Abstract
Nicotiana benthamiana plants were agroinfiltrated with an infectious clone of the Turnip mosaic virus (TuMV) that was engineered to tag replication vesicles with either GFP or mCherry fluorescent proteins. Punctuate vesicle structures were observed in the cytoplasm of infected cells corresponding to viral replication factories. The vesicles were highly motile and co-aligned with the microfilaments. Utilization of latrunculin B, an inhibitor of microfilament polymerization, reduced accumulation of the virus, suggesting that microfilaments are necessary during infection. To investigate biogenesis of the vesicles, leaves were infected simultaneously with two recombinant TuMV infectious clones, one that labeled vesicles in red and one that labeled them in green. We observed cell with green only and red only vesicles indicating a single viral genome origin. In some cases, vesicles exhibited sectors of green, red and yellow fluorescence were also observed, demonstrating that fusion among individual vesicles is possible. Based on those results we propose a model for the biogenesis of viral factory, where viral translation and replication are tightly coupled within virus-induced vesicles.
Key words: plant virus, positive-sense RNA virus, potyvirus, virus factories, viral replication, viral translation, translation initiation factors, vesicle trafficking
Over the past years, a large number of investigations have been devoted to understanding virus-host interactions at the molecular level. Concurrent to these molecular studies, an avenue of investigation at the interface of molecular virology and cell biology has emerged. These studies have shown that animal as well as plant RNA viruses induce substantial cellular remodeling during infection.1 Many of these virus-induced structures are organelles that house the RNA replication complex, and for that reason are given the generic term of replication factories. These factories contain positive and negative strand viral RNAs and the viral RNA-dependent RNA polymerase (RdRp), along with non-structural viral and host proteins. It has been proposed that virus-induced factories increase the local concentration of components required for replication, protect viral RNA from degradation and prevent the activation of host defense functions. Current questions centre on the membrane origins that give rise to the virus-induced factories and the molecular motors as well as pathways that are involved in their trafficking from their site of origin to their final destination. The origin and nature of these virus-induced membrane structures differ according to the virus families.1 In most cases, the modifications involve the formation of spherules, vesicles or multivesicular bodies derived from membranes of the endoplasmic reticulum (ER), mitochondria, peroxisomes, lysomes or chloroplasts. Electron tomography has recently been used for the generation of three-dimensional imaging of Dengue virus- and coronavirus-induced membrane alterations at high resolution. These alterations resulted in a reticulovesicular network of modified ER that integrates convoluted membranes, numerous interconnected double membrane vesicles and “vesicle packets”.2,3 The three-dimensional renderings of these membrane networks show a spatio-temporal platform for the virus replication cycle, and suggest that not only RNA replication but also translation and virion assembly are associated with these virus-induced structures.
Turnip mosaic virus (TuMV) is an excellent model to study plant virus replication factory formation. TuMV has a positive-strand RNA genome of approximately 10 kb long, with a viral genomelinked protein (VPg) covalently linked to its 5′ end and a poly(A) tail at its 3′ end. The genomic RNA is translated into one polyprotein of 358 kDa that is processed into at least 10 mature proteins by viral proteases. Viral replication takes place in virus-induced vesicles derived from the ER (Fig. 1A) and the expression of a single viral protein (i.e., 6K2-VPg-Pro) is sufficient to induce vesicle formation (4). However, many questions are unresolved concerning TuMV vesicle biogenesis and content.
Figure 1.
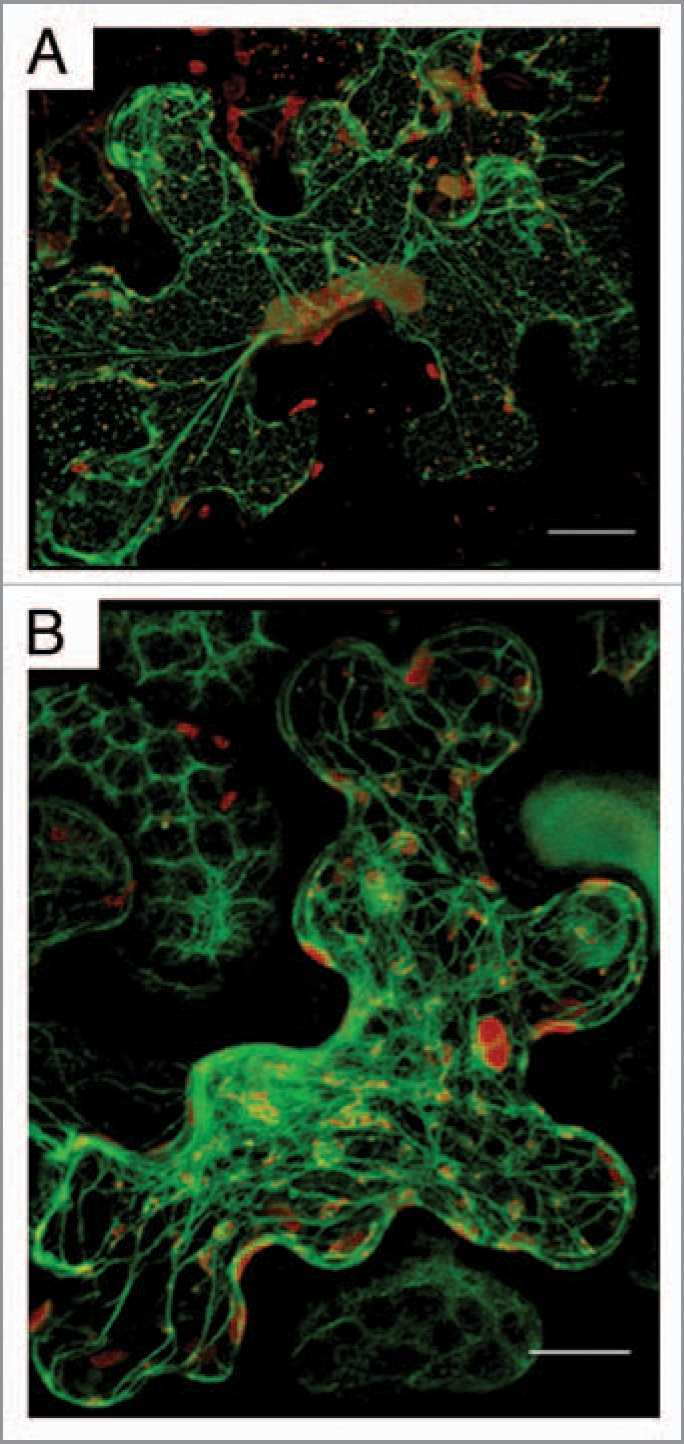
TuMV replication factories are associated with ER and co-allign with microfilaments. Nicotiana benthamiana cells expressing mCherry-tagged TuMV-induced replication factories and ER -resident GFP (A) or the actin domain of fimbrin fused to GFP (B) observed by confocal microscopy at 4 days post-agroinfiltration. Photographs are a three-dimensional rendering of 40 1-µm thick slices that overlap by 0.5-µm. Scale bar, 10 µm.
Vesicle Movement on Microfilaments
Virus replication factories are dynamic structures.5–7 We thus investigated the trafficking of TuMV-induced vesicles by confocal microscopy with an infectious clone that was engineered to tag replication vesicles with either GFP or mCherry fluorescent proteins. The observed vesicles were irregular in shape and varied in size, ranging from 0.6 to 4.3 µm in diameter. Interestingly, some vesicles were highly motile with an average velocity of 0.45 µm/s. Their movement was unidirectional and was characterized by a stop and go activity. Occasionally, fusion was observed between vesicles in the perinuclear zone. Because of the high viscosity of the cytoplasm, movement of large complexes requires an active transport with implication of cytoskeleton elements. When an actin marker fused to GFP was co-expressed, it was observed that the TuMV vesicles co-aligned with the microfilaments (Fig. 1B). When a low concentration (5 µM) of Latrunculin B (latB), which inhibits microfilament polymerization, was applied factory movement was stopped and virus production was significantly decreased.
Each Vesicle Derived from a Single Genome
One may also ask how viral proteins are imported within the replication factories. It is generally assumed that viral RNA translation is taking place in the cytoplasm and the newly synthesized proteins are exported in trans to virus-induced, pre-formed, vesicles. Since many translation factors have been found within the TuMV-induced vesicles,8–10 it is possible that translation instead occurs within the factories or is tightly associated with them. To resolve this issue, leaves were infected simultaneously with two recombinant TuMV infectious clones, one that labeled vesicles in red and one that labeled them in green. Following agro-infection, individual cells were screened for the expression of both green and red vesicles. The rational is that in a cell infected by both viruses, if translation happens in the cytoplasm and proteins are exported randomly to the vesicles, both green and red fluorescing vesicles should be observed.
However, if translation occurs within the vesicle, green- and red-only vesicles should be detected. What was observed were cells with individual green-only and red-only vesicles, suggesting a single-genome origin for each vesicle. Interestingly, vesicles exhibiting sectors of green, red and yellow colors were also observed, possibly resulting from a fusion between vesicles, a phenomenon that was noticed previously during vesicle trafficking. Formation of vesicles derived from a single viral genome indicates the existence of a cis mechanism that incorporates the proteins synthesized from a same viral RNA into the same vesicle. A mechanistic explanation is that viral RNA translation and replication occurs within the factories, and this was shown by the co-localization of several host translation factors with viral double-stranded RNA, a marker of viral RNA replication. This close coupling between viral replication and translation was recently suggested by Hafren and co-authors.11
Factory Biogenesis—A Model
Based on the above results and those of others, a model where viral translation and replication is coupled within TuMV-induced vesicles is being proposed. The sequential steps can be schematized as follows (Fig. 2). Upon release of the genomic RNA into the cytoplasm, the host protein synthetic machinery is usurped for the production of viral proteins, on ER-associated ribosomes (step I in the figure). After several rounds of viral RNA translation, viral proteins accumulate in patches on the outer ER membrane and initiate membrane curvature (II). During this process, the translation machinery through protein interaction with viral factors and viral RNA are trapped within the vesicles. Translation keeps going on within the vesicles and the newly produced proteins contribute to the size increase and maturation of the vesicles. A switch then happens from translation to replication to allow the viral RNA synthesis within in the same vesicles. Membrane curvature increases with accumulating replication components, which ultimately leads to the formation of single-membrane vesicles within the organelle lumen, which may or may not have a pore-like connection to the exterior (III). The clusters of large vesicles observed within the cytoplasm are produced during a second budding from the ER (IV-V).
Figure 2.
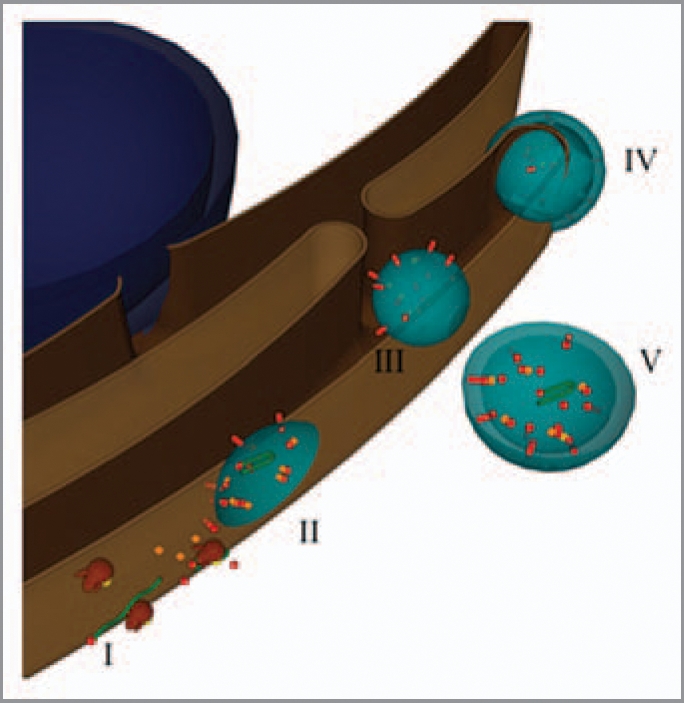
Model for the formation of virus-induced vesicles. The blue sphere represents the nucleus, while the brown structure the ER. Partially transparent virus-induced vesicles are in light blue. Green ribbons and red spheres and rods depict viral RN As and proteins, respectively. Host proteins are the orange cubes, and the brown and yellow structures are ribosomes.
Conclusion
Future issues will be to obtain a more refined 3-dimensional view of TuMV factories in order to better understand the interplay between virus RNA replication and various viral processes, such as translation and encapsidation. A corollary will be to determine the full content of host proteins and the mechanistic role for their presence in virus factories. Finally, trafficking and the fate of virus factories in cell-to-cell and long distance transport need to be investigated.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/11968
References
- 1.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchemin C, Boutet N, Laliberté J-F. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J Virol. 2007;81:775–782. doi: 10.1128/JVI.01277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. The Cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 2009;149:1005–1016. doi: 10.1104/pp.108.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J-Z, Blancaflor EB, Nelson RS. The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone of within the complex aligns with and traffics along microfilaments. Plant Physiol. 2005;138:1853–1865. doi: 10.1104/pp.105.065722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai C-K, Jeng K-S, Machida K, Lai MMC. Association of Hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J Virol. 2008;82:8838–8848. doi: 10.1128/JVI.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beauchemin C, Laliberté J-F. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J Virol. 2007;81:10905–10913. doi: 10.1128/JVI.01243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, et al. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology. 2008;374:217–227. doi: 10.1016/j.virol.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Thivierge K, Cotton S, Dufresne PJ, Mathieu I, Beauchemin C, et al. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology. 2008;377:216–225. doi: 10.1016/j.virol.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Hafren A, Hofius D, Ronnholm G, Sonnewald U, Makinen K. HSP70 and its cochaperone CPIP promote Potyvirus infection in Nicotiana benthamiana by regulating viral coat Protein functions. Plant Cell. 2010;22:523–535. doi: 10.1105/tpc.109.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]


