. (a) Bottom: Schematic of the smFRET imaging set-up. Doubly-labeled Syt3 molecules are coupled to the surface via streptavidin and biotinylated bovine serum albumin (biotin-BSA). Syt3 is shown in the Ca2+-free (left) and SNARE-induced Ca2+-bound (right) conformations (green and red spheres represent Ca2+ ions). Red and green spheres indicate the calculated average positions of the fluorophore centers linked to residues Q410C and N554C. Top: examples of low and high smFRET states of Syt3 corresponding to the Ca2+-free crystal structure (left, FRET ~ 0.1) and the SNARE-induced Ca2+-bound structure (right, FRET ~ 0.8); fluorescence intensity (arbitrary units) traces are shown as a function of time for single Syt3 molecules (donor in green, acceptor in red). The laser illumination sequence is indicated on top of the graphs as a colored bar (red for 635 nm and green for 532 nm illumination). (b) smFRET efficiency histograms of Syt3 without (black) and with Ca2+ (red). The arrows indicate the FRET efficiencies values calculated from the Ca2+-free and SNARE induced Ca2+-bound Syt3 crystal structures. All experiments were performed in the presence of 100 μM Ca2+ or 100 μM EGTA in 50 mM KCl, 100 mM Hepes-Na, pH 7.2 and oxygen scavengers, as described in the Supplementary Methods. (c) Predicted FRET efficiency histogram of isolated Syt3 molecules calculated from a molecular dynamics simulation.
Figure 4.
Syt3 C2AB exhibits a variety of FRET states and dynamic behaviors. Representative single molecule FRET (smFRET) traces (intensity in arbitrary units) as a function of time for Syt3 (donor is shown in green and acceptor in red). The laser illumination sequence is indicated on top of each graph as a colored bar (red for 635 nm and green for 532 nm illumination). Shown are examples of (a) low static FRET, (b) highly dynamic FRET, and (c) high static FRET states. We calculated corresponding FRET efficiency values (shown in separate panels as black lines) as described in the Supplementary Methods; after start of illumination, photobleaching of acceptor or donor dyes occurs at various times as indicated by the subsequent absence of a calculated FRET efficiency value.
SNARE complex interaction changes Syt3 dynamics
Interaction with SNARE complex dramatically affects the Syt3 smFRET distribution by inducing a high FRET population in both the absence and presence of Ca2+, with the effect being more pronounced with Ca2+ (Fig. 5). The observed effect of adding Ca2+ to Syt3 in the presence of SNARE complex (Fig. 5c) is larger than adding Ca2+ to Syt3 only (Fig. 3b) illustrating that the effect of SNARE complex and Ca2+ on Syt3 is not simply additive, but rather that the presence of SNARE complex enhances the influence of Ca2+ on Syt3. At higher ionic strength the effects are similar (Supplementary Fig. 6). The effect is SNARE specific since interactions with anionic lipid containing liposomes, while inducing a slight shift towards higher FRET states in the presence of Ca2+, do not induce the high FRET states induced by the SNARE complex (Supplementary Fig. 5c, left panel). The ratio of low and high smFRET populations in the presence of SNARE complex remains relatively constant even at high SNARE to Syt3 molar ratios (Supplementary Fig. 7). This bimodal character of the smFRET efficiency distribution can be explained by the presence of SNARE interactions with two distinct Syt3 conformations, however a more complex kinetic scheme is also possible.
To characterize the interaction between Syt3 and minimal SNARE complex with another method, we performed biosensor experiments with Syt3 bound to the sensor (Supplementary Fig. 8). The sensorgrams show that the SNARE complex binds to Syt3 both in the presence and absence of Ca2+. The binding appears heterogeneous and suggests the presence of at least two binding modes. While the apparent KD values are similar in the presence and absence of Ca2+ (Supplementary Fig. 8b,d), the kinetics of the interaction is dramatically different (Supplementary Fig. 8a,c) with Syt3 binding more SNARE complex in the presence of Ca2+. The biosensor data are consistent with the observed smFRET distributions (Fig. 5), suggesting a complex dynamic system that does not conform to a simple kinetic binding model. At higher ionic strength, the affinities of the two binding modes in the presence of Ca2+ decrease (Supplementary Fig. 8e,f), again consistent with the pull-down (Supplementary Fig. 1c) and the single-molecule smFRET data (Supplementary Fig. 6).
Determination of conformations with high FRET state
To determine what conformations are consistent with the experimentally observed high FRET state at 0.8, we carried out an exhaustive molecular dynamics simulation with pseudo-atoms at positions 410 and 554 constrained to the observed FRET-derived distance of 44.1 Å (Supplementary Fig. 9 and Online Methods). While there is some variation among the sampled conformations, they all have similar features as the SNARE-induced Ca2+-bound crystal structure of Syt3. The relative orientation of C2A and C2B is similar, the Ca2+-binding loops are on the same side of the molecule, and the linker samples conformations that would allow it to reach the membrane surface when the Ca2+-binding loops bind to the membrane. We conclude that the conformation of Syt3 that is observed in the SNARE-induced Ca2+-bound crystal structure (Fig. 1a) is a good representation of the high-FRET efficiency state that is induced by the SNARE–Syt3 interaction in solution.
DISCUSSION
Members of the synaptotagmin family are the Ca2+ sensor for fast synchronous neurotransmitter release 4. Together with SNAREs and other auxiliary proteins, synaptotagmins trigger fusion of synaptic vesicles with the active zone in the presynaptic terminal upon Ca2+ influx. Despite a large body of physiologic, genetic, biochemical, and structural data, the molecular mechanism of this highly regulated fusion machinery is still unknown. In part this is due to the dynamic nature of the protein–protein (Figs. 3b, 4 and 5) and protein–lipid interactions involved, and the sparseness of the available structural information about these interactions. This work provides fundamental new insights into the dynamics of synaptotagmin and how its conformations are influenced by interaction with SNARE complex.
Synergism between synaptotagmins and SNARE complex
The interaction of synaptotagmins and SNARE complexes is synergistic. Syt1 stabilizes the binary (syntaxin–SNAP-25) complex by elimination of partially assembled states of the binary complex, setting the stage for efficient formation of the trans-SNARE complex28. Conversely, interaction of Syt3 with the SNARE helix bundle dramatically affects the dynamics of Syt3 by amplifying the high FRET conformation of Syt3 (Fig. 5) that is similar to viral fusion loops (Fig. 2). Ca2+ binding to Syt3 further stabilizes the interaction with the SNARE complex (Supplementary Fig. 1c, Supplementary Fig. 8), and the SNARE’s effect on Syt3 dynamics (Fig. 5b).
Conservation of synaptotagmin function
Syt3 C2AB domains can adopt various conformations relative to one another, where none of them are particularly stable (Figs. 3b and 4). We have observed very similar behavior for Syt1 and the interaction with SNARE complex enhances a particular C2AB conformation for both Syt3 (Figs. 5 and Supplementary Fig. 9) and Syt118. Although the exact orientation of the β-sheets of the C2AB domains are somewhat different between SNARE-bound Syt3 and Syt1, the spatial arrangement of their Ca2+-binding loops is similar to each other (Supplementary Fig. 10) and to fusion/membrane interacting loops of viral fusion proteins (Fig. 2). The structural similarities between Syt1 and Syt3 and their interactions with the SNARE complex presented here, together with the published characterization of multiple Ca2+-responsive synaptotagmin isoforms13, suggest a conserved mechanism by which SNARE interactions with Ca2+-responsive synaptotagmins catalyze Ca2+-triggered vesicle fusion.
A general model of the SNARE–synaptotagmin fusion machine
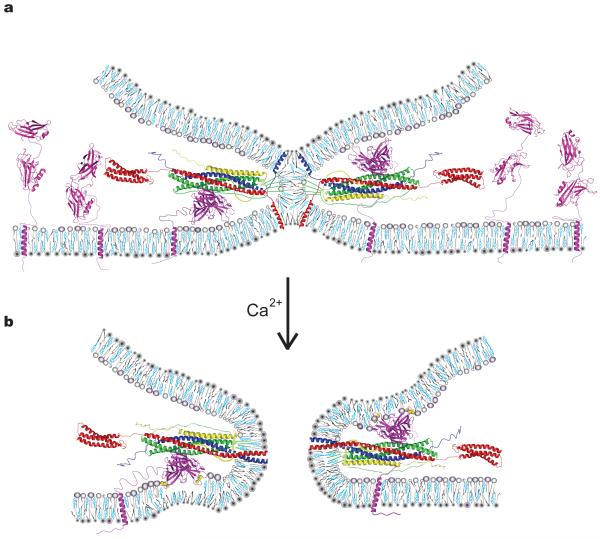
We propose a general model that begins with a SNARE-induced stalk state (Fig. 6a). There is a general notion in the recent literature that the neuronal SNARE-induced membrane juxtaposition may not proceed to full fusion by the clamping action of complexin29,30 or by the intrinsic properties of the SNARE complex31. Although it is possible that membranes do not interact at this SNARE-induced state, it is likely that a membrane stalk has formed based on three arguments: first, in reconstituted assays of biological fusion, lipid mixing readily occurs prior to content mixing32-34; second, Monte Carlo simulations of membrane fusion suggest that the free energy required for stalk formation is lower than that to proceed to full fusion35,36; and third, SNAREs can induce a stable hemifused state37.
Figure 6.
Model of a general Ca2+–SNARE–synaptotagmin fusion-triggering mechanism. (a) Membrane stalk induced by SNARE complex formation prior to Ca2+ arrival between a vesicle (top membrane) and the plasma membrane (bottom membrane). All components are drawn to scale. For details about the generation of the model see Supplementary Methods. Both SNARE-bound and uncomplexed Syt3 molecules residing in the plasma membrane are shown. The model also applies to Syt1 except that the transmembrane domain of Syt1 would reside in the synaptic vesicle. We drew SNARE-bound Syt3 poised to interact with either plasma or synaptic vesicle membrane; it is unknown to which membrane (or both) synaptotagmins bind. Note that in our model simultaneous binding of both complexin and synaptotagmin to SNARE complex is sterically permissible although (partial) competition has been observed in solution17 (syntaxin–red, synaptobrevin–blue; SNAP25-green; complexin-yellow; synaptotagmin-magenta). Phospholipids are shown as gray cartoons, cholesterol in cyan. (b) Fusion pore after Ca2+ (yellow spheres) binding to Syt3. The color code is identical to panel (a). Upon Ca2+-influx Syt3 Ca2+-binding loops coordinate Ca2+ with anionic phospholipids, penetrating the lipid headgroup region8, and may induce positive curvature10.
In our model we therefore assumed that a membrane stalk has formed (Fig. 6a), although this is not absolutely necessary for the proposed role of synaptotagmin in our model. Trans-SNARE complex would recruit synaptotagmins to the stalk and amplify the conformation of synaptotagmin with Ca2+-binding loops of both C2 domains positioned in the vicinity of the same membrane (but yet not penetrating the membrane) (Fig. 6a). Also shown are uncomplexed synaptotagmins whose C2 domains undergo large fluctuations. Upon Ca2+ influx, the high-FRET conformation of complexed synaptotagmins is further enhanced (Fig. 5b) and their Ca2+-binding loops insert into the membrane (Fig. 6b). Simultaneously, the synaptotagmin–SNARE interaction is strengthened (Supplementary Fig. 1c, Supplementary Fig. 8), releasing the complexin “clamp”29,30 and/or SNARE complex assembly “brake”31, leading to full zippering of the SNARE complex. The transition from the hemifused stalk to the fusion pore could be helped by this additional zippering of the SNARE complex. In concert, the interaction of the synaptotagmin Ca2+-binding loops with either the plasma membrane or synaptic vesicle membrane, in the conformation analogous to post-fusion conformation of viral fusion loops (Fig. 2), may perturb membrane regions near the stalk (Fig. 6b). Indeed, based on computer simulations, regions close to the stalk play a critical role in stalk-fusion pore transitions35. Computer simulations suggest that fusion can proceed via small hole formation in semi-stable hemifusion stalk intermediate states via different pathways35,38-40. However, regardless of the pathway additional energy is required, and both computer simulations and experiments suggest that the amount of energy and therefore fusion probability depend on the properties of lipids that comprise the stalk39,41,42. Ca2+-mediated insertion of synaptotagmin’s Ca2+-binding loops into the bilayer along with zippering of SNAREs transmembane helices change local lipid structure, possibly forcing lipid fatty-acid chains to change the volume they occupy and/or their curvature preference43-45, thus lowering the energy barrier for stalk-fusion pore transition. Model fusion systems, in the absence of fusion proteins, suggest that bringing two membranes in close proximity of one another is sufficient for content mixing albeit with lower efficiency than lipid mixing32. Furthermore, in eukaryotic membranes where fusion proteins are utilized, bringing membranes into close proximity is not necessarily sufficient. For example, mutations in viral fusion loops, along with replacement of transmembrane helices with GPI-linkers, block fusion24,25,46. Therefore, we propose that the combined effects of perturbations in the stalk regions, caused by insertion of synaptotagmin’s Ca2+-binding loops into the membrane, and the simultaneous zippering of SNARE transmembrane helices triggers fusion.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Südhof and J. Rizo for discussions, Y. Zhang, B. Cui, S. Solomatin, TH. Lee and A. Persitidinis for discussions of single-molecule experimental set-up and data analysis, Y. Abdiche and K. Lindquist for technical assistance and discussions of biosensor experiments, and the National Institutes of Health for support to A.T.B (RO1-MH63105). A.T.B. presented this work at the first Paul B. Sigler lecture at Yale University on March 23, 2009.
Appendix
ONLINE METHODS
Protein expression and purification
For crystallization we used the C2AB fragment of Syt3 from R. norvegicus (residues 292-587), and for single-molecule FRET experiments we used residues 232-588, C298H Q410C C438S C523S C533V N554C. For SNARE complexes we used the “minimal” SNARE core complex (synaptobrevin[28-89], syntaxin[191-256], SNAP-25[7-83] and SNAP25[141-204]) and the more “extended” SNARE complex (synaptobrevin[25-96], syntaxin[28-262], SNAP-25[1-83], and SNAP-25[120-206]). We expressed and purified all constructs using standard methods. See Supplementary Methods.
Crystallization, data collection, and refinement
We grew crystals of the C2AB fragment of Syt3 by the hanging drop method at 4 °C using a 1:1 molar ratio of C2AB with the extended SNARE complex48 at a total protein concentration of 12–15 mg ml−1 and a precipitant solution containing 50 mM MES, pH 5.4, 17 (v/v) % MPD, 50 mM CaCl2, 100 mM NaCl. Bipyramid shaped crystals appeared after a few days and were 150 μm × 150 μm × 50 μm. We achieved cryoprotection by stepwise transfer of crystals into mother liquor supplemented with 25–60 (v/v) % MPD. We collected diffraction data at the Lawrence Berkeley National Laboratory ALS on beamline 5.0.2. The crystals belong to the space group P41212 with unit cell dimensions a = 205.7 Å, b = 205.7 Å, c = 143.1 Å (Table 1). We processed all data with DENZO and SCALEPACK49. Based on the Matthews coefficient, we expected six C2AB molecules to be present in the asymmetric unit. Only three C2AB molecules, however, form the crystal lattice, resulting in a very high solvent content (~84%) of the crystals (Supplementary Fig. 4). For details of the molecular replacement see Supplementary Methods.
Alternate cycles of manual model building using the program COOT50 positional, B-factor, simulated annealing, and TLS refinements with the program CNS 1.251 and PHENIX52, and addition of ions, reduced the R and Rfree values to 22.9 % and 25.5 %, respectively, for all observed reflections (Table 1). We used tight NCS restraints in the refinement as appropriate for a structure solved at 3.5 Å resolution. Release of the NCS restraints did not show any improvement in Rfree and did not result in any significant deviations from NCS mates.
Single-molecule FRET data collection and analysis
We collected single-molecule FRET (smFRET) data with a prism-based total internal reflection (TIR) fluorescence microscope. We used co-localized Alexa555/Cy3 and Alexa647/Cy5 emission spots to calculate the FRET efficiency. We used an alternating laser illumination sequence to determine the number of acceptors per single spot and if photobleaching occurred. We analyzed only traces containing single acceptor and single donor, as judged by the fluorescence intensity (examples are in Figs. 3a and 4). For details, see Supplementary Methods.
Syt3–SNARE interactions studies by bio-layer interferometry
We studied SNARE–Syt3 interaction in the absence and presence of Ca2+ using Octet Red (ForteBio, Inc.) equipped with a streptavidin SA biosensors (ForteBio, Inc.). For details, see Supplementary Methods.
Simulation of dye positions and expected FRET efficiencies
For the calculation of FRET efficiency-derived distances, we built atomic models of the Cy3/Alexa555 and Cy5/Alexa647 fluorophores attached by malemide linkers to the mutated cysteine residues of Syt3 (Q410C and N554C). While chemical structures of Alexa555 are not available, we were able to obtain the linker structures of Alexa555 from Invitrogen. Considering the similar molecular structure of the Alexa647 dye and the identical linkers of the Cy3/Cy5/Alexa647/Alexa555 dyes, we used the known structures of Cy3 and Cy5 dyes for all simulations (Supplementary Fig. 11). We performed 500 trials of torsion angle molecular dynamics using the Crystallography and NMR System (CNS)51 where we fixed the coordinates of Syt3 in either the SNARE-induced Ca2+-bound or Ca2+-free conformation, and we allowed the attached fluorophores and their linkers to freely rotate and to change their conformation53. We used a repulsive force field without electrostatics and solvent54. For details see Supplementary Methods.
Molecular dynamics simulation of conformations of Syt3
We performed extensive (2 nsec) reduced variable molecular dynamics of the C2A-C2B fragment of Syt3 with the two C2 domains treated as rigid bodies (residues 295-421 and 431-569, respectively), while we kept the torsion angles of the linker (residues 422-430) connecting the two domains variable. We rigidly associated pseudoatoms (simulated points that are treated similar to “physical atoms” in the molecular dynamics calculation) with the C2 domains at the labeled residue positions 410 and 554. We chose the positions of these pseudoatoms as the average positions of fluorophore centers, i.e., the average positions of the corresponding CAO atoms, from two separate simulations with the fluorophore attached to one of the two labeling sites (using the same molecular dynamics protocol described in the previous section). We included only repulsive van der Waals energy terms in the simulation54. We performed the simulations in vacuum for 2 nsec with a timestep of 0.005 psec at 300K. We converted the distances between the pseudo atoms during the simulation into FRET efficiencies using a Förster radius (R0) of 55.5 Å18 and plotted as a histogram (Fig. 3c). This is not meant to be a realistic simulation, which would require inclusion of solvent and electrostatics but rather to provide information about the range of possible conformations of Syt3.
Molecular dynamics simulation of the high FRET conformation of Syt3
We performed reduced variable molecular dynamics of the C2A-C2B fragment of Syt3 (as described above) but with a distance restraint distance restraint of 44.1 Å between the pseudo-atoms at positions 410 and 554 (corresponding to the dye labeling sites used for smFRET). We derived this distance from the observed smFRET efficiency value of the high-FRET state (0.8), using a Förster radius of 55.5 Å18. The distance restraint consisted of a harmonic square well-potential function restraining the distance between the pseudoatoms to 44.1 +/− 0.1 Å55. We added this distance restraint function to the repulsive van der Waals energy terms54 and we set the energy constant of the restraint term to 50 Kcal/mol/Å2. The resulting ensemble of structures is shown in Supplementary Fig. 9.
Footnotes
Present address: Department of Energy, 1000 Independence Avenue SW, Washington, DC 20585
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb.
Accession numbers. Atomic coordinates and structure factors for the SNARE-induced Ca2+-bound C2AB structure of Syt3 have been deposited with the Protein Data Bank under accession code 3HN8.
Note: Supplementary Information is available on the Nature Structural & Molecular Biology website.
References
- 1.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–47. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–9. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 5.Pang ZP, Shin OH, Meyer AC, Rosenmund C, Sudhof TC. A gain-of-function mutation in synaptotagmin-1 reveals a critical role of Ca2+-dependent soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex binding in synaptic exocytosis. J Neurosci. 2006;26:12556–65. doi: 10.1523/JNEUROSCI.3804-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–72. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 7.Sugita S, Shin OH, Han W, Lao Y, Sudhof TC. Synaptotagmins form a hierarchy of exocytotic Ca(2+) sensors with distinct Ca(2+) affinities. Embo J. 2002;21:270–80. doi: 10.1093/emboj/21.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrick DZ, Sterbling S, Rasch KA, Hinderliter A, Cafiso DS. Position of synaptotagmin I at the membrane interface: cooperative interactions of tandem C2 domains. Biochemistry. 2006;45:9668–74. doi: 10.1021/bi060874j. [DOI] [PubMed] [Google Scholar]
- 9.Hui E, Bai J, Chapman ER. Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys J. 2006;91:1767–77. doi: 10.1529/biophysj.105.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–8. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 11.Arac D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–17. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 12.Stein A, Radhakrishnan A, Riedel D, Fasshauer D, Jahn R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat Struct Mol Biol. 2007;14:904–11. doi: 10.1038/nsmb1305. [DOI] [PubMed] [Google Scholar]
- 13.Bhalla A, Chicka MC, Chapman ER. Analysis of the synaptotagmin family during reconstituted membrane fusion. Uncovering a class of inhibitory isoforms. J Biol Chem. 2008;283:21799–807. doi: 10.1074/jbc.M709628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittelsteadt T, et al. Differential mRNA expression patterns of the synaptotagmin gene family in the rodent brain. J Comp Neurol. 2009;512:514–28. doi: 10.1002/cne.21908. [DOI] [PubMed] [Google Scholar]
- 15.Li C, et al. Ca(2+)-dependent and -independent activities of neural and non-neural synaptotagmins. Nature. 1995;375:594–9. doi: 10.1038/375594a0. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Mashimo T, Sudhof TC. Synaptotagmin-1, −2, and −9: Ca(2+) sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron. 2007;54:567–81. doi: 10.1016/j.neuron.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–87. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 18.Choi U, et al. Single molecule FRET derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1763. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–8. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton RB, Ernst JA, Brunger AT. Crystal structure of the cytosolic C2A-C2B domains of synaptotagmin III. Implications for Ca(+2)-independent snare complex interaction. J Cell Biol. 1999;147:589–98. doi: 10.1083/jcb.147.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons DL, et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–5. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 22.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–8. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarousse N, Wilson JD, Arac D, Rizo J, Kelly RB. Endocytosis of synaptotagmin 1 is mediated by a novel, tryptophan-containing motif. Traffic. 2003;4:468–78. doi: 10.1034/j.1600-0854.2003.00101.x. [DOI] [PubMed] [Google Scholar]
- 24.Kielian M, Klimjack MR, Ghosh S, Duffus WA. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–72. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowling W, Denisova E, LaMonica R, Mackow ER. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J Virol. 2000;74:6368–76. doi: 10.1128/jvi.74.14.6368-6376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 27.Huang H, Cafiso DS. Conformation and Membrane Position of the Region Linking the Two C2 Domains in Synaptotagmin 1 by Site-Directed Spin Labeling. Biochemistry. 2008 doi: 10.1021/bi801470m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–20. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraudo CG, et al. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–6. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–21. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorensen JB, et al. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–66. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan YH, van Lengerich B, Boxer SG. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci U S A. 2009;106:979–84. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun Y, Wickner W. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc Natl Acad Sci U S A. 2007;104:13010–5. doi: 10.1073/pnas.0700970104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci U S A. 2008;105:15382–7. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katsov K, Muller M, Schick M. Field theoretic study of bilayer membrane fusion: II. Mechanism of a stalk-hole complex. Biophys J. 2006;90:915–26. doi: 10.1529/biophysj.105.071092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmerberg J, Gawrisch K. The physical chemistry of biological membranes. Nat Chem Biol. 2006;2:564–7. doi: 10.1038/nchembio1106-564. [DOI] [PubMed] [Google Scholar]
- 37.Giraudo CG, et al. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J Cell Biol. 2005;170:249–60. doi: 10.1083/jcb.200501093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasson PM, et al. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc Natl Acad Sci U S A. 2006;103:11916–21. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsov K, Muller M, Schick M. Field theoretic study of bilayer membrane fusion. I. Hemifusion mechanism. Biophys J. 2004;87:3277–90. doi: 10.1529/biophysj.103.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller M, Katsov K, Schick M. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys J. 2003;85:1611–23. doi: 10.1016/S0006-3495(03)74592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chernomordik LV, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melia TJ, You D, Tareste DC, Rothman JE. Lipidic antagonists to SNARE-mediated fusion. J Biol Chem. 2006;281:29597–605. doi: 10.1074/jbc.M601778200. [DOI] [PubMed] [Google Scholar]
- 43.Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J. 2006;91:487–95. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian A, Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys J. 2009;96:2676–88. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, et al. Bilayer edge and curvature effects on partitioning of lipids by tail length: atomistic simulations. Biophys J. 2008;95:2647–57. doi: 10.1529/biophysj.108.131409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–91. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert JM, Greenberg HB. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–63. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ernst JA, Brunger AT. High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex. J. Biol. Chem. 2003;278:8630–8636. doi: 10.1074/jbc.M211889200. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski ZM. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. W. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 53.Wozniak AK, Schroder GF, Grubmuller H, Seidel CA, Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc Natl Acad Sci U S A. 2008;105:18337–42. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engh R, Huber R. Accurate bond and ange parameters for X-ray protein structure refinement. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 55.Brunger AT. X-PLOR, version 3.1. A system for X-ray crystallography and NMR. Yale Unversity Press; New Haven, CT: 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.