Abstract
Supra-molecular assembly is a powerful strategy used by nature for building nano-scale architectures with predefined sizes and shapes. Numerous challenges remain however to be solved in order to demonstrate precise control over the synthesis, folding and assembly of rationally designed three-dimensional (3D) nano-objects made of RNA. Using the transfer RNA molecule as a structural building block, we report the design, efficient synthesis and structural characterization of stable, modular 3D particles adopting the polyhedral geometry of a non-uniform square antiprism. The spatial control within the final architecture allows precise positioning and encapsulation of proteins. This work demonstrates that a remarkable degree of structural control can be achieved with RNA structural motifs to build thermostable 3D nano-architectures that do not rely on helix bundles or tensegrity. RNA 3D particles can potentially be used as carriers or scaffolds in nano-medicine and synthetic biology.
Classification: SELF-ASSEMBLY, SUPRA-MOLECULAR CHEMISTRY, BIOCHEMISTRY, BIOPOLYMERS
INTRODUCTION
RNA molecules are extremely versatile functional biopolymers that participate in key informational pathways that characterize living organisms on Earth1. Far from being simply coding for proteins as mRNA, a plethora of non-coding RNAs are actively contributing to proteins translation as ribosomal RNAs and transfer RNAs, and various catalytic and regulatory functions as ribozymes, riboswitches and regulatory elements. The remarkable functions of RNA stem from the ability of RNA to fold and assemble into exquisite modular and hierarchical three-dimensional (3D) shapes2–4 that, in a cell, are only surpassed by proteins. Understanding how RNA folding and assembly can be controlled in order to create any desired functional shapes made of RNA is therefore an important and challenging problem, with numerous implications in the developments of therapeutics and tools for nano-medicine and synthetic biology applications5–7.
By taking advantage of RNA architectonics, an approach for rationally designing 3D RNA architectures6, we previously showed that the structural information encoding specific conformation of natural RNAs could be implemented within artificial RNA sequences to control both 3D shape and self-assembling interfaces. For instance, several modular RNA units were designed to construct particles8–14, filaments8,15–17 and a variety of self-assembling programmable planar nano-structures12,14. Recently, the potential of RNA assemblies for nanobiotechnology was best exemplified by functionalized dimeric and trimeric nanoparticles based from natural Phi29 RNA molecules able to deliver siRNA to induce apoptosis in cancer cells13,18,19. Herein, we report a versatile strategy to synthesize thermally stable, self-assembling RNA particles with non-uniform antiprism polyhedral shape. A series of 11 tRNA squares (TS1–TS11) were prepared from folded tRNA units bearing programmed kissing loop and tail connectors, as detailed in the Supplementary Information (Table S1). These were assembled into tRNA octamers (TO1–2 being constructed from TS1 and TS2, for example) whose structures were characterized by atomic force microscopy (AFM) and cryogenic electron microscopy (cryo-EM) with single particle image reconstruction. Fully addressable RNA antiprisms were further functionalized to immobilize molecules inside or outside the cage. This study incorporates complex RNA structural motifs and spatial addressability to generate well-defined 3D polyhedral RNA nanoparticles with full control over composition and stoichiometry. As such, it paves the way towards RNA-based delivery systems of therapeutic molecules for nano-medicine and in vivo multifunctional complexes for synthetic biology.
RESULTS AND DISCUSSION
Rational design of spatially addressable 3D tRNA particles
tRNAs are classified primarily according to the length of the variable arm region, where Class I tRNAs correspond to the canonical tRNA shape such as tRNA(Phe), and Class II tRNAs differ by exhibiting an elongated variable arm that branches from the vertex of the tRNA core. Class II tRNAs are particularly attractive as vertices for designing artificial 3D polyhedron architectures. Their five-way helix junction motifs fold into stable 3D junctions, with three arms extending in the x, y and z dimensions. The helical axes of the amino acceptor (aa) and anticodon (ac) arms are perpendicular to one another and can be superimposed to the x and y axes (Figure 1a). According to the crystallographic structure of the class II tRNA(Ser)20, the variable arm is oriented in the z direction, out of the xy plane formed by the aa and ac arms, by making angles of 120°, 70°, and 25° with the x, y and z axes, respectively (Figure 1a). However, as the variable arm is not as structurally constrained as the two other arms, its orientation in relation to the xy plane can eventually be changed by ± 15° without affecting the structural integrity of the motif21.
Figure 1.
Structure and design principles of tRNA-based architectures. (a) 3D model of the tRNA unit: the variable arm points out of the plane defined by the anticodon (ac) and amino-acid (aa) arms that are perpendicular to each other. Angle values between arms are indicated. (b) Generic secondary structure diagram for self-assembling tRNA units derived from the structure of class II tRNA(Ser). The strand topology of the tRNA unit is designed so that the 5' and 3' ends are localized at the tip of the variable arm. Kissing-loops (KL) (in green) are inserted at the extremities of the ac and aa arms. Nucleotides in blue are those corresponding to the Class II tRNA(Ser) motif. N: indicates any paired nucleotide; X indicates nucleotide involved in intermolecular KL or tail-tail bps. Dashed lines indicate tertiary interactions. (c) Schematic indicating the various intermolecular connectors. KL complex (i) and tail-tail connectors without (ii) and with (iii) major triple base pairs. Blue and black colors indicate the two different interacting RNAs. (d) Cis and trans orientations of tRNA squares resulting from tail-tail edges connectors of 21bp or 26 bp, respectively. (e) 3D model of the non-uniform square antiprism. Three-different views are shown. The tRNA units entering into the composition of the fully programmable octamer (TO1–2) are indicated (see also Tables S1 and S2 in Supplementary Information for other TO combinations).
Using an inverse folding design strategy6,7, we previously used the class II tRNA(Ser) motif as a building block for constructing tetrameric particles with square shape14. Its fold allows relocation of the 5’/3’ termini from the aa arm to the variable arm, and insertion of kissing-loop motifs (KL) at the ends of extended aa and ac arms (Figure 1b). By taking advantage of different, selective kissing-loop interactions (Figure 1c), four addressable tRNA units can be programmed to assemble into stable, tRNA squares with 14 nm edges. As demonstrated for other tetrameric RNA particles12, more complex architectures based on tRNA squares can potentially be generated via assembly through programmed tail-tail connectors located at the level of their variable arms (Figure 1c)14. For instance, possible control over the final supramolecular architecture can be achieved by varying the base pair length of tail-tail edges in order to position two adjacent tRNA squares in cis (21bp) or trans (26bp) configurations, leading to either 3D particles or planar arrays, respectively (Figure 1d). Based on this approach, we designed an octameric tRNA particle with the shape of a non-uniform square antiprism by combining two tRNA squares through four tail-tail edges (Figure 1e). Square antiprisms are symmetrical objects with d4 symmetry. They typically consist of eight vertices that define two parallel copies of square connected by an alternating band of eight triangles (Figure S1). By contrast to a perfectly uniform square antiprism with 16 identical edges defining two squares and eight equilateral triangles, the RNA antiprism is non-uniform as its eight triangles are non-equilateral (Figure S1). Rather than taking advantage of pure tensegrity, the RNA octamer relies on structural constraints imposed by the tRNA motif for twisting the two squares by 30° relative to each other. This is achieved through only four tail-tail edges of 8.5 nm, the other four triangle edges remaining virtual (Figure S1). While keeping this distance constant, the positioning of the 3' and 5' ends with respect to the RNA antiprism can be modulated based on the length of the interacting variable arms. Additionally, the presence of two extra nucleotides at the tail 3' end can also enhance the stability of the tail-tail connector by formation of local triple base pairs with the first two base pairs of the variable arm (see Figure 1c).
Self-assembly and stability of tRNA nano-particles
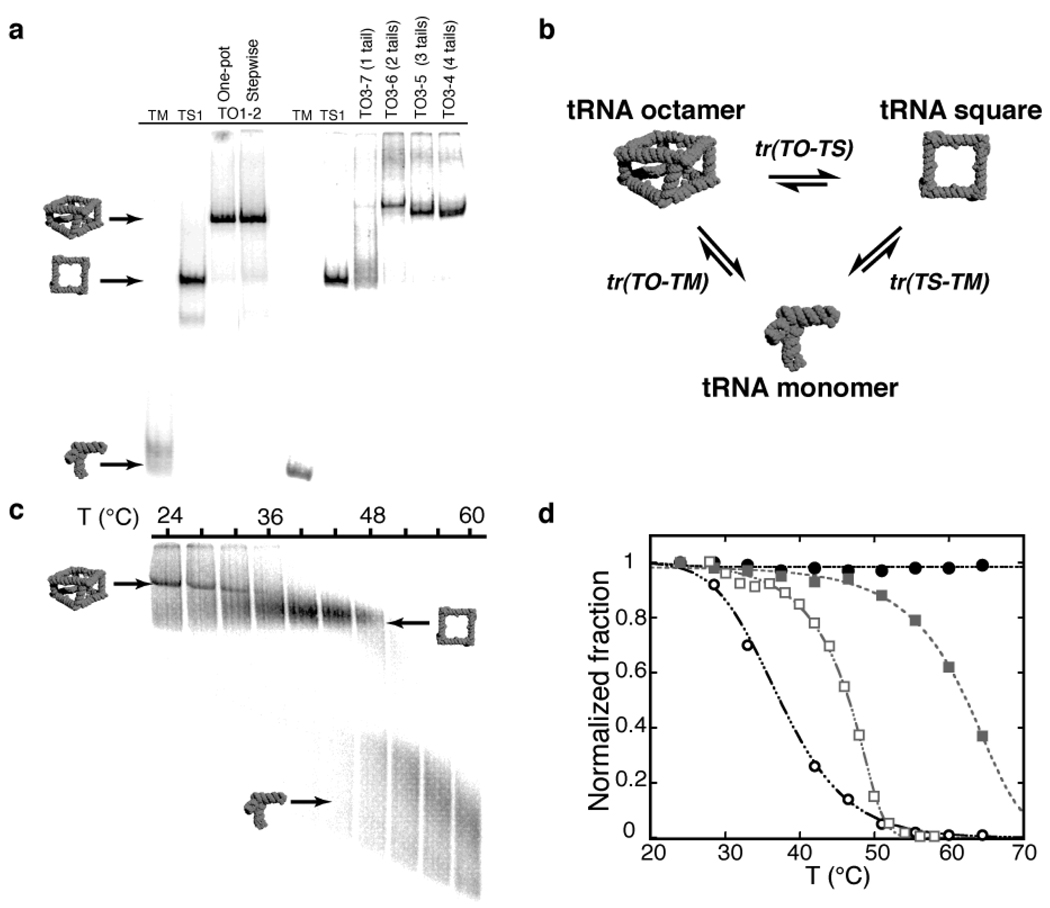
Several sets of tRNA units were designed and synthesized by run-off transcription from PCR generated templates to make a variety of finite size tRNA octamer (TO) particles. First, we designed eight different tRNA units able to assemble through selective kissing-loops and four tail-tail connectors of different base pair sequences into a fully addressable octamer (TO1–2), with the spatial position of each unit being known into the final architecture. When all mixed in one-pot (Materials and Methods and Supplementary Information), these tRNA units lead to octamers with ~90% yields as estimated by native polyacrylamide gel electrophoresis (PAGE) assays (Figure 2a). Because kissing-loop connectors efficiently assemble at ten-fold lower Mg2+ concentrations than tail-tail connectors, a stepwise assembly strategy at different Mg2+ concentration can be used to generate octamers with fewer distinct kissing-loop or tail-tail connectors (Materials and Methods and Supplementary Information). While tRNA squares (TS1 to TS11 in Supplementary Information) are typically obtained with ~80% yield at RNA concentrations ranging from 10 nM to 2 µM14, stepwise assembly of non-fully programmable octamers lead to 60% to 80% yields, slightly lower than those for fully programmable octamers. In the range of conditions tested, we do not observe any exchange of tRNA units between assembled squares or octamers with identical kissing-loop connectors (data not shown).
Figure 2.
tRNA octamer self-assembly and thermal stability. (a) Characterization of tRNA supramolecular assembly of various tRNA octamers (TO) by non-denaturing PAGE at 2 mM Mg(OAc)2: lane M is for the monomer tRNA unit control (400 nM), lane TS1 are for a control tRNA square (TS1, 100 nM), lanes TO1–2 show octamer products (50 nM) after one pot or stepwise assembly. Lanes TO3–4 to TO3–7 (50 nM) correspond to octamers with a varying number of tail connectors. (b) Schematic of the three major transitions of TO disassembly observed by TGGE analysis (Materials and Methods and Supplementary Information). (c) Example of TGGE gel showing the cooperative biphasic dissociation of TO3–4 (20 nM) into squares and monomers at 0.2 mM Mg(OAc)2 (see also Table 1). (d) Comparison of melting curves for TS3 (grey squares) and TO3–4 (black circles) obtained from TGGE gels at 0.2 mM (empty symbols) and 15 mM (solid symbols) Mg(OAc)2.
The thermal stability of tRNA octamers (TO) versus tRNA squares (TS) was studied at various Mg2+ concentrations by temperature gradient gel electrophoresis (TGGE), an efficient method to separate different supra-molecular assemblies based on temperature dependent conformational changes12,14 (Figure 2b). Initial thermal stability measurements indicate that the transition of disassembly of tRNA squares into monomers, tr(TS-TM), is a cooperative process with the 90° tRNA motif contributing significantly to the overall stability of the tRNA square14 (Figure 1b). Transition tr(TS-TM) is Mg2+ concentration dependent, with observed melting temperatures (Tm) increasing by 20°C from 0.2 to 15 mM Mg2+ (Figure 2).
The dissociation of the TO3–4 octamer, which is formed of two squares (TS3+TS4) with identical kissing-loop edges and connected through four identical tail-tail edges, was also shown to be highly magnesium dependent (Figure 2). At 0.2 mM Mg2+, it occurs in two transitions where the octamer disassembles first into two tetramers through a transition tr(TO-TS) at 34°C, followed by the tr(TS-TM) transition of disassembly of tetramers into monomers at 46°C. This is expected as the kissing-loop connectors are known to be more stable than tail-tail connectors at low Mg2+ concentrations. However, at Mg2+ concentrations equal or above 2 mM, the TO3–4 octamer is more stable than its constitutive squares (Figure 2b–d) and directly disassembles in one unique transition into monomers (transition tr(TO-TM)). At 15 mM Mg2+, while tr(TS-TM) occurs at ~65°C, the Tm for tr(TO-TM) is above 70°C, the upper temperature limit for TGGE (Figure 2 c–d, Table 1). At 2 mM Mg2+, tr(TO-TM) for TO3–4 is 62°C. By contrast, the removal of one and two tail-tail edges results in octamers TO3–5 and TO3–6 that disassemble through biphasic processes with tr(TO-TS) occurring at 54°C and 43°C, respectively, below the same transition tr(TS-TM) of 58°C. Octamer TO3–7 that lacks three tail-tail edges is only observed at temperature below 15°C. The degree of compactness (and rigidity) of octamers seem to be directly proportional to the number of connectors as suggested by the decreasing electrophoretic mobility of octamers with three (TO3–5), two (TO3–6) and one (TO3–7) tail-tail edges versus octamers (TO3–4). The remarkable thermal stability (and rigidity) of the TO3–4 with four tail-tail edges results from the cooperative structural stabilization of kissing-loop and tail-tail connectors that enhance the stability and rigidity of the square structure upon formation of tail-tail connectors at Mg2+ concentrations equal or above 2 mM. In these conditions, the penalty for bringing two adjacent tRNA squares in close proximity, which likely results from electro-negative repulsions, can be reduced by counter ions.
Table 1.
Melting temperature (Tm) values measured by TGGE for various transitions between tRNA squares (TS), tRNA octamers (TO) and tRNA monomers (TM) architectures at 0.2, 2 and 15 mM Mg(OAc)2 (see Materials and Methods and Supplementary Information).
| Mg2+(mM) | 0.2 | 2 | 15 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| transition | TO-TS | TS-TM | TO-TS | TS-TM | TO-TM | TS-TM | TO-TM | ||
|
|
TS3 | - | 46 | - | 58 | - | 65 | - | |
| TO3-4 (4 tails) |
34 | 46 | - | - | 62 | - | >70 | ||
| TO3-5 (3 tails) |
n.d. | n.d. | 54 | 58 | - | n.d. | n.d. | ||
| TO3-6 (2 tails) |
n.d. | n.d. | 43 | 58 | - | n.d. | n.d. | ||
Like protein complexes that are typically more resistant to protease degradation than monomeric proteins, RNA units within octamers are more resistant to nuclease degradation than monomers or those within tetramers (Figure S2). By comparing the extent of ribonuclease T1 degradation of monomeric radio-labeled tRNA unit to those of similar units in both TO3–4 octamer and TS3 tetramer contexts (Materials and Methods), we show that a significant amount of RNA units in TO3–4 (25%) and to a lesser extent in TS3 (5%) are still intact while the monomer is completely degraded after 1 hour incubation (Figure S2). This correlates with the higher thermal stability of RNA octamers versus tetramers and monomers.
Characterization of the tRNA anti-prism architecture
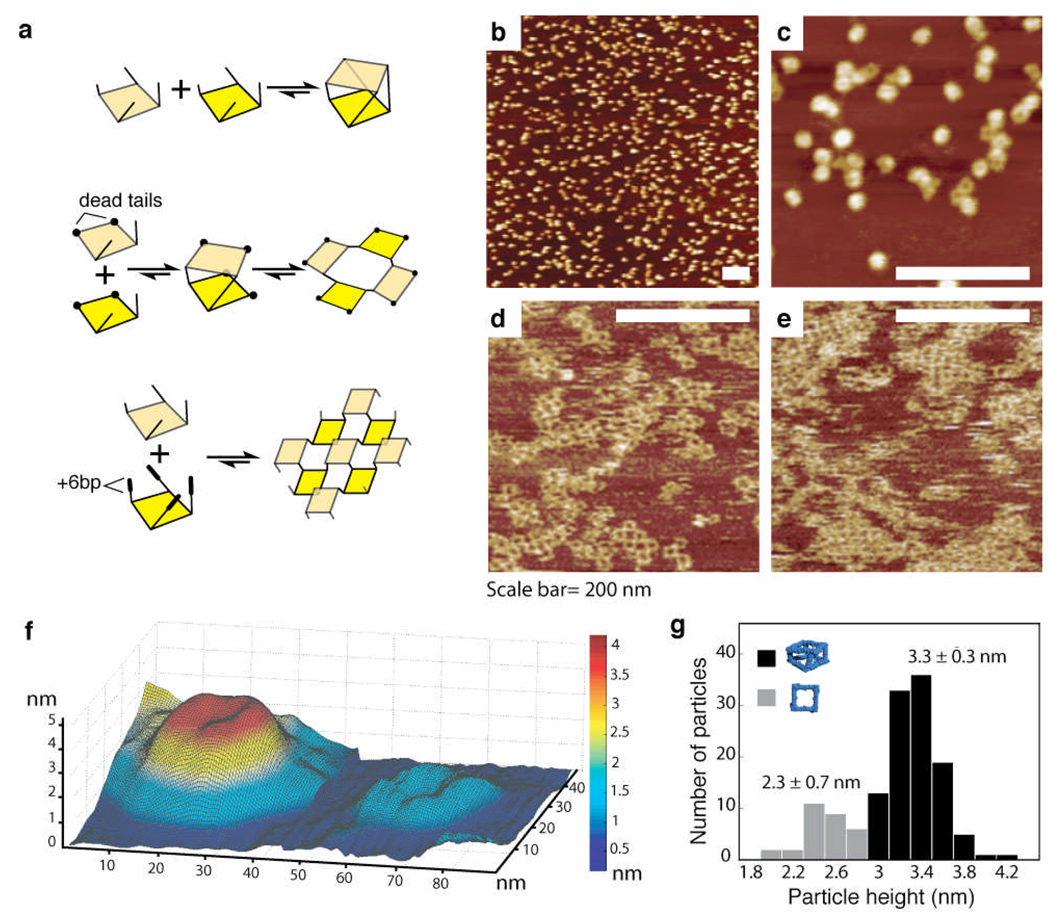
To provide direct evidence for the self-assembly of tRNA units into square antiprism shapes, the octamer samples were imaged by atomic force microscopy (AFM)12,14,22 and cryogenic transmission electron microscopy (cryo-EM)23–25 (Materials and Methods). AFM images reveal a rather uniform and mono-dispersed population of TO3–4 particles (Figure 3). More than 70% of them have an averaged size of 25.2 ± 3.6 nm (at mid height) and height of 3.3 ± 0.3 nm (Figure 3g). The remaining 30% particles essentially correspond to squares with height of 1.5 nm and average width of ~14 nm (Figure 3). These data are in good agreement with the ~80–90% yield of octamer formation estimated by native PAGE analysis. Most of the particles visualized by AFM correspond to flattened octamers likely caused by strong electrostatic interactions with the substrate and dehydration24. Their observed height corresponds to two superimposed squares and their width corresponds to the measured diagonals of 2D objects resulting from two squares sliding on top of each other (Figure S3). Interestingly, the squares lying on the surface are essentially square dimers resulting from the "opening" of octamers onto the surface. Visualization of the TO3–6 octamer, which lacks two adjacent tail-tail edges, revealed an interesting rearrangement into small RNA nano-grids (Figure 3d). This pattern is similar to the one adopted by tRNA squares (TS10 +TS11) that are programmed with tail-tail edges in the trans configuration to assemble into 2D arrays (Figure 3e). The strong electrostatic interactions with the substrate can significantly alter the preferred orientation of tail-tail connectors, forcing the "open" tRNA octamer, and to a lesser extent the tRNA antiprism, to open or rearrange on the mica surface. While the tail-tail connectors offer good geometrical control over the assembly of particular set of tRNA squares, destabilization of some of these connectors can therefore lead to shape-shifting transitions, especially when RNA assemblies interact with a surface.
Figure 3.
Structural characterization of tRNA architectures by AFM. (a) Top-to-bottom: Hierarchical stepwise assembly schemes for tRNA squares designed to assemble into (b–c) the antiprism TO3–4, (d) the "open" octamer TO3–6 and (e) the planar TS10–11 array (TS10 + TS11). TO3–4 and TS10–TS11 have tail-tail connectors in cis and trans configuration, respectively. TO3–6 has two tail-tail connectors missing with respect to TO3–4. Corresponding AFM images were obtained in air. (f) 3D rendering of AFM image data to compare the relative height of "closed" octamer versus square. (g) Height distribution of octamers obtained from AFM analysis indicates that more than 70% of supra-molecular assemblies correspond to two superimposed squares.
Overall, the AFM data strongly suggest that the various structural components of the tRNA octamer favor formation of a closed, compact molecular complex. However, the most convincing structural evidence for TO1–2 antiprism formation comes from cryo-EM imaging with single particle reconstruction23–25. The cryo-EM images show that most particles have the expected size (Figure 4a). The yield of correctly assembled antiprism is ~84%, in good agreement with AFM and native PAGE analysis. However, since particles apparently adopt a preferred orientation, particles were classified to obtain an approximately equal distribution of particles in all orientations. A reconstruction of the RNA particle 3D structure was obtained at 24.5 Å resolution from 1,570 particles using the EMAN reconstruction packages26 (Figure S4, Materials and Methods and Supplementary Information). The computed projections from this 3D reconstruction match well with the class averages of observed particles with similar views (Figure 4b–c). The antiprism reconstruction is in excellent agreement with the predicted 3D model displayed in Figure 1. As predicted, the length for the kissing-loop edges and side tail-tail edges are 14 nm and 8.5 nm, respectively. Most remarkably, the arms of the tRNA ser motifs at the vertices of the tRNA octamer particle adopt similar orientations as those from the tRNA(Ser) crystallographic structure20 used for building the model. All the DNA particles that have been synthesized take advantage of helix bundles or tensegrity to fold. By contrast, the tRNA octamer relies on the structural constraints imposed by the tRNA motif for twisting the two squares by ~30° relative to each other. As the orientation of the variable arms varies within class II tRNAs21, it is anticipated that different geometrical objects could be obtained from other tRNA motifs.
Figure 4.
Structural characterization of tRNA architectures by Cryo-EM with single image particle reconstruction. (a) Typical cryo-EM image of antiprism particles. More than 85% of the objects seen are well-formed antiprisms. (b) Reconstructed 3D model of the non-uniform square antiprism TO1–2 at 24.5 Å resolution. The view orientations are similar to those in Figure 1e. (c) Class averages of particles with similar views observed by cryo-EM (EM) with the corresponding projections of the RNA antiprism 3D structure reconstructed from the cryo-EM images (recon) and the corresponding view of the initial antiprism model (model). These particle views are selected from different image frames to represent views at different orientations (Figure S3).
Spatial control of the tRNA antiprism
To illustrate the spatial control of the tRNA antiprism as well as its potential as a carrier for proteins, some of its tRNA units were biotinylated at their 5' end in order to be conjugated to streptavidin, a globular tetrameric protein complex of 5 nm in diameter27,28. By keeping the overall length of a tail-tail edge constant (21 bp) but varying the length of its two variable arm components, the precise positioning of the 5' end of each constituting tRNA unit can be controlled due to the helicity of RNA duplexes. Therefore, with both a "short variable arm" tRNA unit and "long variable arm" tRNA unit, the 5' end can be oriented either toward the inside or the outside of the antiprism (TO8–9), respectively (Figure S1d). Streptavidin is able to simultaneously bind up to four biotin molecules. Therefore, the stoichiometry of streptavidin versus functionalized antiprism and the number and location of 5'-biotinylated tectoRNAs within the antiprism are variables allowing various distinct supramolecular architectures to be obtained (Figures 5a and S5). As the central cavity of the antiprism particle can potentially accommodate globular proteins up to 8–9 nm in diameter, inward "encapsulation" of two streptavidins is possible after biotinylation of two "short" tRNA units localized on two opposite corners of the antiprism (Figure S1e). At saturating concentration of streptavidin versus antiprism, up to 50% of antriprisms are conjugated to encapsulated streptavidins and blocked from aggregating. By contrast, outward positioning of "long" 5' biotinylated tRNA units within the antiprism (Figures S1d) leads to the preferential formation of supra-molecular filaments and higher order structures when mixed with streptavidin.
Figure 5.
Coupling streptavidin to spatially addressable antiprims (TO8–9). (a) The location of the 3’ tail-tail connector in the variable arm determines the orientation of the iodoacetyl-biotin linker, and thus can be used to control the positioning of streptavidin with respect to the antiprism. (b–c) Antiprisms with biotin oriented inward form small antiprism-streptavidin complexes. (d–e) Antiprisms with biotin oriented outward form antiprism-streptavidin filaments. Corresponding AFM images of the streptavidin-antiprism conjugates, formed at ratio of streptavidin vs antiprism is 1:1 (50 nM), are shown (see Figure S5, Supplementary Information and Materials and Methods). (f) Distribution of the population of antiprisms in function of antiprism-streptavidin chain lengths and inward or outward orientation of conjugated biotin. The frequency (in percent) stands for the total occurrence of antiprisms in an antiprism-streptavidin complex of specific length (indicated in number of constituent antiprisms). These experiments were reproduced at least twice.
As demonstrated by AFM imaging, at streptavidin vs antiprism ratio of 1:1, "outward" antiprism-streptavidin conjugates assemble mostly into filaments with an average of 7 ± 3 antiprisms and a periodicity of ~26 nm (Figure 5b–c–f). Small circular arrangements and larger aggregates corresponding to branched structures can also be occasionally observed. By contrast, in similar experimental conditions, ~17% of "inward" antiprisms-streptavidin conjugates do not form any aggregate while the remaining ~80% assemble in small aggregates of mostly two to three antiprisms (Figure 5d–e–f). The observed height of antiprisms with inward streptavidin is significantly higher than the one of uncoupled antiprisms (4.1 nm with streptavidin versus 3.3 nm) suggesting that streptavidin slightly contribute to rigidifying the antiprism. These results clearly demonstrate that the tRNA antiprisms offer spatial control in the positioning of streptavidin. That two inward antiprisms can be bridged by streptavidin is expected considering that each biotin is attached to the RNA through a flexible linker of 2.7 nm that allows a significant degree of freedom of movement for the bound protein. Rather than blocking multimerization through streptavidin bridges, the inward orientation of biotin restrains the movement of the protein at the level of the RNA cage so that it cannot form in-line interactions as easily as when biotins are positioned outwards. By contrast, the outward positioning of biotin on the antiprism facilitates the assembly into controlled chains of RNA-protein filaments.
CONCLUSION
The current study clearly demonstrates the application of RNA architectonics to the construction of 3D polyhedral objects. The versatile tRNA fold can be used as a structural building block for efficient synthesis of stable, programmable all-RNA NPs with antiprism polyhedral shape defining an inside and outside. By contrast to DNA design strategies that essentially rely on classic Watson-Crick base pairings29–31, tensegrity32–35 or helical bundles35–39, our RNA architectonics design strategy takes advantage of natural structural motifs to build rigid and stable self-assembling RNA nanostructures6,7. For instance, the non-uniform RNA antiprism relies on the particular structural constraints of the tRNA(Ser) motif for twisting the two squares by ~30° relative to each other. As the orientation of the variable arms varies within class II tRNAs21, other geometrical 3D objects could potentially be obtained from other tRNA motifs (Figure S6). RNA antiprism particles offer spatial control for the positioning of other molecules such a proteins, have the possibility to protect encapsulated molecules against the surrounding, and can be functionalized to comprise novel properties or exhibit specific targeting against different molecules. For instance, the information coding for therapeutic siRNAs, functional aptamers or ribozymes can directly be embedded within the sequences of these all-RNA NPs for acting as all-RNA cargos, scaffoldings for delivery vehicles of other molecules or multifunctional nano-factories within the cell (Figure S6). While these RNA NPs could be chemically modify to increase their stability towards ribonucleases, they could also be synthesized in vivo. Indeed, the in vivo availability of RNA molecules that can be directly incorporated into nanostructures makes RNA self-assembly particularly appealing for in vivo nanoconstruction. While the RNA 3D antiprism is thermostable at magnesium concentrations above 2 mM, its ability to open up on a surface and forms planar arrays also illustrates its potential as controllable shape-shifting nano-structures. As being constituted of tRNAs, its assembly could possibly be modulated by tRNA modifying enzymes within the cell.
Because of the rich treasure trove of small RNA motifs available in nature, RNA architectonics offers the possibility to generate any arbitrary shapes with sizes ranging between 5 and 25 nm, complementing well 2D and 3D DNA origami30,35,37,39–42, which are better suited for the design of larger nanostructures. Considering the strategic role of RNA in multiple cellular functions, synthetic approaches for generating stable 3D RNA NPs with full control over the stoichiometry of their multiple components have a broad range of potential applications in synthetic biology and nano-medicine5,7,43,44.
MATERIALS AND METHODS
RNA 3D Modeling and Sequence Design
The antiprism atomic model was manually constructed “in silico” with the software Swiss Pdbviewer by combining RNA fragments extracted from known x-ray crystallographic structures. Four Thermus thermophilus class II tRNA(Ser) (1SER) were arranged into a square-shaped tetramer and connected through four kissing-loop edges built after the DIS HIV-1 kissing complex structure (1JJM). The antiprism is formed by combining two square models through tail-tail edges resulting from the formation of 6 bps duplex between complementary tails localized at the 3' end of the variable arms of the tRNA(ser) motif. The optimal length for the tail-tail edge is 21bp. By choosing variable arms of length 5bp and 10bp, the arm 5' position can be directed inwards or outwards relative to the tRNA motif, respectively. According to consensus secondary structures specifying invariant nucleotide positions, sequences were optimized with the mFold program45 to maximize thermodynamic stability, minimize the occurrence of alternative secondary structure folds and minimize non-cognate cross-pairings through loop and tail connectors (Table S1).
RNA synthesis
All RNA molecules were prepared by in vitro T7 RNA polymerase run-off transcription from PCR generated templates as previously described9,14. RNA assembly was typically monitored by PAGE using either 32P-alpha-ATP labeled RNA or 3’-[32P]pCp labeled RNA as described previously (see Supplementary Information). RNA-5’-phosphorothioates (GMP(S)-RNA) transcripts were synthesized by T7 RNA run-off transcription in presence of guanosine mono-phosphorothioate (GMP(S)) as previously described46. GMP(S)-RNA (Bs2, Ds4, Bs4’, Ds2’) were coupled to biotin using EZ-Link PEO-Iodoacetyl Biotin (PIAB) (Pierce Biotechnology) according to the manufacturer protocol and purified by denaturing PAGE followed by HPLC (see Supplementary Information).
RNA self-assembly, native PAGE and TGGE assays
RNA antiprisms were assembled from tRNA units (Tables S1 and S2) either using one-pot (TO1–2) or stepwise (TO3–4 to TO8–9 and arrays) assembly protocols. When all mixed in one-pot at equimolar concentration [100 nM – 200 nM], heated at 90°C for 3 mins, and slowly cooled from 60°C to 10°C after addition of magnesium buffer to a final concentration (10 mM Tris-borate pH 8.2 (TB), 50 mM KCl, and 15 mM Mg(OAc)2,), tRNA units lead to octamers with ~90% yields as estimated by native PAGE assays. For stepwise assembly, tRNA squares (TS1 to TS11 in Supplementary Information) were first separately assembled at low Mg2+ concentration (10 mM Tris-borate pH 8.2 (TB), 50 mM KCl, and 0.2 mM Mg(OAc)2) according to a procedure similar to the "one pot" assembly. Sets of two squares were then mixed at higher Mg2+ concentration (2–15 mM) to assemble further into octamers by slow annealing from 50°C to 10°C (Figure 2a and Supplementary Information). Native PAGE and TGGE experiments were performed as described9,12,14. For TGGE analysis, a temperature gradient typically set up from 25 to 65°C was applied perpendicular to the electric field. Mg(OAc)2 was present at the specified concentration in both the gel and running buffer (TB).
RNase T1 probing
RNase T1 degradation was performed on 3'-32P labeled tRNA units as monomers (800 nM) or as parts of octamer (TO3–4, 100 nM) and tetramer (TS3, 200 nM) assemblies at various enzyme concentrations (0.01 u/µl to 5 u/µl) and times in presence of 15 mM Mg2+ and 25°C (Figure S2). T1 nuclease cuts single stranded RNA regions at the level of unpaired Gs. RNA degradation products were visualized on 15% / 8 M urea PAGE.
AFM characterization
RNA architectures were assembled in solution, deposited on a mica surface in 15mM Mg2+ buffer and visualized either under solution (TO3–6 and TS10–TS11 arrays) or dried under nitrogen before AFM imaging in air (TO3–4 and TO8–9/streptavidin complexes). AFM images were acquired at room temperature in tapping mode using a Multimode microscope equipped with a Nanoscope IIIa controller (Veeco, Santa Barbara) (see Supplementary Information).
Cryo-EM and single particle reconstruction
TO1–2 samples for cryo-EM were prepared as described above. Micrographs were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 kV, a nominal magnification of 80,000X, and a dose of ~30 e−/Å2. 764 images were automatically collected by the Leginon system47. All images were recorded with a Tietz F415 4k × 4k pixel CCD camera. Experimental data were processed using the Appion software package48. 23,387 particles were automatically selected taking special care to pick oblique views of the particles. After classification, the number of particles was reduced to 1,570 to ensure an approximately equal distribution of particles in all orientations (Figure S4d). The 3D reconstruction was carried out using the EMAN reconstruction package26. A resolution of 24.5 Å was determined by Fourier Shell Correlation (FSC) at a cutoff of 0.5 (see Supplementary Information).
Streptavidin-antiprism conjugates
Biotinylated antiprisms were obtained by incorporating two biotinylated-tRNA units in two opposite corners (Bs2 and Ds4 of TS9 for inward orientation and Bs4’ and Ds2’ of TS8 for outward orientation) (see Supplementary Information). "Outward" or "inward" biotinylated-antiprisms (50 nM of TO8–9) and streptavidin (strep) were mixed in varying molar ratios and incubated for 2 hrs at 25 °C before being analyzed by AFM or PAGE. Best discrimination between inward and outward antiprism-streptavidin conjugates is obtained at strep: TO8–9 molar ratios of 1:1. At higher ratio, AFM and native PAGE indicate that the number of antiprisms involved in chains decreases (data not shown).
Supplementary Material
ACKNOWLEDGEMENTS
L.J wishes to dedicate this paper to St. Luke and St. Marguerite Bourgeoys. Research funding to LJ was provided by the National Institutes of Health (R01 GM079604). Cryo-EM experiments were conducted at the National Resource for Automated Molecular Microscopy which is supported by the National Institutes of Health though the "National Center for Research Resources" P41 program (RR17573). We thank Dr. Helen Hansma for the generous use of her AFM and laboratory facilities. We also thank Dr. Clint Potter and Dr. Bridget Carragher for their scientific input regarding Cryo-EM and reconstruction.
Footnotes
AUTHOR CONTRIBUTIONS
I.S. and C.G. contributed equally to this work; I.S., C.G. and L.J. conceived and designed experiments; I.S., C.G., and L.J analyzed data; C.G. and L.J. designed the RNA particle and 3D model; I.S. performed PAGE experiments; A.C. performed AFM characterization; N.V. and E.J. performed the Cryo-EM characterization and reconstruction; L.J., C.G and I.S. co-wrote the paper; All authors discussed the results and commented on the manuscript.
ADDITIONAL INFORMATION
Supplementary information file with an extended Materials and Methods section, two supporting tables (S1 and S2) and six additional Figures (S1–S6).
REFERENCES
- 1.Gesteland RF, Cech TR, Atkins JF. The RNA world. Third Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 2.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J. Nanosci. Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Severcan I, et al. Computational and Experimental RNA Nanoparticle Design. In: Alterovitz G, Ramoni M, Mary Benson R, editors. Automation in genomics and proteomics: An engineering case based approach. New York: Wiley; 2009. pp. 193–220. [Google Scholar]
- 8.Jaeger L, Leontis NB. Tecto-RNA: One-dimensional Self-assembly through Tertiary Interactions. Angew. Chemie. Int. Ed. 2000;14:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu D, Huang LP, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J. Nanosci. Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 11.Horiya S, et al. RNA LEGO: magnesium-dependent formation of specific RNA assemblies through kissing interactions. Chem. Biol. 2003;10:645–654. doi: 10.1016/s1074-5521(03)00146-7. [DOI] [PubMed] [Google Scholar]
- 12.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 13.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koyfman AY, et al. Controlled spacing of cationic gold nanoparticles by nanocrown RNA. J. Am. Chem. Soc. 2005;127:11886–11887. doi: 10.1021/ja051144m. [DOI] [PubMed] [Google Scholar]
- 17.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene. Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, et al. Evaluation of specific delivery of chimeric phi29 pRNA/siRNA nanoparticles to multiple tumor cells. Mol. Biosyst. 2009;5:1361–1368. doi: 10.1039/b903428e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser) Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 21.Perona JJ, Hou YM. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007;46:10419–10432. doi: 10.1021/bi7014647. [DOI] [PubMed] [Google Scholar]
- 22.Hansma HG, Oroudjev E, Baudrey S, Jaeger L. TectoRNA and 'kissing-loop' RNA: atomic force microscopy of self-assembling RNA structures. J. Microsc. 2003;212:273–279. doi: 10.1111/j.1365-2818.2003.01276.x. [DOI] [PubMed] [Google Scholar]
- 23.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 24.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 25.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 27.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 28.Le Trong I, Aubert DG, Thomas NR, Stenkamp RE. The high-resolution structure of (+)-epi-biotin bound to streptavidin. Acta Crystallogr. D Biol. Crystallogr. 2006;62:576–581. doi: 10.1107/S0907444906011887. [DOI] [PubMed] [Google Scholar]
- 29.Seeman NC. DNA enables nanoscale control of the structure of matter. Q. Rev. Biophys. 2006:1–9. doi: 10.1017/S0033583505004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeman NC. An overview of structural DNA nanotechnology. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J. Am. Chem. Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 32.Liu D, Wang M, Deng Z, Walulu R, Mao C. Tensegrity: construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 33.Goodman RP, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 34.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathieu F, et al. Six-Helix Bundles Designed from DNA. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 38.Wang R, Liu W, Seeman NC. Prototyping nanorod control: A DNA double helix sheathed within a DNA six-helix bundle. Chem. Biol. 2009;16:862–867. doi: 10.1016/j.chembiol.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 41.Ke Y, et al. Scaffolded DNA Origami of a DNA Tetrahedron Molecular Container. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 42.Kuzuya A, Komiyama M. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun. (Camb) 2009:4182–4184. doi: 10.1039/b907800b. [DOI] [PubMed] [Google Scholar]
- 43.Saito H, Inoue T. RNA and RNP as new molecular parts in synthetic biology. J. Biotechnol. 2007;132:1–7. doi: 10.1016/j.jbiotec.2007.07.952. [DOI] [PubMed] [Google Scholar]
- 44.Saito H, Inoue T. Synthetic biology with RNA motifs. Int. J. Biochem. Cell Biol. 2009;41:398–404. doi: 10.1016/j.biocel.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Baudrey S, Jaeger L, Bazan GC. Characterization of tectoRNA assembly with cationic conjugated polymers. J. Am. Chem. Soc. 2004;126:4076–4077. doi: 10.1021/ja031552v. [DOI] [PubMed] [Google Scholar]
- 47.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.