Abstract
A conserved mitogen-activated protein kinase (MAPK) cascade orthologous to the mating/filamentation MAPK pathway in yeast is required for fungal pathogenicity on plants. One of the key targets of this signaling pathway is the homeodomain transcription factor Ste12. Mutational analysis of ste12 orthologues in a variety of plant pathogenic fungi suggests that Ste12 functions as a master regulator of invasive growth. In this mini-review we highlight recent progress in understanding the role of Ste12 in filamentous fungi and discuss future challenges of unravelling the mechanisms by which Ste12 controls fungal virulence downstream of the Pathogenicity MAPK cascade.
Key words: Ste12, signaling, virulence, plant pathogen, fungal pathogenicity, MAPK cascade
Introduction
Virulence in plant pathogenic fungi is controlled by a network of cellular pathways that respond to signals encountered during host infection. In spite of the broad diversity of fungal lifestyles and modes of infection, the signaling components that regulate pathogenic development are largely conserved. MAPK cascades have attracted considerable attention, because their core elements are essential for virulence in a wide array of fungal pathogens of plants and mammals.1–4
MAPK cascades are characterized by a three-tiered signaling module comprising a MAPK kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK) and the MAPK which is activated by dual phosphorylation of conserved threonine and tyrosine residues within the activation loop.5 Fungal MAPK cascades are triggered by an array of stimuli and regulate a wide range of processes including cell cycle, reproduction, morphogenesis, stress response and virulence.2,6 The model fungus Saccharomyces cerevisiae has five MAPK cascades controlling mating, filamentous growth, cell wall integrity, cell adaptation to stress and sporulation.6 Several of these MAPK cascades have been associated with virulence in fungal pathogens.4,7 This mini-review focuses on the so-called Pathogenicity MAPK (PMK) cascade which has a broadly conserved role in plant infection, with particular emphasis on the homeodomain transcription factor (TF) Ste12, an emerging key regulator of invasive growth in fungi.
Two Saccharomyces cerevisiae MAPK Cascades Recruit the Homeodomain Transcription Factor Ste12
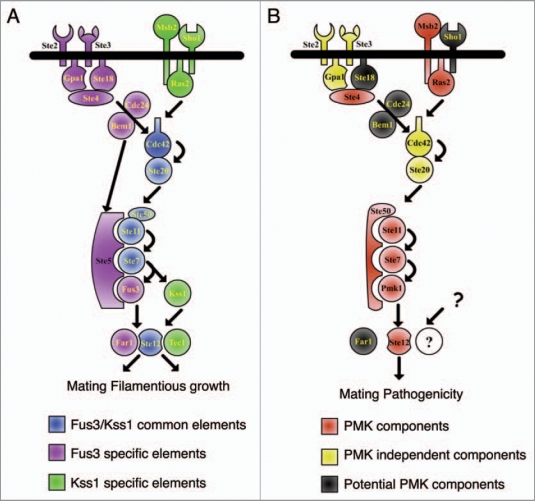
In S. cerevisiae, mating and filamentous growth are controlled by two structurally related MAPKs, Fus3 and Kss1, respectively (Fig. 1).8 Despite sharing many pathway components, the two MAPKs have distinct activation mechanisms and signaling outputs.6 Fus3 signaling is initiated by pheromone binding to the cognate G protein-coupled receptors Ste2 and Ste3, triggering dissociation of the G α subunit Gpa1 from the G βγ subunits Ste4 and Ste18 and allowing signal transmission to the guanine nucleotide exchange factor Cdc24.9 The Kss1-mediated filamentation response is initiated by the mucin-type protein Msb2 and the tetraspan protein Sho1,10 and propagated through the small GTP-binding protein Ras2.11 Both pathways eventually converge on the small Rho-type G protein Cdc42 and the PAK-like protein kinase Ste20.6 The latter activates the MAPK module composed of MAPKKK Ste11, MAPKK Ste7 and MAPKs Fus3 or Kss1.6,9 Correct activation of Fus3 by pheromone requires the scaffold protein Ste5 which recruits the Ste11-Ste7-Fus3 complex to the plasma membrane,12 while activation of Kss1 by Ste7 does not require Ste5.13 Phosphorylated Fus3 and Kss1 MAPKs stimulate a number of downstream TFs that function as regulators of pathway-specific genes (Fig. 1). Among these, the homeodomain TF Ste12 acts as a central node in both mating and invasive growth response.14 The dual function of this Ste12 has received considerable attention as a basic model for understanding how eukaryotic cells maintain signaling specificity by discriminating between different inputs, i.e., pheromone and nutrient limitation, to generate the appropriate outputs, mating and filamentation, without allowing leakage from one pathway into the other.15 These studies revealed that Ste12 is under complex regulation involving several regulatory proteins and co-factors that are tightly controlled by each MAPK.
Figure 1.
Schematic view of the Fus3/Kss1 MAPK cascade in S. cerevisiae (A) and of the orthologous Pathogenicity MAPK cascade in plant pathogenic fungi (B).
S. cerevisiae Ste12 is a 688-amino acid protein characterized by a helix-turn-helix DNA-binding homeodomain at the N terminal region. Depending on the interaction partner, the Ste12 homeodomain binds either to pheromone response elements (PREs; TGAAACR) in mating gene promoters16 or to filamentation response elements (FREs) on filamentous growth-specific target genes.14 The homeodomain of Ste12 is also required for binding to the regulatory protein Dig2 and the cofactor Tec1.17,18 No functional domain was described so far in the central and C terminal regions of Ste12, although the latter was found to promote formation of the Ste12 homodimer, as well as binding to cofactors such as the TF Mcm1 or the negative regulator Dig1.18,19 S. cerevisiae Ste12 is a target of several protein kinases including the MAPKs Fus3 and Kss1, as well as the cyclin-dependent kinase Srb10.15,20 Although phosphorylation is not essential for transcriptional activity of Ste12, it impacts on its regulation by controlling protein stability.19–21
Ste12 was recently shown to be present in two protein complexes composed of Ste12, Dig1 and either Dig2 or Tec1.17 The Dig2/Ste12/Dig1 complex mainly binds to the PRE motif, whereas the Tec1/Ste12/Dig1 complex preferentially targets the FRE motif.17 In the absence of stimulating signal, both complexes remain inactive by means of the repressors Dig1 and Dig2.17 Upon pheromone stimulation, Fus3 MAPK phosphorylates members of the Ste12 complex as well as the Far1,15 protein, leading to dissociation and/or conformational change of the complex and to relief of Ste12 repression.22 Fus3-mediated phosphorylation also promotes Tec1 degradation, thus ensuring that only mating-specific genes are transcribed in this context.22 Activated Ste12 binds to mating gene-specific promoters either as a homodimer or as a heterodimer with Mcm1, stimulating transcription of mating genes and of FAR1 which in turn induces cell cycle arrest in G1, essential for mating. Under prolonged pheromone stimulation, phosphorylation of Ste12 by Fus3 can accelerate Ste12 turnover in a Far1-dependent manner by targeting ubiquitination and degradation of Ste12, providing an additional layer of control.21
Kss1 MAPK, activated through nutrient limitation, also can phosphorylate Ste12, Dig1 and Dig2, but not Far1 or Tec1. This results in the activation of the protein complex by dissociation of the inhibitor Dig2, leading to transcription of filamentation genes such as TEC1, itself. The increase in cellular Tec1 levels favors the formation of a Tec1/Ste12 heterodimer,17 resulting in even higher transcript levels of filamentous growth genes. Intriguingly, transcriptional activation of filamentation-specific genes by Ste12 is also controlled by the nutrientsensing cyclin-dependent kinase Srb10. Cellular levels of Srb10 were depleted under nutrient-limiting conditions but increased under nutrient-rich conditions. Accumulating Srb10 phosphorylates two serine residue of Ste12 (Ser261 and Ser451), leading to destabilization and inactivation of Ste12 under nutrient rich conditions.20
Conserved Role of the Pathogenicity MAPK Cascade in Plant Pathogenic Fungi
The Fus3/Kss1 MAPK cascade is highly conserved in filamentous fungi. A recent comparative genomic analysis in nine fungal species with different lifestyles ranging from saprophytic to pathogenic on human or plants, and including both asco- and basidiomycetes, revealed structural conservation of all Fus3/Kss1 pathway components of S. cerevisiae, except Ste5, Dig1 and Dig2.3 One of the major differences between filamentous fungi and yeast is the presence of a single MAPK instead of the two MAPKs Fus3 and Kss1. At present it is not clear how a single MAPK integrates the different signaling functions carried out by Fus3 and Kss1 in S. cerevisiae.
The essential role of a Fus3/Kss1 MAPK orthologue in plant infection was first described in the rice blast fungus Magnaporthe oryzae (Fig. 2). Mutants lacking the PMK1 MAPK failed to cause disease symptoms on rice leaves.7 PMK orthologues were subsequently found to be required for virulence in a large number of biologically and taxonomically diverse plant pathogens, suggesting an ancient evolutionary role of this MAPK cascade in fungal pathogenicity on plants.4 Loss of PMK orthologues invariably leads either to a drastic reduction or complete loss of pathogenicity, associated with a strong defect in penetration and invasive growth.4 The penetration defect was initially attributed to the inability of the MAPK mutants to differentiate specialized infection structures called appressoria.4 However, M. oryzae pmk1 mutants were unable to colonize rice plant tissue even when inoculated through wound sites,7 suggesting that the role of this MAPK in pathogenicity extends beyond appressorium differentiation. Even in non-appressorium-forming pathogens such as the soilborne vascular wilt fungus Fusarium oxysporum, deletion of the PMK orthologue Fmk1 leads to complete loss of pathogenicity and defects in virulence-related functions such as adhesion to tomato roots, secretion of pectinolytic enzymes, invasive growth and production of wilt symptoms (Fig. 2).23,24
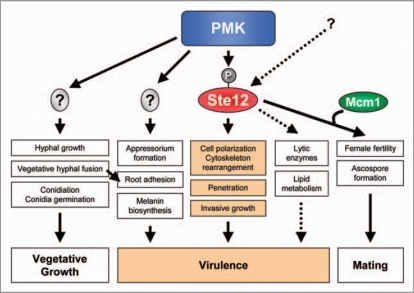
Figure 2.
Model for the role of Ste12 in fungal plant pathogens. Ste12 regulates mating, penetration and invasive growth downstream of PMK, whereas the remaining PMK-controlled functions are mediated by other unknown transcriptional regulators. Additional pathways may also regulate Ste12 activity. Invasive growth and penetration functions mediated by Ste12 play key roles in fungal virulence on plants.
Ste12 is a Regulator of Fungal Mating and Virulence
Ste12 proteins from filamentous fungi share structural features with their orthologue in S. cerevisiae. Similar to yeast Ste12, they are around 700 amino acids in length and contain a homeodomain at the N terminus.3 Indeed, the ste12 gene from the plant pathogen Colletotrichum lindemuthianum was able to restore invasive growth when expressed in a yeast ste12 mutant.25
Similar to S. cerevisiae, Ste12 orthologues of filamentous fungi are required for mating. Loss of Ste12 led to defects in the sexual cycle of the plant pathogens Cryphonectria parasitica26 and Botrytis cinerea,27 the human pathogens Candida albicans28 and Cryptococcus neoformans,29 as well as the saprophytes Neurospora crassa,30 Aspergillus nidulans31 and Sordaria macrospora.32 By contrast, ste12 mutants were generally not affected in vegetative growth and conidiation, except for B. cinerea, N. crassa and Mycosphaerella graminicola.27,30,33,34 These results suggest that the essential role of Ste12 in mating is conserved between S. cerevisiae and filamentous fungi (Fig. 2).
The structural conservation of MAPK cascades between S. cerevisiae and filamentous fungal pathogens suggested a possible role of Ste12 as a downstream target of the Pathogenicity MAPK. In support of this idea, mutants lacking Ste12 were either nonpathogenic25,35–37 or strongly attenuated in virulence26,27,33,38 in all plant pathogens studied so far. Similarly to deletion of PMK, loss of Ste12 led to defects in penetration and invasive growth.25,36,37,39 Collectively, the results from these studies provide strong circumstantial evidence suggesting that Ste12 is a direct downstream target of the PMK MAPK pathway (Fig. 2). However, this hypothesis has not been confirmed yet experimentally.
Phenotypes of ste12 mutants are generally less pleiotropic than those of the MAPK mutants, suggesting that Ste12 only regulates a subset of the MAPK-dependent virulence functions (Fig. 2). For example, MAPK-deficient strains of the airborne plant pathogens M. oryzae,35 C. lagenarium,37 C. lindemuthianum25 and B. cinerea27 were unable to differentiate appressoria, whereas ste12 mutants still produced these infection structures. However, ste12 mutant appressoria failed to develop penetration pegs and to invade the host tissue, suggesting a role of Ste12 in polarity establishment during invasive growth.37,39 Similarly, Δste12 mutants of F. oxysporum were still able to undergo vegetative hyphal fusion, adhere to host roots or secrete pectinolytic cell-wall degrading enzymes, a set of virulence-related functions controlled by the MAPK Fmk1. However, both Δste12 and Δfmk1 mutants failed to penetrate cellophane membranes, colonize living plant tissue and kill tomato plants.36 These results suggest that Ste12 controls a major pathogenicity function, invasive growth, while additional TFs regulate other MAPK-dependent processes such as appressorium differentiation, hyphal fusion or root adhesion.36 Besides its major role downstream of the Pathogenicity MAPK, Ste12 may also be targeted by additional upstream pathways. For example, F. oxysporum Δfmk1Δste12 double mutants showed a partly restored capacity to secrete pectinases compared to Δfmk1 single mutants, suggesting a possible repressing role of Ste12.36
Future Challenges: Understanding the Role of Ste12 in Plant Pathogenicity
Increasing evidence suggests that the homeodomain TF Ste12 controls two complex developmental processes in filamentous fungi, mating and invasive growth. This role parallels that in S. cerevisiae where Ste12 functions as the key TF downstream of the mating and filamentous growth MAPK pathways.8 However, beyond this apparent analogy many open questions remain. S. cerevisiae Ste12 receives and processes specific signaling inputs from two different upstream MAPKs, Fus3 and Kss1, whereas only one orthologous MAPK is present in filamentous fungi. How does this a single MAPK integrate the different upstream signal inputs through Ste12 in a way that ensures specificity of the transcriptional readouts? In yeast, a key mechanism ensuring specificity between mating and invasive growth responses involves selective interaction of Ste12 with different cofactors such as Mcm1, Far1 or Tec1. Some of these cofactors, such as the MADS box TF Mcm1, are conserved in filamentous fungi.3 Indeed, Mcm1 was shown to interact with the Ste12 homeodomain in S. macrospora.32 By contrast, most filamentous fungi, including plant pathogens, lack structural orthologues of Tec1, the key interaction partner of Ste12 for filamentous growth in yeast. Similarly, Dig1 and Dig2, two negative regulators of Ste12 activity in S. cerevisiae were not found in the genomes of filamentous fungi.3 It remains to be determined if other TFs or regulatory partners have taken over the role of these proteins in mediating Ste12 activity during invasive growth in fungal plant pathogens.
In yeast Ste12 activity is largely controlled at the post-translational level via phosphorylation, protein stability and interaction with other partner proteins. It is likely that regulation of Ste12 functions in a similar way in filamentous fungi, yet there are currently few experimental data available. Ste12 proteins from yeast and filamentous fungi display a number of structural discrepancies that may affect the regulatory mechanisms. For example, Ste12 proteins from filamentous fungi contain two C2H2 zinc fingers in the C terminal region, which are lacking in yeast. The exact role of the zinc finger domain is still unclear. In the Ste12 orthologue of M. oryzae, this region was essential for virulence.39 However, the zinc finger domain appears to be dispensable for DNA binding, in contrast to the homeodomain which mediates both DNA binding and interaction with other TFs such as MCM1.32 The central region of Ste12 proteins from filamentous fungi is also divergent from yeast, since it contains several stretches of highly conserved amino acid residues that are absent in S. cerevisiae Ste12.34
Phosphorylation provides an important mechanism for regulation of Ste12. However, a number of key phosphorylation sites identified in S. cerevisiae Ste12 are not conserved in filamentous fungi.20,31,35,40 Inspection of the Ste12 amino acid sequences in filamentous fungi showed the presence of predicted phosphorylation sites for different kinases, including cAMP- and cGMPdependent kinases, PKC and casein II protein kinase. Several of these putative phosphorylation sites were conserved among Ste12 orthologues of plant pathogenic ascomycetes suggesting that they may be involved in regulation of Ste12 activity.3,34 So far, however, site directed mutagenesis of predicted phosphorylation sites in M. oryzae indicated that they were dispensable for virulence.39 Clearly, more studies are needed to define the role of phosphorylation, either by the MAPK or by other protein kinases, as a mechanism of controlling Ste12 activity in filamentous fungi.
Alternatively, regulation of Ste12 activity may also occur at the post-transcriptional level. In C. lindemuthianum and B. cinerea, the presence of an alternative splicing form of ste12 was described, in which the third exon is skipped leading to a truncated version of the protein that lacks the second zinc finger domain.25,27 While this truncated version was able to complement ste12-deficient mutants,27 its overexpression in a wild type strain led to reduced virulence indicating a possible repressing function of the truncated Ste12 version.25,27 Interestingly, the exon/intron structure in this region of the ste12 gene is highly conserved between filamentous fungi, suggesting that the mode of control by alternative splicing might also be functional in other species. Presence of both the full length and the truncated transcript version of F. oxysporum ste12 was detected during growth in vitro and in planta (Lopez-Berges et al., unpublished). However, only the full length transcript form was so far detected in M. oryzae and Mycosphaerella graminicola despite the fact that M. oryzae has the potential to produce an alternatively spliced transcript according to its DNA intron/exon structure.33 The relevance of alternative splicing as a major mechanism for controlling Ste12 activity needs to be corroborated by further studies.
In S. cerevisiae a large number of Ste12 target genes, were identified by different genome-wide approaches, transcriptomics41 and chromatin-immunoprecipitation.42 These include both matingspecific and filamentation-specific targets.42 By contrast, only few Ste12 target genes have so far been identified in plant pathogens. A recent microarray analysis in C. parasitica identified 152 genes that were either down or upregulated in a ste12 deletion mutant, but their role in plant infection is not known. Interestingly, a significant number of Ste12-regulated genes were also responsive to hypovirus infection, suggesting that Ste12 may be one of the hypovirus targets.26 In C. lindemuthianum, a comparative analysis of cell surface proteins in the wild type and the ste12 mutant led to the identification of a major protein of unknown function which was absent in the mutant.25 The availability of genomewide arrays for an increasing number of fungal phytopathogens will contribute to the identification of new transcriptional targets controlled by Ste12 during plant infection.
Outlook
Plant pathogenic fungi have evolved a stunning variety of infection mechanisms to achieve colonization of their hosts. Strikingly, these diverse plant pathogens all require a highly conserved MAPK pathway, whose terminal component is the TF Ste12, making this signaling cascade an attractive target for antifungal control strategies. While Ste12 has emerged as a master switch of fungal virulence, little is known on the regulatory mechanisms and the transcriptional targets of this TF in filamentous fungi. The complexity of its regulation and its conserved role as a key regulator of pathogenicity will undoubtedly attract further research efforts. Unravelling Ste12 function in invasive growth should significantly advance our understanding on how fungal pathogens cause disease on plants.
Acknowledgements
N.R. acknowledges financial support by the Marie Curie Training Network SIGNALPATH (MRTN-CT-2005-019277). Research in the group of A.D.P. is supported by projects SIGNALPATH (MRTN-CT-2005-019277) and ARIADNE (FP7-PEOPLEITN-2008-237936) from the European Commission and by grant BIO2007-62661 from the Spanish Ministerio de Ciencia e Innovación (MICINN).
Abbreviations
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- MAPKKK
MAPK kinase kinase
- TF
transcription factor
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/11908
References
- 1.Lee N, D’Souza CA, Kronstad JW. Of smuts, blasts, mildews and blights: cAMP signaling in phytopathogenic fungi. Annu Rev Phytopathol. 2003;41:399–427. doi: 10.1146/annurev.phyto.41.052002.095728. [DOI] [PubMed] [Google Scholar]
- 2.Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC, Wang P, et al. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, Huguet R, et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 2009;46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhao X, Mehrabi R, Xu JR. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell. 2007;6:1701–1714. doi: 10.1128/EC.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 6.Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 7.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 8.Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Dohlman HG. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science. 2004;306:1508–1509. doi: 10.1126/science.1104568. [DOI] [PubMed] [Google Scholar]
- 10.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, Neal C, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elion EA. Routing MAP kinase cascades. Science. 1998;281:1625–6. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- 14.Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 15.Elion EA, Qi M, Chen W. Signal transduction. Signaling specificity in yeast. Science. 2005;307:687–688. doi: 10.1126/science.1109500. [DOI] [PubMed] [Google Scholar]
- 16.Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- 17.Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson KA, Nelson C, Tai G, Hung W, Yong C, Astell C, et al. Two regulators of Ste12p inhibit pheromoneresponsive transcription by separate mechanisms. Mol Cell Biol. 2000;20:4199–4209. doi: 10.1128/mcb.20.12.4199-4209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkman-Correia C, Stroke IL, Fields S. Functional domains of the yeast STE12 protein, a pheromoneresponsive transcriptional activator. Mol Cell Biol. 1993;13:3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 21.Esch RK, Wang Y, Errede B. Pheromone-induced degradation of Ste12 contributes to signal attenuation and the specificity of developmental fate. Eukaryot Cell. 2006;5:2147–2160. doi: 10.1128/EC.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou S, Zhao S, Song Y, Liu H, Nie Q. Fus3-triggered Tec1 degradation modulates mating transcriptional output during the pheromone response. Mol Syst Biol. 2008;4:212. doi: 10.1038/msb.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delgado-Jarana J, Martinez-Rocha AL, Roldan-Rodriguez R, Roncero MI, Di Pietro A. Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development and virulence through multiple signalling pathways. Fungal Genet Biol. 2005;42:61–72. doi: 10.1016/j.fgb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Di Pietro A, Garcia-MacEira FI, Meglecz E, Roncero MI. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol. 2001;39:1140–1152. [PubMed] [Google Scholar]
- 25.Hoi JW, Herbert C, Bacha N, O’Connell R, Lafitte C, Borderies G, et al. Regulation and role of a STE12-like transcription factor from the plant pathogen Colletotrichum lindemuthianum. Mol Microbiol. 2007;64:68–82. doi: 10.1111/j.1365-2958.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 26.Deng F, Allen TD, Nuss DL. Ste12 transcription factor homologue CpST12 is downregulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot Cell. 2007;6:235–244. doi: 10.1128/EC.00302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schamber A, Leroch M, Diwo J, Mendgen K, Hahn M. The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol Plant Pathol. 2010;11:105–119. doi: 10.1111/j.1364-3703.2009.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 29.Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, et al. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D, Bobrowicz P, Wilkinson HH, Ebbole DJ. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics. 2005;170:1091–1104. doi: 10.1534/genetics.104.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallim MA, Miller KY, Miller BL. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol Microbiol. 2000;36:290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- 32.Nolting N, Poggeler S. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol Microbiol. 2006;62:853–868. doi: 10.1111/j.1365-2958.2006.05415.x. [DOI] [PubMed] [Google Scholar]
- 33.Kramer B, Thines E, Foster AJ. MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet Biol. 2009;46:667–681. doi: 10.1016/j.fgb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Wong Sak Hoi J, Dumas B. The STE12 and STE12-like proteins: fungal transcription factors regulating development and pathogenicity. Eukaryot Cell. 2010. [DOI] [PMC free article] [PubMed]
- 35.Park G, Xue C, Zheng L, Lam S, Xu JR. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 2002;15:183–192. doi: 10.1094/MPMI.2002.15.3.183. [DOI] [PubMed] [Google Scholar]
- 36.Rispail N, Di Pietro A. Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol Plant Microbe Interact. 2009;22:830–839. doi: 10.1094/MPMI-22-7-0830. [DOI] [PubMed] [Google Scholar]
- 37.Tsuji G, Fujii S, Tsuge S, Shiraishi T, Kubo Y. The Colletotrichum lagenarium Ste12-like gene CST1 is essential for appressorium penetration. Mol Plant Microbe Interact. 2003;16:315–325. doi: 10.1094/MPMI.2003.16.4.315. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Sanchez AM, Martin-Rodrigues N, Ramos B, de Vega-Bartol JJ, Perlin MH, Diaz-Minguez JM. fost12, the Fusarium oxysporum homolog of the transcription factor Ste12, is upregulated during plant infection and required for virulence. Fungal Genet Biol. 2010;47:216–225. doi: 10.1016/j.fgb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 2004;53:1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 40.Pi H, Chien CT, Fields S. Transcriptional activation upon pheromone stimulation mediated by small domain of Saccharomyces cerevisiae Ste12p. Mol Cell Biol. 1997;17:6410–6418. doi: 10.1128/mcb.17.11.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]