Abstract
The current paper describes a line of cultured rat hepatoma cells (McA-RH7777 cells) that mimics the behavior of rat liver by producing an excess of mRNA for sterol regulatory element-binding protein 1c (SREBP-1c) as opposed to SREBP-1a. These two transcripts are derived from a single gene by use of alternative promoters that are separated by many kilobases in the genome. The high level of SREBP-1c mRNA is abolished when cholesterol synthesis is blocked by compactin, an inhibitor of 3-hydroxy-3-methylglutaryl CoA (HMG CoA) reductase that inhibits cholesterol synthesis. Levels of SREBP-1c mRNA are restored by mevalonate, the product of the HMG CoA reductase reaction, and by ligands for the nuclear hormone receptor LXR, including 22(R)-hydroxycholesterol and T0901317. These data suggest that transcription of the SREBP-1c gene in hepatocytes requires tonic activation of LXR by an oxysterol intermediate in the cholesterol biosynthetic pathway. Reduction of this intermediate lowers SREBP-1c levels, and this in turn is predicted to lower the rates of fatty acid biosynthesis in liver.
Keywords: sterol regulatory element-binding proteins, statins, oxysterols, fatty acid synthesis, nuclear receptors
Sterol regulatory element-binding proteins (SREBPs) are a family of three transcription factors that stimulate the synthesis of sterols and unsaturated fatty acids in animal cells (1). The SREBPs are synthesized as membrane-bound proteins, the active portions of which must be released proteolytically to enter the nucleus and activate transcription. Recent data indicate that SREBPs are regulated at multiple levels including (i) the rate of proteolytic cleavage (2) and (ii) the level of mRNA abundance (3–10).
The SREBPs are tripartite proteins that are synthesized on endoplasmic-reticulum (ER) membranes (1). The NH2-terminal segment of ≈480 amino acids is a transcription factor of the basic helix–loop–helix–leucine zipper family. This domain is followed by a membrane-attachment domain of ≈90 amino acids consisting of two membrane-spanning segments, separated by a short hydrophilic loop. The COOH-terminal domain of ≈600 amino acids performs a regulatory function. The NH2-terminal and COOH-terminal domains of SREBPs project into the cytosol, and only the short hydrophilic-loop projects into the ER lumen. The SREBPs form complexes with SREBP cleavage-activating protein, a polytopic membrane protein that escorts the SREBPs from the ER to the Golgi complex (11–13). There, each SREBP is cleaved sequentially by Site-1 protease and Site-2 protease, which release the NH2-terminal fragment so that it can enter the nucleus, in which it binds to sterol regulatory elements in the promoters of multiple genes encoding enzymes required for the synthesis of cholesterol and unsaturated fatty acids. When excess cholesterol accumulates in cells, the movement of SREBPs from ER to Golgi is blocked, proteolysis cannot occur, and the transcription of the target genes declines (2, 13).
The three SREBPs are encoded by two genes designated SREBP-1 and SREBP-2. The SREBP-1 gene gives rise to two transcripts, SREBP-1a and SREBP-1c (1) that are produced from alternate promoters separated by ≈14 kb in the human genome (14) and ≈10 kb in the mouse genome (G. Liang, J.L.G., and M.S.B., unpublished observations). The two promoters give rise to two different first exons that are spliced into a common second exon (14–16). Thereafter, the two transcripts are identical. The first exon of the SREBP-1a gene encodes a 42-residue acidic sequence at the NH2 terminus of the protein that renders it a strong transcriptional activator that induces mRNAs for genes encoding the enzymes for cholesterol and fatty acid biosynthesis as well as the low-density lipoprotein receptor. The SREBP-1c transcript has a shorter acidic activation domain (24 amino acids), and the protein acts selectively to increase mRNAs for enzymes involved in the synthesis of unsaturated fatty acids. The SREBP-2 transcript has a long activation domain (48 amino acids). It is a potent activator of cholesterol synthesis and a weaker activator of fatty acid biosynthesis (1, 17).
In permanent lines of nonhepatic cells such as mouse NIH 3T3 cells, human embryonic kidney (HEK)-293 cells, and Chinese hamster ovary (CHO) cells, the SREBP-1a transcript is much more abundant than the SREBP-1c transcript (16, 18; J.O., M.S.B., and J.L.G., unpublished observations). However, in livers of animals and humans, in which fatty acid synthesis is active, the SREBP-1c transcript is up to 10-fold more abundant than the SREBP-1a transcript (16).
Early evidence indicated that nuclear SREBP-1 and SREBP-2 are regulated independently in liver. Thus, when hepatic cholesterol levels were lowered in hamsters (19, 20) and mice (16) by feeding a diet containing a cholesterol synthesis inhibitor (lovastatin) and a bile acid binding resin (colestipol), the amount of nuclear SREBP-2 increased. This is the same response that is observed in cultured nonhepatic cells, and it leads to increased transcription of the genes encoding the cholesterol biosynthetic enzymes and the low-density lipoprotein receptor. In cultured cells, cholesterol deprivation also increases the amount of nuclear SREBP-1, but the opposite occurred in liver where nuclear SREBP-1 actually fell after treatment with lovastatin and Colestipol (16, 19).
A possible explanation for the discrepant regulation of SREBP-1 in liver and nonhepatic cells emerged from the recent finding that transcription from the SREBP-1c promoter but not the SREBP-1a promoter is stimulated by the nuclear hormone receptor LXR (7, 8). LXR was identified originally as an orphan nuclear hormone receptor that acts by forming heterodimers with the ubiquitous dimerizing partner, RXR (21). Janowski et al. (22) showed that LXR is activated by a variety of sterols including 22(R)-hydroxycholesterol and 24(S),25-epoxycholesterol. The latter is an intermediate in the synthesis of cholesterol (23). In mice lacking one of the two isoforms of LXR (α isoform), the level of total SREBP-1 mRNA in liver was reduced moderately (24). In mice homozygous for a knockout of both isoforms of LXR (α and β), the level of SREBP-1c mRNA in liver and intestine was barely detectable (7). In normal mice and hamsters, the level of SREBP-1c mRNA was increased when the animals were treated with T0901317, a nonsterol synthetic ligand of LXR (7, 8). This response did not occur in LXR-α/LXR-β double-knockout mice. The mRNAs for SREBP-1a and SREBP-2 were not affected by these treatments, indicating that the LXR induction is specific for the SREBP-1c promoter. Consistent with these findings, an LXR binding site was identified in the SREBP-1c promoter, and disruption of this binding site abolished the response to T0901317 (7).
The data on SREBP-1c in liver suggest a model in which excess sterols activate the transcription of the SREBP-1c gene through LXR, and this in turn leads to an increase in the synthesis of unsaturated fatty acids, which are esterified to the excess cholesterol, facilitating its storage (7). All the previous studies of LXR have been performed in livers of intact animals. Further understanding of this regulatory mechanism would be facilitated by the availability of cultured-cell lines that mimic the behavior of the intact liver. Human HepG2 cells, which often are used as a tissue-culture model of liver cells, are not useful for this purpose because they produce more SREBP-1a than SREBP-1c mRNA, and thus they resemble nonhepatic cells in tissue culture (16).
In the current paper, we describe a line of cultured rat hepatoma cells designated McA-RH7777 that mimics the liver in producing high levels of SREBP-1c mRNA and protein. We provide evidence that this production depends on the synthesis of an endogenous sterol that activates LXR. When synthesis of this sterol is blocked by an inhibitor of 3-hydroxy-3-methylglutaryl (HMG) CoA reductase, the level of SREBP-1c mRNA falls, again mimicking the behavior of the liver. The SREBP-1c mRNA is restored by treatment of cells with mevalonate, the product of the HMG CoA reductase reaction, and by ligands for LXR, 22(R)-hydroxycholesterol and T0901317.
Materials and Methods
Materials.
We obtained N-acetyl-leucinal-leucinal-norleucinal (ALLN) from Calbiochem, [α-32P]CTP [800 Ci/mmol (1 Ci = 37 GBq)] from Amersham Pharmacia, and cholesterol, 25-hydroxycholesterol, and 22(R)-hydroxycholesterol from Steraloids (Wilton, NH). Sodium mevalonate and sodium compactin were prepared as described (25, 26). The LXR-agonist T0901317 was provided kindly by Bei Shan of Tularik, Inc.. FCS (GIBCO/BRL) was delipidated by a modification of the method of Cham and Knowles (27) as described (18). In 12 preparations of delipidated FCS, the mean concentration of free fatty acids was reduced from 840 to 7.7 μM, the mean concentration of cholesterol was reduced from 280 to 7.5 μg/ml, and the mean concentration of triglycerides was reduced from 600 to 23 μg/ml.
Culture and Fractionation of McA-RH7777 Cells.
The rat hepatoma cell line, McA-RH7777 (CRL-1601; American Type Culture Collection) (28), was maintained in monolayer culture at 37°C in a 5%-CO2 incubator. On day 0, cells were set up at a density of 7 × 105 cells per 100-mm dish in medium A [DMEM (low glucose) containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate] supplemented with 10% (vol/vol) FCS. On day 2, cells were washed with PBS and refed with medium B (medium A supplemented with 10% delipidated FCS) containing the appropriate additions. After incubation for 15 h at 37°C, cells received 25 μg/ml ALLN and were incubated further for an additional 1 h. Cells were harvested and fractionated into nuclear extract and membrane pellet fractions as described (12).
SDS/PAGE and Immunoblot Analysis.
Immunoblot analysis of endogenous rat SREBP-1 and SREBP-2 was carried out with rabbit polyclonal antibodies against the NH2-terminal domains of SREBP-1 (IgG-295) and SREBP-2 (IgG-302), respectively (6). Bound antibodies were visualized with peroxidase-conjugated affinity purified donkey anti-rabbit IgG (Jackson ImmunoResearch) by using the SuperSignal CL-HRP substrate according to the manufacturer's instructions. 8% SDS gels were calibrated with prestained molecular-weight markers (Bio-Rad). Blots were exposed to Kodak X-Omat film at room temperature.
RNase Protection Assay.
A cDNA fragment for rat SREBP-2 was amplified by PCR using rat liver first-strand cDNA (CLONTECH) as template and degenerate primers derived from conserved hamster and human SREBP-2 sequences as follows: 5′-primer (sense), 5′-GAGCTGACTCTCGGGGACAT-3′; and 3′-primer (antisense), 5′-ACTGCCGCCACCACCTCCAG-3′. The amplified cDNA fragment was subcloned into pCRII (Invitrogen). The insert was sequenced to confirm its identity. After linearization of plasmid DNA with XbaI, antisense RNA was transcribed with [α-32P]CTP by using bacteriophage SP6 RNA polymerase (Ambion, Austin, TX). The rat SREBP-1 and β-actin cRNA probes were labeled as described (6). Specific activities of the transcribed RNAs were in the range of ≈2 × 109 cpm/μg for the SREBP-1 and SREBP-2 cRNAs and ≈6 × 108 cpm/μg for the β-actin cRNA.
Total RNA was isolated from duplicate dishes of monolayers of McA-RH7777 cells by using the RNA Stat-60 kit (Tel-Test, Friendswood, TX). Aliquots of total RNA (20 μg) from each sample were hybridized in the same reaction with 32P-labeled probes for rat SREBP-1, SREBP-2, and β-actin cRNAs at 68°C for 10 min by using a HybSpeed RPA kit (Ambion). After digestion with RNase A/T1, protected fragments corresponding to rat SREBP-1a (257 bp), SREBP-1c (160 bp), SREBP-2 (110 bp), and β-actin (75 bp) were separated on 8 M urea/5% polyacrylamide gels. Gels were dried and subjected to autoradiography by using reflection film and BioMax intensifying screens (Kodak) at −80°C. Relative levels of the three SREBP transcripts were quantified with a Fuji Bio-Imaging analyzer, and the data were corrected for differences in the number of 32P-labeled CTP atoms in the protected fragments of SREBP-1a, -1c, and -2 mRNAs (72, 39, and 35 cytidines, respectively).
Results
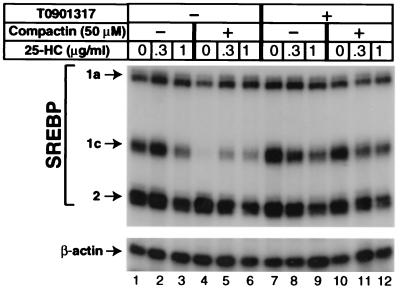
To study the regulation of SREBP-1c in hepatocytes, we selected McA-RH7777 cells, which were derived originally from a rat hepatoma (28). These cells are designated CRL-1601 in the ATCC catalogue. When the McA-RH7777 cells were incubated in delipidated FCS, the amount of SREBP-1c mRNA was ≈3-fold greater than that of SREBP-1a, as determined by an RNase protection assay (Fig. 1, lane 1) and quantified by using a PhosphorImager. This ratio approaches that of the normal rat liver (9:1), and it is strikingly different from the ratio in nonhepatic cultured cells, where the SREBP-1a mRNA markedly exceeds that of SREBP-1c (16). The amount of SREBP-2 mRNA was approximately equal to the sum of SREBP-1a and SREBP-1c (Fig. 1, lane 1). The addition of 25-hydroxycholesterol at 1 μg/ml (2.5 μM) partially reduced the amount of SREBP-1c mRNA but had no major effect on the other two transcripts (Fig. 1, lane 3).
Figure 1.
Reduction of SREBP-1c transcripts after treatment of cultured McA-RH7777 cells with compactin and reversal with LXR-agonist T0901317. On day 0, cells were set up in medium A supplemented with 10% FCS as described in Materials and Methods. On day 2, the cells were washed with PBS and switched to medium B containing 10% delipidated serum with (lanes 4–6 and 10–12) or without (lanes 1–3 and 7–9) 50 μM sodium compactin and 50 μM sodium mevalonate. Certain dishes also received a mixture of cholesterol (10 μg/ml) and increasing concentrations of 25-hydroxycholesterol (25-HC) as indicated. Some of the cells received 10 μM of T0901317. After incubation at 37°C for 16 h, cells were harvested, and 20-μg aliquots of total RNA from pooled dishes were hybridized for 10 min at 68°C to 32P-labeled cRNA probes for rat SREBP-1, SREBP-2, and β-actin as described in Materials and Methods. After digestion with RNase A/T1, protected fragments were separated by gel electrophoresis and exposed to film for 8 h at −80°C as described in Materials and Methods. The relative levels of the SREBP-1a, -1c, and -2 transcripts in lane 1 were 1.0, 2.9, and 4.6, respectively, as quantified by using a PhosphorImager as described in Materials and Methods.
When the McA-RH7777 cells were incubated in the presence of the HMG CoA reductase-inhibitor compactin, the amount of SREBP-1c mRNA fell dramatically (lane 4), and this fall was restored partially by 25-hydroxycholesterol (lanes 5 and 6). Again, there was no effect on the mRNA for SREBP-1a or SREBP-2. The addition of the LXR-agonist T0901317 raised the amount of SREBP-1c mRNA selectively (lane 7), and this result was reversed partially by 25-hydroxycholesterol (lanes 8 and 9). In the presence of T0901317, compactin failed to suppress the SREBP-1c mRNA (compare lanes 10 and 4). Under these conditions, the further addition of 25-hydroxycholesterol partially suppressed the SREBP-1c mRNA. All these changes in SREBP-1c mRNA were selective in that they did not apply to the SREBP-1a transcript, which is derived from the same gene.
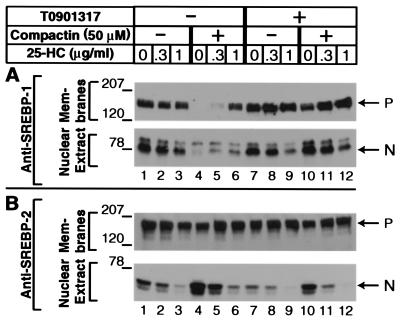
To determine the effects of these reagents on SREBP protein levels, we harvested the cells, prepared nuclear extracts and membrane pellets, and performed SDS/PAGE and immunoblotting with antibodies specific for SREBP-1 or SREBP-2. The SREBP-1 antibody does not distinguish between SREBP-1a and SREBP-1c, which differ by only a few amino acids at their NH2 termini. As shown in Fig. 2, when the McA-RH7777 cells were incubated in delipidated serum, the addition of 25-hydroxycholesterol caused a marked decrease in nuclear SREBP-2, but only a partial decrease in nuclear SREBP-1 (lane 3). The selective suppression of nuclear SREBP-2 by sterols in delipidated serum is similar to the result obtained previously in the nonhepatic human cell line HEK-293 (18). The earlier study showed that this selectivity was caused by the lack of fatty acids in the medium. Suppression of SREBP-1 cleavage but not of SREBP-2 requires the presence of both sterols and unsaturated fatty acids.
Figure 2.
Effect of LXR activation and cholesterol synthesis inhibition on membrane and nuclear SREBPs in McA-RH7777 cells. On day 0, cells were set up and treated identically to those described in Fig. 1. On day 3, after a 16-h incubation at 37°C, cells received ALLN at 25 μg/ml and were harvested 1 h later. Aliquots of membrane and nuclear extract fractions (for A and B: Upper, 60 μg protein per lane; Lower, 80 μg protein/lane) were subjected to SDS/PAGE. Immunoblot analysis was carried out with 5 μg/ml rabbit anti-rat SREBP-1 (A) or anti-rat SREBP-2 (B). Filters were exposed for 5–10 s at room temperature. P, SREBP precursor forms; N, nuclear cleaved forms of SREBP-1c (A) and SREBP-2 (B).
The addition of compactin eliminated both the precursor and nuclear forms of SREBP-1 (Fig. 2A, lane 4), which is consistent with the reduction in the SREBP-1c mRNA (Fig. 1, lane 4). The 25-hydroxycholesterol partially restored the SREBP-1 precursor, and it also increased the nuclear form of SREBP-1 (Fig. 2A, lane 6), which is consistent with its ability to raise the SREBP-1c mRNA (Fig. 1, lane 6). In direct opposition to its negative effects on SREBP-1, compactin raised the amount of nuclear SREBP-2 (Fig. 2B, lane 4) presumably by enhancing proteolytic processing, and this effect was reversed by 25-hydroxycholesterol (Fig. 2B, lane 6). In the absence of compactin, T0901317 had little effect on SREBP-1 levels, and it did not affect the response to 25-hydroxycholesterol (Fig. 2A, compare lanes 7–9 with lanes 1–3). However, T0901317 did prevent the suppression of SREBP-1 levels by compactin (Fig. 2A, compare lanes 10 and 4) as expected from its ability to prevent the compactin-mediated suppression of the SREBP-1c mRNA (see Fig. 1). T0901317 had little effect on SREBP-2 either in the absence or presence of compactin (Fig. 2B, compare lanes 7 and 1; lanes 10 and 4).
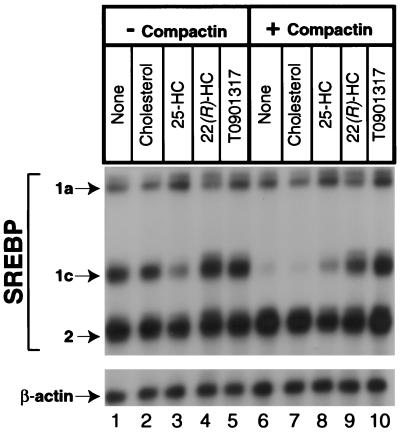
To test further the hypothesis that T0901317 is acting through LXR, we compared its effects with those of 25-hydroxycholesterol, which is a weak ligand for LXR, and 22(R)-hydroxycholesterol, which is a strong ligand (22). In the absence of compactin, 25-hydroxycholesterol partially reduced the amount of SREBP-1c mRNA (Fig. 3, lane 3) as seen in Fig. 1. The 22(R)-Hydroxycholesterol and T0901317 had no such effect (lanes 4 and 5). Cholesterol, which is not a ligand for LXR, also had no effect (lane 2). As expected, compactin markedly lowered the amount of SREBP-1c mRNA (lane 6), and this effect was reversed by T0901317 (lane 10). It also was reversed strongly by 22(R)-hydroxycholesterol (lane 9) and only slightly by 25-hydroxycholesterol (lane 8). Cholesterol had no effect (lane 7). None of these reagents had a significant effect on the SREBP-1a transcript or on the SREBP-2 transcript.
Figure 3.
Induction of SREBP-1c transcripts in McA-RH7777 cells after treatment with LXR ligands. On day 0, cells were set up as described in Fig. 1. On day 2, the cells were washed with PBS and switched to medium B with (lanes 6–10) or without (lanes 1–5) 50 μM sodium compactin and 50 μM sodium mevalonate. The indicated sterol or the LXR-agonist T0901317 was added at a final concentration of 10 μM. After incubation for 16 h at 37°C, cells were harvested, and aliquots of total RNA (20 μg) from pooled dishes were hybridized for 10 min at 68°C to 32P-labeled cRNA probes for rat SREBP-1, SREBP-2, and β-actin. Protected fragments were separated by gel electrophoresis and exposed to film for 8 h at −80°C as described in Materials and Methods. 25-HC, 25-hydroxycholesterol; 22(R)-HC, 22(R)-hydroxycholesterol.
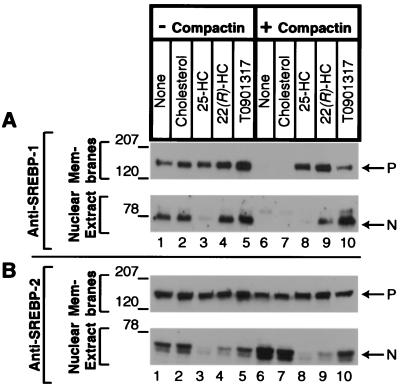
The sterol specificity observed at the mRNA level was reflected at the protein level (Fig. 4). In the absence of compactin, 25-hydroxycholesterol lowered the amount of nuclear SREBP-1 apparently through its ability to block proteolytic processing of the precursor. Cholesterol, 22(R)-hydroxycholesterol, and T0901317 had no such effect. In the presence of compactin, the SREBP-1 precursor and nuclear forms were markedly decreased, and this effect was reversed by 22(R)-hydroxycholesterol and T0901317 (lanes 9 and 10). The 25-hydroxycholesterol restored the SREBP-1 precursor, but there was no nuclear form (lane 8), apparently because 25-hydroxycholesterol blocked proteolytic processing. (Note that the concentration of 25-hydroxycholesterol used in this experiment was 4-fold higher than the highest used in Fig. 2). Cholesterol had no effect. With respect to SREBP-2, 25-hydroxycholesterol reduced the nuclear form in the absence and presence of compactin (lanes 3 and 8). T0901317 had no major effect on SREBP-2 (lanes 5 and 10). The 22(R)-hydroxycholesterol reduced nuclear SREBP-2 partially in the absence (lane 4) and presence (lane 9) of compactin, apparently because this sterol has some ability to block proteolytic processing.
Figure 4.
Effect of various LXR agonists on SREBP processing in McA-RH7777 cells in the absence or presence of compactin. On day 0, cells were set up and treated identically to those described in Fig. 3. On day 3, after a 16-h incubation at 37°C, the cells received ALLN at 25 μg/ml and were harvested 1 h later. Membrane and nuclear extract fractions were prepared, and aliquots (80 μg protein) were subjected to SDS/PAGE. Immunoblot analysis was carried out with 5 μg/ml rabbit anti-rat SREBP-1 (A) or anti-rat SREBP-2 (B). Filters were exposed for 1–10 s at room temperature. P, SREBP precursor forms; N, nuclear cleaved forms of SREBP-1c (A) and SREBP-2 (B); 25-HC, 25-hydroxycholesterol; 22(R)-HC, 22(R)-hydroxycholesterol.
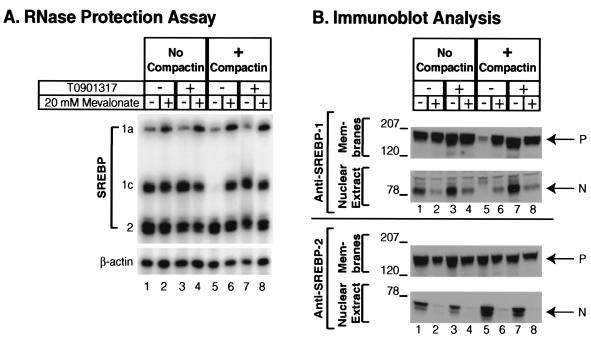
Up to this point, the data are consistent with the possibility that the production of SREBP-1c mRNA requires an endogenous LXR ligand that is produced through the cholesterol biosynthetic pathway. Compactin blocks the synthesis of this ligand and therefore leads to a fall in SREBP-1c mRNA levels, which are restored when cells are supplied with an exogenous ligand, either T0901317 or 22(R)-hydroxycholesterol. If this hypothesis is true, then the compactin-mediated fall in SREBP-1c mRNA should be abrogated if the cells are treated simultaneously with mevalonate, which is the product of the HMG CoA reductase reaction (29). The experiment of Fig. 5A shows that the SREBP-1c mRNA was reduced by compactin (lane 5), and this reaction was prevented when the medium contained 20 mM mevalonate (lane 6). The effect of mevalonate was as great as that of T0901317 (lane 7). The concentration of mevalonate used in this experiment was high, because cultured cells have an inefficient process for mevalonate uptake (30).
Figure 5.
Mevalonate overcomes the inhibitory effect of compactin on SREBP-1c mRNA (A) and protein (B) levels in McA-RH7777 cells. On day 0, cells were set up in medium A supplemented with 10% FCS at 7 × 105 cells per 100-mm dish. On day 2, cells were washed with PBS and switched to medium B with (lanes 5–8) or without (lanes 1–4) 50 μM sodium compactin and 50 μM sodium mevalonate. In addition, cells were treated with 20 mM sodium mevalonate (lanes 2 and 6), 1 μM of the LXR-agonist T0901317 (lanes 3 and 7), or a combination of both (lanes 4 and 8). (A) After a 16-h incubation at 37°C, cells were harvested, and aliquots of total RNA (20 μg) from pooled dishes were hybridized for 10 min at 68°C to the indicated 32P-labeled cRNA probes. RNase-protected fragments were separated by gel electrophoresis and exposed to film for 8 h at −80°C. (B) After a 16-h incubation at 37°C, cells received 25 μg/ml ALLN and were harvested 1 h later. Membranes and nuclear extract fractions were prepared, and aliquots (54 μg protein per lane) were subjected to SDS/PAGE. In A and B, immunoblot analysis was carried out with 5 μg/ml rabbit anti-rat SREBP-1 (Upper) or anti-rat SREBP-2 (Lower). Filters were exposed for 1 s at room temperature. P, SREBP precursor forms; N, nuclear cleaved forms of SREBP-1c and SREBP-2.
At the protein level, mevalonate abolished processing of SREBP-2 and partially inhibited processing of SREBP-1 (Fig. 5B, lanes 2 and 4). In the presence of compactin, mevalonate restored the SREBP-1 precursor (lanes 5 and 6), but it did not restore fully the nuclear form presumably because of a block in proteolytic processing. The mevalonate-mediated block in SREBP-1 processing was evident also in the presence of compactin plus T0901317 (lanes 7 and 8). T0901317 restored both the precursor and nuclear forms of SREBP-1 (lane 7), and the further addition of mevalonate selectively reduced the nuclear form (lane 8).
Discussion
The current results provide evidence that the basal level of transcription of the SREBP-1c gene in cultured rat hepatoma cells depends on tonic activation of LXR by an endogenously synthesized sterol. When synthesis of this sterol is blocked by the HMG CoA reductase inhibitor compactin, the SREBP-1c transcript falls markedly, and it is restored when the cells are supplied with the LXR-ligands T0901317 or 22(R)-hydroxycholesterol. It is restored also by mevalonate, the product of the HMG CoA reductase reaction. The nature of the endogenous LXR ligand is unknown but a candidate is 24(S),25-epoxycholesterol, which is an intermediate in cholesterol synthesis (23) and a potent LXR ligand (22). Although the current experiments were performed with a single line of cultured rat hepatocytes (McA-RH7777), qualitatively similar results have been obtained with another line of cultured rat hepatoma cells (FT02B cells; unpublished observations).
The effects of 25-hydroxycholesterol on SREBP-1c mRNA levels were complex. Three responses were observed: (i) when administered alone, 25-hydroxycholesterol decreased the SREBP-1c mRNA partially (Fig. 1, lanes 1–3); (ii) when administered to compactin-treated cells, 25-hydroxycholesterol increased the SREBP-1c mRNA, partially overcoming the compactin-mediated decrease (Fig. 1, lanes 4–7); and (iii) when administered in the presence of T0901317, 25-hydroxycholesterol blocked the elevation in SREBP-1c mRNA (Fig. 1, lanes 7–9 and 10–12). All three observations can be explained by the previous finding that 25-hydroxycholesterol is a weak activator of LXR (22). At saturation, 25-hydroxycholesterol produces only one-sixth of the activation that is produced by the presumed endogenous ligand 24(S),25-epoxycholesterol (22). In the intact cell, 25-hydroxycholesterol might displace the more active endogenous ligand from LXR and thereby reduce the activation of the SREBP-1c promoter. Because 25-hydroxycholesterol does have some agonist activity, it would restore SREBP-1c mRNA partially when synthesis of the endogenous ligand is blocked by compactin. Finally, 25-hydroxycholesterol should compete with T0901317 for binding to LXR thereby lowering activation in the presence of this compound.
The results in cultured hepatoma cells provide a likely explanation for the previous finding that nuclear SREBP-1 declines in hamster (19) and mouse (20) liver when the livers are depleted of sterols by administration of an HMG CoA reductase inhibitor and a bile acid binding resin. They also explain the finding that SREBP-1c transcripts and nuclear SREBP-1c levels are reduced in livers of mice with a combined knockout of the LXR-α and -β genes (7).
The current results also provide a potential explanation for the fall in plasma triglycerides that occurs when humans are treated with high doses of potent HMG CoA reductase inhibitors (31). If these inhibitors reduce SREBP-1c levels in human livers, then the rate of fatty acid synthesis would be expected to fall, which in turn may lead to a fall in the triglyceride content of newly secreted very low-density lipoproteins (32). It should be possible to test this hypothesis in experimental animals.
Acknowledgments
We thank David Mangelsdorf for helpful comments and critical review of the manuscript, Clinton Steffey and Angela Dietrich for excellent technical assistance, Lisa Beatty, Christine Alvares, and Linda Donnelly for invaluable help with tissue culture, and Bei Shan of Tularik, Inc. for providing T0901317. This work was supported by research funds from the National Institutes of Health (HL20948) and the Perot Family Foundation. R.A.D.-B. is the recipient of a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund.
Abbreviations
- SREBP
sterol regulatory element-binding protein
- HMG
3-hydroxy-3-methylglutaryl
- ALLN
N-acetyl-leucinal-leucinal-norleucinal
References
- 1.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foretz M, Guichard C, Ferre P, Foufelle F. Proc Natl Acad Sci USA. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimomura I, Bashmakov Y, Ikemoto S, Horton J D, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J-M A, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz J R, Tu H, Luk A, Repa J J, Medina J C, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf D J, et al. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasty A H, Shimano H, Yahagi N, Amemiya-Kudo M, Perrey S, Yoshikawa T, Osuga J-I, Okazaki H, Tamura Y, Iizuka Y, et al. J Biol Chem. 2000;275:31069–31077. doi: 10.1074/jbc.M003335200. [DOI] [PubMed] [Google Scholar]
- 10.Osborne T F. J Biol Chem. 2000;276:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 11.Sakai J, Nohturfft A, Goldstein J L, Brown M S. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 12.DeBose-Boyd R A, Brown M S, Li W-P, Nohturfft A, Goldstein J L, Espenshade P J. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 13.Nohturfft A, Yabe D, Goldstein J L, Brown M S, Espenshade P J. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 14.Hua X, Wu J, Goldstein J L, Brown M S, Hobbs H H. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 15.Miserez A R, Cao G, Probst L, Hobbs H H. Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai J, Guryev O, Brown M S, Goldstein J L. J Biol Chem. 1998;273:26138–26148. doi: 10.1074/jbc.273.40.26138. [DOI] [PubMed] [Google Scholar]
- 18.Hannah, V. C., Ou, J., Luong, A., Goldstein, J. L. & Brown, M. S. (2001) J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Sheng Z, Otani H, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimomura I, Bashmakov Y, Shimano H, Horton J D, Goldstein J L, Brown M S. Proc Natl Acad Sci USA. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 22.Janowski B A, Grogan M J, Jones S A, Wisely G B, Kliewer S A, Corey E J, Mangelsdorf D J. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer T A, Gayen A K, Phirwa S, Nelson J A, Taylor F R, Kandutsch A A, Erickson S K. J Biol Chem. 1985;260:13391–13394. [PubMed] [Google Scholar]
- 24.Peet D J, Turley S D, Ma W, Janowski B A, Lobaccaro J-M A, Hammer R E, Mangelsdorf D J. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 25.Brown M S, Faust J R, Goldstein J L, Kaneko I, Endo A. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 26.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 27.Cham B E, Knowles B R. J Lipid Res. 1976;17:176–181. [PubMed] [Google Scholar]
- 28.Becker J E, de Nechaud B, Potter V R. In: Onco-Developmental Gene Expression. Fishman W H, Sell S, editors. New York: Academic; 1976. pp. 259–270. [Google Scholar]
- 29.Goldstein J L, Brown M S. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim C M, Goldstein J L, Brown M S. J Biol Chem. 1992;267:23113–23121. [PubMed] [Google Scholar]
- 31.Blumenthal R S. Am Heart J. 2000;139:577–583. doi: 10.1016/s0002-8703(00)90033-4. [DOI] [PubMed] [Google Scholar]
- 32.Horton J D, Shimano H, Hamilton R L, Brown M S, Goldstein J L. J Clin Invest. 1999;103:1067–1076. doi: 10.1172/JCI6246. [DOI] [PMC free article] [PubMed] [Google Scholar]