Abstract
The cells of the second heart field (SHF) contribute to the outflow tract and right ventricle, as well as to parts of the left ventricle and atria. Isl1, a member of the LIM-homeodomain transcription factor family, is expressed early in this cardiac progenitor population and functions near the top of a transcriptional pathway essential for heart development. Isl1 is required for the survival and migration of SHF-derived cells into the early developing heart at the inflow and outflow poles. Despite this important role for Isl1 in early heart formation, the transcriptional regulation of Isl1 has remained largely undefined. Therefore, to identify transcription factors that regulate Isl1 expression in vivo, we screened the conserved noncoding sequences from the mouse Isl1 locus for enhancer activity in transgenic mouse embryos. Here, we report the identification of an enhancer from the mouse Isl1 gene that is sufficient to direct expression to the SHF and its derivatives. The Isl1 SHF enhancer contains three consensus Forkhead transcription factor binding sites that are efficiently and specifically bound by Forkhead transcription factors. Importantly, the activity of the enhancer is dependent on these three Forkhead binding sites in transgenic mouse embryos. Thus, these studies demonstrate that Isl1 is a direct transcriptional target of Forkhead transcription factors in the SHF and establish a transcriptional pathway upstream of Isl1 in the SHF.
INTRODUCTION
The cardiac progenitors that give rise to the outflow tract, right ventricle, and portions of the atria are distinct from the progenitors that form the left ventricle (Buckingham et al., 2005). Fate-mapping studies in avian and mouse systems have demonstrated that the cells that comprise the outflow tract region and the right ventricle originate from an extra cardiac progenitor population that resides in the pharyngeal mesoderm adjacent to the pharyngeal endoderm (Cai et al., 2003; Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001). This population of cells, referred to as the second heart field (SHF), is progressively added to the growing linear heart tube at the outflow and inflow poles of the heart (Black, 2007; Buckingham et al., 2005). The linear heart tube forms from the classical cardiac crescent, also referred to as the first heart field (FHF), and the existence of an extra cardiac population of progenitor that contributes to the outflow tract and right ventricle provides an embryological explanation for the longstanding observations that many genes and transgenes exhibit regionally restricted expression within the heart along these boundaries (Black, 2007; Buckingham et al., 2005).
A subset of the SHF, referred to as the anterior heart field (AHF), specifically contribute to the outflow tract and right ventricle and is defined in mice by the expression of an Fgf10 transgene, as well as by the expression of a Mef2c-AHF-lacZ transgene (Kelly et al., 2001; Verzi et al., 2005). The broader SHF has been defined by the expression of the Isl1 gene, which encodes a LIM-homeodomain protein essential for cardiac development (Cai et al., 2003). Isl1 expression is detected very early at the cardiac crescent stage, contiguous with, but medial and dorsal to MLC2a-positive cells of the FHF (Cai et al., 2003). Following ventral morphogenesis of the mouse embryo, Isl1+ cells are present in the pharyngeal and splanchnic mesoderm, as well as in the foregut endoderm (Cai et al., 2003). Lineage tracing analyses show that Isl1 descendants contribute extensively to the heart including to parts of the inflow tract region, the atria, and the left ventricle, as well as the outflow tract and the right ventricle (Cai et al., 2003). In addition, Isl1+ splanchnic mesoderm cells contribute to the branchiomeric skeletal muscles in the head (Nathan et al., 2008). Recent lineage analyses using Isl1-Cre activation of a conditional lacZ reporter knocked in to the Gata4 locus suggested a substantially broader Isl1 expression domain than previously reported using standard Rosa26R Cre-activated reporters (Ma et al., 2008).
Inactivation of Isl1 in mice results in growth arrest at embryonic day (E) 9.5, and Isl1 knockout mice display profound defects in heart development, including a failure to undergo looping morphogenesis and the apparent absence of complete segments of the heart (Cai et al., 2003). It was suggested that defective heart formation in Isl1 knockout mice is at least partially caused by aberrant proliferation of the pharyngeal mesoderm, ultimately resulting in failed migration of Isl1-expressing cells into the heart (Cai et al., 2003). Recent studies using genetic marking of Isl1+ progenitors demonstrated that embryonic Isl1-expressing cells are multipotent cardiac progenitors that give rise to three cardiac lineages, including cardiomyocytes, smooth muscle, and endothelial cells (Laugwitz et al., 2005; Moretti et al., 2006). In addition to those major cardiac lineages, it was recently suggested that Isl1 and Nkx2.5-expressing progenitors also contribute to proepicardium during heart morphogenesis (Zhou et al., 2008). However, in spite of its essential role in cardiac development, the regulation of Isl1 transcription in the SHF remains largely undefined.
Several transcription factors in addition to Isl1 have also been implicated in the transcriptional regulatory program controlling SHF development (Black, 2007). These include the MADS domain transcription factor MEF2C, the bHLH transcription factor HAND2, and the T-box transcription factors TBX20 and TBX1 (Black, 2007; Takeuchi et al., 2005; Xu et al., 2004). Several members of the Forkhead transcription factor family have also been implicated in early cardiovascular development (Papanicolaou et al., 2008). The Forkhead family of transcription factors consists of 39 distinct members in humans, and members of this family are involved in many diverse biological processes (Carlsson and Mahlapuu, 2002; Lehmann et al., 2003). Foxa2 is expressed in the anterior primitive streak and the node as early as E6.5 in the mouse (Monaghan et al., 1993; Sasaki and Hogan, 1994). In addition, Foxa2 regulates Tbx1 in the SHF, suggesting a role for this Forkhead factor in the transcriptional program controlling the development of the SHF and its derivatives (Hu et al., 2004; Maeda et al., 2006; Yamagishi et al., 2003). Foxf1 mutant mice die by E8.0 due to extraembryonic mesoderm defects, consistent with the observation that Foxf1 expression is induced during gastrulation at E6.5 in the mesoderm in mice (Kalinichenko et al., 2003; Mahlapuu et al., 2001; Peterson et al., 1997). The broad expression of Foxf1 in the early mesoderm is consistent with a possible role for this transcription factor in establishing cardiovascular progenitor identity in the early lateral mesoderm. Likewise, Foxc1 and Foxc2 are expressed in the SHF at the cardiac crescent stage (E7.75 in mouse) and may play a role in establishing identity of cardiovascular progenitors (De Val and Black, 2009; Seo and Kume, 2006). Knockout of either Foxc1 or Foxc2 in the mouse results in perinatal lethality with a spectrum of cardiovascular defects, including aberrant formation of the aortic arches, ventricular septal defects, and abnormal semilunar valves (Winnier et al., 1999). Importantly, compound Foxc1/c2 homozygous mutants die much earlier with more severe cardiovascular defects, including lack of the outflow tract and right ventricle, suggesting that these two factors have overlapping and dose-dependent roles in cardiovascular development (Kume et al., 2001; Seo and Kume, 2006). FoxH1 has also been demonstrated to play a key role in SHF development (von Both et al., 2004). Foxh1 null mice fail to form AHF-derived structures, such as the outflow tract and right ventricle, but normally form inflow and atrial regions (von Both et al., 2004), suggesting a role restricted to anterior regions of the SHF for this member of the Forkhead factor family.
Despite the identification of multiple transcription factors involved in SHF development, few regulatory relationships among those factors have been identified. It has been established that Mef2c functions as a downstream transcriptional target of Isl1 in the SHF, but the upstream regulators of Isl1 in the SHF are essentially unknown. In the present study, we identified an enhancer from the mouse Isl1 gene that is sufficient to direct expression to the SHF. This evolutionarily conserved Isl1 enhancer is active in the cardiac crescent by E7.0 and in the pharyngeal mesoderm and outflow tract by E8.5 in the mouse. The Isl1 SHF enhancer contains three evolutionarily conserved Forkhead binding sites that are efficiently bound by several relevant candidate Forkhead proteins in vitro, and the Isl1 Forkhead sites are required for enhancer activity in the pharyngeal mesoderm in transgenic mouse embryos. Thus, these studies demonstrate an important role for Forkhead transcription factors in the regulation of Isl1 in the SHF.
MATERIALS AND METHODS
Cloning and mutagenesis
The 3642-bp Isl1-F fragment was generated by PCR from SV129Ev mouse genomic DNA using the following primers: 5′-cctctcgagactcagttttgagca-3′ and 5′-ctcaggggatgtaaaagcttttc-3′. This fragment was then cloned as an XhoI-SspI fragment into the XhoI and SmaI sites of the transgenic reporter plasmid HSP68-lacZ (Kothary et al., 1989). Deletion fragments of the Isl1-F enhancer were generated by digestion and subsequent subcloning back into the corresponding sites of HSP68-lacZ, using the following restriction sites: Isl1-F1, XhoI-EcoRV; Isl1-F2, EcoRV-DraI. For Isl1-F3, Isl1-F-HSP68-lacZ was cleaved with AseI and SalI and the Isl1-F3-HSP68-lacZ transgene fragment was directly purified for oocyte microinjection. To generate the Isl1-F2Δ298 fragment, the Isl1-F fragment was mutated to create a PstI restriction site at nucleotides 2,077-2,082 of the Isl1-F fragment, followed by cleavage with NsiI and PstI and subsequent re-ligation to excise 298 bp in between the native NsiI site and the engineered PstI site. The following mutant sequences were introduced sequentially into Isl1-F2 to create the triple FOX mutant: mFOX1, 5′-gggctaatccctgcagggcgcttgac-3′; mFOX2, 5′-cagaccgcacaggagctccagcaccttt-3′; mFOX3-3, 5′-tccctttggatggtacctcttgtttcaa-3′. The entire sequence of the triple FOX mutant Isl1-F2 fragment was confirmed by sequencing on both strands.
Generation of transgenic mice
Transgenic reporter fragments were digested from the hsp68-lacZ plasmid backbone, gel purified, and suspended in 5 mM Tris-HCl, 0.2 mM EDTA, pH 7.4, at a concentration of 2 μg/ml for pronuclear injection. Transgenic embryos or stable lines were generated as described previously (Rojas et al., 2005). DNA was extracted from the yolk sac of embryos or from tail biopsies from mice by digestion in tail lysis buffer (100 mM NaCl, 25 mM EDTA, 1% sodium dodecyl sulfate, 10 mM Tris-Cl, 200 μg/ml proteinase K, pH 8.0) at 56°C overnight, followed by genomic DNA extraction, restriction digestion, and Southern blotting using standard methods. Embryos collected at E8.5 and earlier were genotyped by PCR, using the lacZ primers LACZ5 (5′-cggtgaatggtgctgcgttgga-3′) and LACZ3 (5′-accaccgcacgatagagattc-3′).
X-gal staining, in situ hybridization, and immunofluorescence
β-Galactosidase expression in transgenic embryos was detected by X-gal staining, performed as described previously (Dodou et al., 2003). Sections from X-gal-stained embryos and tissues were prepared and counterstained with Neutral Fast Red, as described previously (Anderson et al., 2004). Whole-mount in situ hybridization of mouse embryos was performed exactly as described previously (Rojas et al., 2005). Following hybridization, embryos were washed and treated with RNaseA, as described previously (Wilkinson and Nieto, 1993). Signal was detected using an alkaline phosphatase-conjugated anti-digoxigenin antibody and BM purple alkaline phosphatase substrate (Roche Pharmaceuticals). Isl1 antisense probe was generated from a pBluescript(SK)II+ plasmid containing the mouse Isl1 cDNA linearized with XhoI and transcribed with T3 polymerase.
For detection of β-galactosidase and Isl1 proteins in mouse embryos by immunofluorescence, lacZ transgenic embryos were collected at E8.5 and fixed overnight in 4% paraformaldehyde in phosphate buffered saline, pH 7.4. Fixed embryos were infused in series of sucrose solutions (15% and 30% sucrose in PBS) for 30 minutes each, and then were frozen in 100% OCT compound (Tissue Tek II) on dry ice and cryosectioned at a thickness of 10 μm. Sections were stored at -80°C until use. Sections were analyzed for Isl1 and β-galactosidase protein expression using rabbit anti-β-galactosidase (ICN) diluted 1:500 and mouse anti-Isl1 (Developmental Studies Hybridoma Bank, Iowa City) diluted 1:100 in blocking solution (5% normal goat serum and 5% non-fat powdered milk). Binding of anti-β-galactosidase and anti-Isl1 primary antibodies was detected using Oregon Green-conjugated goat anti-rabbit and Alexa Fluor 594-conjugated goat anti-mouse secondary antibodies (Molecular Probes), each diluted 1:200 in blocking solution.
Electrophoretic mobility shift assay (EMSA)
Double-stranded oligonucleotides were labeled by filling in overhanging 5′ ends with Klenow in the presence of 32P-dCTP. Labeled probes were then purified on a nondenaturing polyacrylamide-TBE gel. Binding reactions were conducted with recombinant Forkhead proteins at room temperature in 1× binding buffer (40 mM KCl, 15 mM HEPES, pH 7.9, 1 mM EDTA, 0.5 mM DTT, 5% glycerol) with 1 μg of poly dI-dC as a non-specific competitor, and competitor DNA as described previously (Rojas et al., 2005). Recombinant FoxF1, FoxA2, FoxC2, and FoxH1 were synthesized using the TNT Quick Coupled Transcription/Translation Systems, as described in the manufacturer’s directions (Promega).
FoxF1 protein was generated from plasmid pCITE-FoxF1, which has been described previously (Rojas et al., 2005). FoxA2 protein was generated from plasmid pcDNA1-FoxA2, which was made by cloning the mouse Foxa2 cDNA as an EcoRI-XbaI fragment into the EcoRI and XbaI sites of pcDNA1/amp (Invitrogen). FoxC2 and FoxH1 proteins were generated from their respective mouse cDNAs cloned into plasmids pCR2.1-TOPO-FoxC2 and pCR2.1-TOPO-FoxH1. The sequences of the control FoxF1, FoxA2, FoxC2, and FoxH1 binding sites have been described previously (Overdier et al., 1994; Peterson et al., 1997; Pierrou et al., 1994).
The sense-strand sequences of the oligonucleotides encompassing the Forkhead sites from Isl1-F2 used for EMSA were: FOX1, 5′-ggggagggctaatccaaacaaggcgcttgacttg-3′; mFOX1, 5′-gggctaatccctgcagggcgcttgac-3′; FOX2, 5′-ggtgcgcagaccgcacagcaaacacagcacctttagc-3′; mFOX2, 5′-cagaccgcacaggagctccagcaccttt-3′, FOX3, 5′-ggtcattccctttggatttgttttcttgtttcaaaaaa-3'; mFOX3, 5′-tccctttggatggtacctcttgtttcaa-3′.
RESULTS
Identification of an enhancer in the Isl1 locus with activity in the SHF
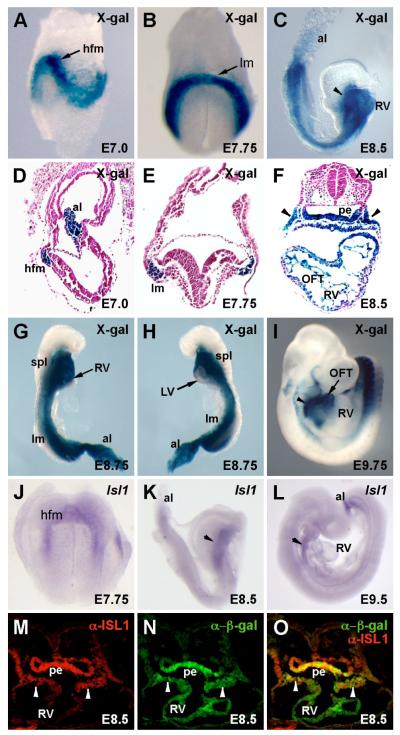
To identify enhancers within the mouse Isl1 locus, we compared the sequences of human, mouse, and chicken Isl1 for conserved regions using BLAST and VISTA analyses (Altschul et al., 1990; Mayor et al., 2000) over a region spanning from -300 kb to +300 kb of the mouse Isl1 locus. These analyses resulted in the identification of ten deeply conserved noncoding regions, which were each tested for enhancer activity in transgenic embryos using the transgenic reporter plasmid HSP68-lacZ (Kothary et al., 1989). Several of these fragments directed consistent patterns of expression in transgenic embryos between E8.5 and E9.5. One of the regions, referred to as Isl1-F, was located between 3324 and 6965 bp downstream of the translational stop codon in the mouse Isl1 gene (Fig. 1). Isl1-F was selected for further detailed analysis because it was sufficient to direct β-galactosidase expression to the SHF, including the pharyngeal mesoderm and its derivatives in outflow tract and right ventricular myocardium during early heart development (Fig. 2).
Fig. 1.
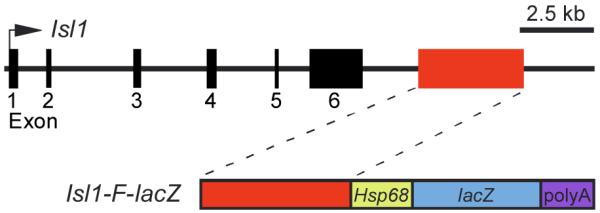
Schematic representation of part of the Isl1 locus and the Isl1-F transgene. The top line represents an approximately 18-kb region of the mouse Isl1 locus, including its six exons (black boxes). Exon 1 is the first coding exon. Exon 6 contains the translational stop codon and 3′ untranslated region. The Isl1-F enhancer is represented as a red box in the upper diagram. The lower schematic depicts the transgene construct Isl1-F-lacZ, which contains the 3642-bp Isl1-F fragment in the transgenic reporter plasmid HSP68-lacZ.
Fig. 2.
The Isl1-F-lacZ transgene is expressed broadly in the mesoderm and in the SHF during early mouse embryonic development. Representative whole-mount (A-C, G-I) and sagittal (D) or transverse (E, F) sections of X-gal-stained Isl1-F-lacZ transgenic embryos are shown. (A, D) Isl1-F directed strong lacZ expression to the mesoderm at E7.0, including heart-forming anterior lateral mesoderm (hfm) and in the extraembryonic mesoderm in the allantois (al). (B, E) Isl1-F activity became more robust and more refined to the lateral mesoderm (lm) at E7.75. (C, F) At E8.5, expression of Isl1-F-lacZ was evident in the SHF and its derivates in the pharyngeal mesoderm (arrowheads), pharyngeal endoderm (pe), outflow tract (OFT), and right ventricle (RV). (G, H) β-galactosidase activity was restricted to the RV and was largely absent from the left ventricle (LV) at E8.75. spl, splanchnic mesoderm. (I) At E9.75, β-galactosidase activity from the Isl1-F-lacZ transgene expression remained strong in the pharyngeal mesoderm (arrowhead) and was still visible in the OFT at this stage. (J-L) For comparison, whole mount in situ hybridization with an Isl1 antisense probe was performed at E7.75 (J), E8.5 (K), and E9.5 (L). Arrowheads mark the pharyngeal mesoderm in K and L. (M-O) Immunofluorescence analyses on transverse sections of Isl1-F-lacZ transgenic embryos at E8.5 using anti-Isl1 and anti-β-galactosidase antibodies shows that endogenous Isl1 protein (red) colocalizes with β-galactosidase (green) in SHF-derived tissues, including the pharyngeal mesoderm (arrowheads) and pharyngeal endoderm (pe).
To define in detail the expression directed by the Isl1-F enhancer in vivo, we examined X-gal-stained transgenic embryos by whole mount and transverse section at various developmental stages. Expression of β-galactosidase was first seen throughout the embryonic mesoderm and in the allantois at E7.0 (Fig. 2A, D). Expression directed by the Isl1-F enhancer was strong in the precardiac anterior lateral mesoderm and continued to be apparent in most of the embryonic mesoderm at E7.75 (Fig. 2B, E). Strong expression of β-galactosidase continued in the pharyngeal mesoderm and endoderm and in the outflow tract and right ventricle at E8.5 (Fig. 2C, F). Examination of X-gal-stained transverse sections from E8.5 transgenic embryos indicated that the majority of cells in the outflow tract myocardium and the right side of the single common ventricle were positive for β-galactosidase, whereas most of cells on the left side of the ventricle were negative for transgene expression (Fig. 2F), which is consistent with the contribution of Isl1+ cells to the developing heart (Cai et al., 2003). Similarly, at E8.75, Isl1-F transgenic embryos showed robust β-galactosidase expression in the right ventricle but essentially no expression in the left ventricle (Fig. 2G, H). At E9.75, expression directed by the Isl1-F enhancer continued to be robust in the pharyngeal mesoderm and was also visible in the outflow tract and right ventricle (Fig. 2I). This pattern of lacZ expression directed by Isl1-F was consistent with the pattern of endogenous Isl1 mRNA expression by in situ hybridization, albeit somewhat broader (Fig. 2J-L). Immunofluorescence anayses of β-galactosidase and Isl1 protein expression also showed that β-galactosidase colocalized with endogenous Isl1 protein within the pharyngeal mesoderm and endoderm in Isl1-F transgenic embryos at E8.5 (Fig. 2M-O). The strong β-galactosidase activity in the heart at E8.5 and E9.75 was probably due, at least in part, to the long half-life of the β-galactosidase protein (Thompson et al., 1991; Voon et al., 2005), allowing cells in which Isl1-F was active to continue to be marked even after endogenous expression of Isl1 had been extinguished (Fig. 2M-O). Taken together, these observations demonstrate the early and specific activation of the Isl1-F enhancer in the SHF and its derivatives in the developing heart.
A small, evolutionarily conserved fragment of the Isl1 SHF enhancer is required and sufficient for function in vivo
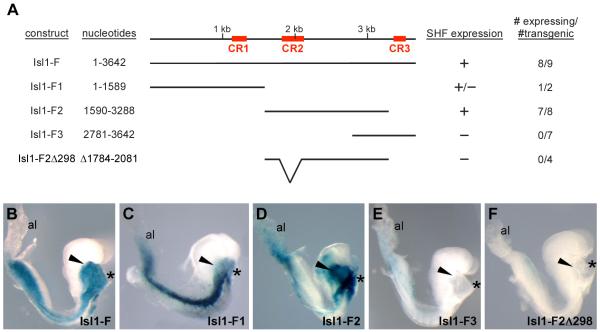
Within the conserved Isl1-F enhancer fragment, there are three segments of deep evolutionary conservation, which we denoted as CR1, CR2, and CR3 (Fig. 3A). We hypothesized that one or more of these conserved regions were likely to be important for Isl1-F function in vivo. Therefore, we performed a series of deletion analyses to determine if these three conserved regions were important for SHF activity at E8.5 (Fig. 3).
Fig. 3.
Deletion analyses of the Isl1-F enhancer identify smaller, conserved regions that are required and sufficient for enhancer function in the SHF in vivo. (A) Schematic representation of deletion constructs of the Isl1-F SHF enhancer examined in these studies. The top line depicts the organization of Isl1-F. The red boxes indicate regions of high sequence homology between the mouse, human, and chick Isl1 genes, denoted as CR1, CR2, and CR3. The nucleotide positions of each deletion construct, relative to Isl1-F, are indicated on the left. The mesoderm activity of each of the deletion constructs and the number of independent transgenic lines that displayed SHF activity over the total number of lacZ+ lines are indicated in the columns to the right. (B-F) Representative transgenic embryos collected at E8.5 for each of the deletion constructs. Isl1-F (B) and Isl1-F2 (D) directed strong and specific expression of lacZ to the pharyngeal mesoderm (arrowheads). Isl1-F1 (C) directed weak expression to the pharyngeal mesoderm compared to strong expression to the lateral mesoderm and Isl1-F3 (E) displayed no detectable expression in the pharyngeal mesoderm. Deletion of CR2 (Δ1784-2081) in the context of Isl1-F2 to generate fragment Isl1-F2Δ298 resulted in a complete loss of enhancer activity (F). Asterisks denote the heart in all panels.
We created three deletion constructs designed to test smaller fragments that each contained only a single conserved region (Fig. 3A). The CR1 region was contained within Isl1-F1 (nucleotides 1-1589) and showed variable degrees of expression in transgenic embryos (Fig. 3C). Although Isl1-F1 was sufficient to direct expression of lacZ to some regions of the mesoderm, it was not sufficient for pharyngeal mesoderm expression (arrowhead in Fig. 3C). Isl1-F2 encompassed CR2 and extended from nucleotides 1590 to 3288 within Isl1-F. Importantly, Isl1-F2 consistently directed robust expression to the pharyngeal mesoderm in 7 of 8 transgenic embryos (Fig. 3D). Isl1-F2 also showed activity in the lateral mesoderm, albeit less than full length Isl1-F (Fig. 3B, D). In contrast, deletion of the 5′ end of Isl1-F to create Isl1-F3 (nucleotides 2781-3642), which included all of CR3, completely abolished transgene activity in vivo in all embryos examined (Fig. 3E).
To determine whether CR2 was necessary for enhancer activity of Isl1-F2, a small region (nucleotides 1784-2081) encompassing CR2, was deleted. This Isl1-F2Δ298 transgene lacked any detectable expression of β-galactosidase in the pharyngeal mesoderm (Fig. 3F), indicating that CR2 was required for the activity of Isl1-F2 in the pharyngeal mesoderm in vivo. We next wanted to determine whether the CR2 region was sufficient for pharyngeal mesoderm activity in vivo. Therefore, we further deleted both the 5′ and 3′ ends from Isl1-F2 to create a smaller 516 bp fragment (nucleotides 1686-2201), which included all of CR2. Interestingly, this smaller region of Isl1-F2 did not have complete enhancer activity in the pharyngeal mesoderm at E8.5 (data not shown), suggesting that essential functional elements reside within Isl1-F2 both inside and outside of CR2.
Isl1-F2 contains multiple, conserved Forkhead-binding sites
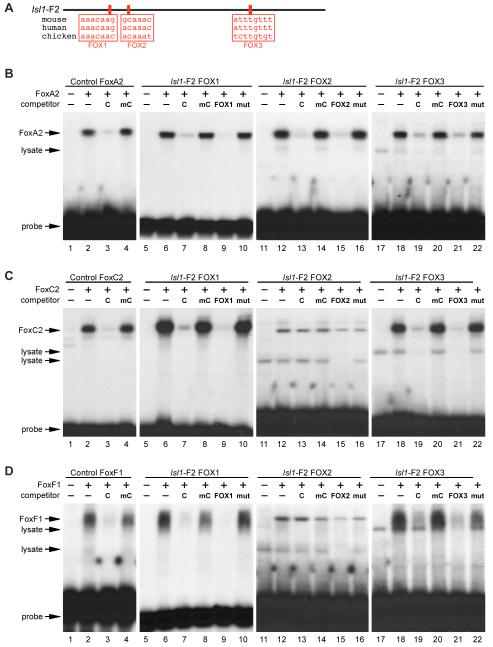
The deletion analyses shown in Fig. 3 identified the Isl1-F2 fragment as sufficient and required for enhancer activity in the pharyngeal mesoderm in vivo. Therefore, to help identify specific transcription factor binding sites, we analyzed the Isl1-F2 fragment for candidate transcription factor binding sites by comparing sequences from multiple species. Analyses by BLAST (Altschul et al., 1990), ClustalW (Thompson et al., 1994), and TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess) identified three candidate binding sites for Forkhead (FOX) transcription factors, which were highly conserved among human, mouse, and chicken (Fig. 4A). The first two sites (FOX1 and FOX2) were present in CR2, while the third site (FOX3) was located outside CR2 but still within Isl1-F2 (Fig. 4A). The presence of multiple conserved consensus Forkhead sites was interesting since several members of the Forkhead family are known to be involved in early heart and mesoderm development (Kume et al., 2001; Papanicolaou et al., 2008; Seo and Kume, 2006; von Both et al., 2004), suggesting that Isl1 might be a direct target of one or more Forkhead transcription factors through the putative FOX sites in the Isl1-F2 enhancer.
Fig. 4.
The Isl1-F2 enhancer contains three high affinity Forkhead binding sites. (A) ClustalW and TESS analyses, comparing the sequence of the conserved enhancer region from mouse, human, and chick, identified three conserved candidate binding sites (red boxes) for Forkhead transcription factors, denoted as FOX1, FOX2, and FOX3. (B-D) Recombinant FoxA2, FoxC2, and FoxF1 proteins were transcribed and translated in vitro and used in EMSA with a radiolabeled double-stranded oligonucleotide encompassing the three Isl1 FOX sites as well as control FOX sites. Lanes 1, 5, 11, and 17 in panels B-D contains reticulocyte lysate without any recombinant protein. (B) Recombinant FoxA2 efficiently bound to all three Isl1 FOX sites (lanes 6, 12, and 18). Binding was specifically competed by excess unlabeled FoxA2 control probe and by unlabeled self probe, but not by a mutant versions of the probes (FOX1, lanes 7-10; FOX2, lanes 13-16; FOX3, lanes 19-22). (C) Recombinant FoxC2 bound to the FOX1 and FOX3 sites but not to the FOX2 site (lanes 6, 12, and 18) and binding to the FOX1 and FOX3 sites was specifically competed by excess control or self probe, but not by a mutant probes (FOX1, lanes 7-10; FOX3, lanes 19-22). (D) Similarly, recombinant FoxF1 protein bound to the FOX1 (lanes 6-10) and FOX3 (lanes 18-22) sites but not to the FOX2 site (lanes 12-16). Each protein bound efficiently to its bona fide specific control probe and was competed for binding by an excess control probe, but not by a mutant version of the probe (lanes 2-4 in panels B-D). Free probe and lysate-derived mobility shifts are indicated.
To determine whether the candidate FOX sites in the Isl1-F2 enhancer might represent bona fide cis-acting elements, we tested each of the three sites for binding to candidate general mesoderm and cardiac Forkhead factors by EMSA. FoxA2 bound to all three FOX sites in the Isl1-F2 enhancer (Fig. 4B; lanes 6, 12, and 18) as well as to a consensus FoxA2 control site (Fig. 4B, lane 2; Overdier et al., 1994). Binding to all four sites was efficiently competed by excess unlabeled FoxA2 control probe and its unlabeled self probe, but not by mutant versions of those same unlabeled competitors (Fig. 4B, lanes 3-4 for control site, lanes 7-10 for FOX1, lanes 13-16 for FOX2, and lanes 19-22 for FOX3). All three Isl1-F2 FOX sites bound to FoxA2 in EMSA as well as the FoxA2 control site, but the FOX1 site bound particularly well (note the relative amounts of FoxA2 retarded probe compared to free probe in each panel in Fig. 4B).
FoxC2 bound specifically to the FOX1 and FOX3 sites from Isl1-F2 (Fig. 4C, lanes 6 and 18) and this binding was efficiently competed by excess unlabeled control and self probe but not by mutant versions of those competitors (Fig. 4C; lanes 7-10 for FOX1 and lanes 19-22 for FOX3). The FOX1 probe was bound about as efficiently by FoxC2 as the control FoxC2 site, whereas the FOX3 site was bound relatively more weakly by this Forkhead protein in EMSA (compare bound probe to free probe in each of the panels in Fig. 4C). In contrast to the FOX1 and FOX3 sites, the FOX2 site was not specifically bound by FoxC2 since binding to that element was not competed by excess unlabeled probe (Fig. 4C, lanes 13-16). Similarly, FoxF1 bound specifically to the FOX1 and FOX3 sites in the Isl1-F2 enhancer and to a control FoxF1 site (Fig. 4D, lanes 2, 6, and 18) but FoxF1 did not bind specifically to the FOX2 site since binding to that site was not competed by unlabeled control or self probe (Fig. 4D, lanes 13-16). Taken together, the EMSA data in Fig. 4 show that the FOX1 and FOX3 sites from Isl1-F2 are bound specifically by FoxA2, FoxC2, and FoxF1 and that binding to the FOX1 site was particularly strong, at least in vitro. By contrast, the FOX2 site showed selectivity among Forkhead proteins since this site was efficiently and specifically bound by FoxA2 but not by FoxC2 or FoxF1.
The Forkhead (FOX) binding sites are required for Isl1-F2 enhancer function in vivo
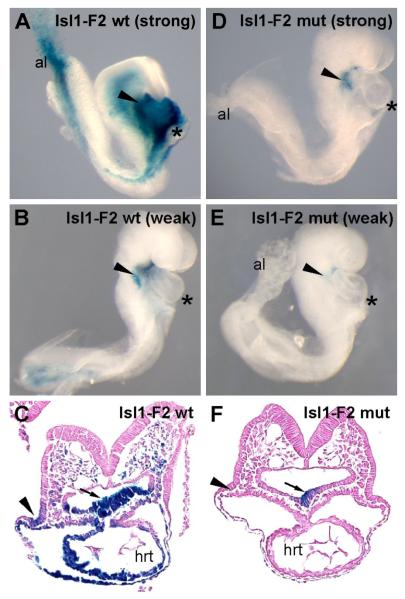
To test the function of the FOX sites from the Isl1-F2 enhancer in vivo, we introduced mutations into all three FOX sites in the context of the Isl1-F2-lacZ transgene and determined the effect of those mutations on enhancer function in transgenic embryos at E8.5 (Fig. 5). The introduced mutations were identical to those used in the EMSA analyses, which were demonstrated to completely block the ability of those sites to compete for Forkhead protein binding even when present in the EMSA reactions at ~50-fold excess (Fig. 4). As expected, the wild type Isl1-F2 enhancer directed strong (Fig. 5A, 50% of independent transgenic events) or modest (Fig. 5B, 50% of independent transgenic events) expression to the SHF. X-gal-staining of transverse sections of embryos bearing the wild type Isl1-F2 transgene showed that the wild-type enhancer directed specific expression to the pharyngeal mesoderm and endoderm as well as to the distal outflow tract (Fig. 5C). In contrast, the mutant transgene had greatly diminished or absent β-galactosidase activity (Fig. 5D-F). Among the five transgenic embryos that harbored the FOX mutant transgene, one showed modest expression (Fig. 5D), two showed very weak expression (Fig. 5E) and two showed no detectable β-galactosidase activity (not shown). Interestingly, transverse sections of the strongest of the triple FOX mutant Isl1-F2 transgenic embryos showed that expression of β-galactosidase was completely abolished in the pharyngeal mesoderm and the outflow tract, although some expression remained in the pharyngeal endoderm (Fig. 5F). These results indicate that the Isl1 SHF enhancer identified in these studies requires the conserved Forkhead binding sites for activity in the pharyngeal mesoderm and its derivatives in vivo and strongly support the notion that Isl1 is a direct transcriptional target of Forkhead transcription factors in the SHF.
Fig. 5.
The Isl1-F2 SHF enhancer is dependent on Forkhead transcription factor binding sites for enhancer activity in vivo. Transgenic embryos harboring Isl1-F2 wild-type (wt) transgenes (A-C) or Isl1-F2 triple FOX mutant (mut) transgenes (D-F) were collected at E8.5 and X-gal-stained, and the lateral right-sided views of whole-mount embryos (A, B, D, E) and transverse sections (C, F) are shown. Examples of strong (A) and weak (B) transgenic lines harboring the wild type Isl1-F2 transgene exhibited specific expression to the pharyngeal mesoderm and pharyngeal endoderm, as well as in the outflow tract myocardium with variable degrees of expression strength depending on the specific transgenic line. (D, E) By contrast, embryos transgenic for the triple FOX mutant Isl1-F2 construct showed either weak or no expression in SHF derivatives and showed no expression in the outflow tract. (C, F) Transverse sections of transgenic embryos with strong expression of the wild type Isl1-F2-lacZ transgene (C) or strong expression of the triple FOX mutant Isl1-F2-lacZ (mut) transgene (F) show that while the wild type transgene was active in the pharyngeal mesoderm (arrowheads), pharyngeal endoderm (arrows) and SHF mesoderm-derived lineages in the heart (hrt), the mutant transgene was only weakly active in pharyngeal endoderm and showed no activity in SHF-derived mesoderm or any of its mesodermal derivatives. Asterisks denote the heart in panels A, B, D, and E.
DISCUSSION
Isl1 is among the earliest marker of the developing cardiac mesoderm, and mouse embryos lacking Isl1 have profound defects in heart development (Cai et al., 2003). Numerous studies in recent years have highlighted the essential role of Isl1 in the transcriptional program for SHF development (Cai et al., 2003; Cohen et al., 2007; Dodou et al., 2004; Klaus et al., 2007; Lin et al., 2006; Lin et al., 2007; Park et al., 2006; Takeuchi et al., 2005). Several direct transcriptional targets of Isl1 have been identified, including other essential cardiac transcription factor genes such as Mef2c and Nkx2-5 (Dodou et al., 2004; Takeuchi et al., 2005). However, the regulation of the Isl1 gene itself, particularly in cardiac lineages, has remained largely unresolved. An important recent study by Evans and coworkers showed that Isl1 requires β-catenin for expression in cardiovascular progenitors, suggesting a role for canonical Wnt signaling upstream of Isl1 (Lin et al., 2007). In addition, it was shown by chromatin immunoprecipitation that the Isl1 proximal promoter was bound directly by β-catenin (Lin et al., 2007). However, other than the proximal upstream region, these studies did not identify any cis-regulatory enhancers, and Wnt and β-catenin signaling alone is unlikely to account for the cardiac progenitor expression of Isl1 during development. In the present study, we identified an enhancer from the mouse Isl1 gene with activity in the SHF, which allowed us to identify Isl1 as a direct transcriptional target of Forkhead transcription factors in SHF-derived mesoderm.
The activity of the Isl1 enhancer described here closely matched the endogenous pattern of Isl1 expression in vivo, although the pattern of β-galactosidase expression presented here is somewhat broader than the pattern of endogenous Isl1 mRNA or Isl1 protein (Fig. 2), which is probably due to the longer half-life of β-galactosidase (Thompson et al., 1991; Voon et al., 2005). Importantly, a previously described Isl1-nlacZ knock-in mouse showed a similarly broad pattern of nlacZ expression when compared to the pattern of expression of the endogenous Isl1 transcript (Sun et al., 2007). The expanded Isl1 expression domain defined by the knock-in studies included the outflow tract, aorta, pulmonary artery, atrial septum, venous valves, cardiac ganglia, and the sinoatrial and atrioventricular nodes (Sun et al., 2007). Isl1-F also shows expression in the outflow tract, aorta, pulmonary artery, and atrial septum, but it does not have activity in all the domains (e.g. the motor neurons in the neural tube) seen with the Isl1-nlacZ knock-in allele (Sun et al., 2007). Isl1 is expressed broadly in many regions other than the SHF, including the ventral neural tube and the visceral endoderm (Pfaff et al., 1996), and the expression of the lacZ knock-in allele reflects this broad pattern of expression (Sun et al., 2007). By contrast, the activity of the enhancer described here is restricted to the SHF, including pharyngeal and splanchnic mesoderm and pharyngeal endoderm. The function of Isl1-F as a modular enhancer with activity restricted to the SHF is consistent with the more restricted expression pattern of Isl1-F-lacZ enhancer compared to Isl1-nlacZ knock-in allele. Thus, our results demonstrate that the Isl1-F enhancer faithfully recapitulates a subset of the endogenous Isl1 pattern and suggest that this enhancer, or the smaller Isl1-F2, may be a useful tool for marking the SHF component of the Isl1 expression pattern.
Evolutionary conservation of Isl1-F2 sequence
Evolutionary pressure has produced many radical changes in the cardiopulmonary system of vertebrates, resulting in addition of several components to the primitive chordate ancestor to make a vertebrate heart (Fishman and Chien, 1997). One essential addition is a second chamber that is designed for generating high systemic blood pressure. Further organization of the original tubular heart into separate systemic and pulmonary circulations was accomplished by separation of ventricles and atria by septa (Bishopric, 2005). Fish have a simpler, two-chambered heart that provides both oxygenation and systemic circulation, while birds and mammals have a four-chambered heart including two atria and two ventricles that allow separate flow of oxygenated and deoxygenated blood, and a complete division of the systemic and pulmonary circulations (Bishopric, 2005). Indeed, the right ventricle and corresponding separate outflow tracts are relatively recent additions to the vertebrate heart, and these structures are largely derived from cells of SHF origin, particularly from the anterior regions in the pharyngeal mesoderm (Abu-Issa and Kirby, 2007; Abu-Issa et al., 2004; Black, 2007; Buckingham et al., 2005; Kelly and Buckingham, 2002). Therefore, it is interesting to note that the Isl1-F2 enhancer, which is active in the SHF, is highly conserved among amniotes but not conserved in lower vertebrates such as frogs and fishes (Supplemental Material, Fig. S1).
The conservation of the Isl1-F2 sequence between birds and mammals, together with the lack of sequence conservation with frogs and fishes, suggests that the Isl1-F2 enhancer may have evolved in amniotes as Isl1 function was co-opted for heart development. Consistent with this notion, no cardiac phenotype was reported when Isl1 expression was abolished by morpholino knockdown in zebrafish, and the expression of Isl1 in zebrafish does not occur in heart-forming regions (Korzh et al., 1993; Segawa et al., 2001). Likewise, a scan for ultra-conserved enhancers from the zebrafish Isl1 locus, resulted in the identification of three distal enhancers with activity in motor and sensory neurons, but no cardiac or cardiac progenitor enhancers were identified (Uemura et al., 2005). While these observations do not rule out a role for Isl1 in heart development in lower vertebrates, together, they support the idea that a role for Isl1 in heart development may have co-evolved with the development of the complex cardiopulmonary system found in birds and mammals. If this is the case, then the evolutionary adaptation of Isl1 to heart development would likely have occurred through changes in noncoding sequences, resulting in the acquisition of enhancer elements with activity in the SHF in birds and mammals. It is attractive to speculate that the evolution of the Isl1-F2 enhancer, and its regulation by Forkhead transcription factors, may have played an important role in the evolution of the right ventricle. Future studies focused on the deletion of the Isl1-F2 enhancer from the germline of mice will be important to determine if this cis-acting element is required for heart development in vivo.
Forkhead-Dependent Activation of Isl1 in the SHF
The studies presented here demonstrate a clear requirement for conserved consensus Forkhead binding sites for Isl1-F2 enhancer function in SHF-derived mesoderm in transgenic embryos (Fig. 5). Our results show that each of the three conserved Forkhead (FOX) transcription factor binding sites in Isl1-F2 were efficiently bound in EMSA by at least one of the Forkhead proteins examined, including FoxA2, FoxC2, and FoxF1 (Fig. 4). Each of these members of the Forkhead family is an excellent candidate for direct regulation of Isl1 in vivo, based on expression and functional data.
Mice lacking both the Foxc1 and Foxc2 genes show severe defects in right ventricle and outflow tract formation, and expression of both genes overlaps the expression of Isl1 in the SHF beginning at the cardiac crescent stage at E7.75 (Seo and Kume, 2006). Importantly, Isl1 expression in Foxc1/Foxc2 double mutants was reduced in the pharyngeal and splanchnic mesoderm at E8.5 but was unaffected in the pharyngeal endoderm (Seo and Kume, 2006). These previous results are consistent with our observation that mutation of the FOX sites in the Isl1-F2 enhancer completely abolished enhancer activity in the pharyngeal mesoderm, but did not completely eliminate expression in the pharyngeal endoderm (Fig. 5).
Foxa2 is expressed in the pharyngeal endoderm and the phenotypes in Foxa2 null mice demonstrate an essential role for this factor in endoderm development (Ang and Rossant, 1994; Weinstein et al., 1994). Interestingly, Foxa2 mutant chimeras in which Foxa2 expression was rescued in the visceral endoderm survive longer than Foxa2 null embryos and died around E9.5 with severe heart defects even though expression of Foxa2 was not detected in cardiac mesoderm (Dufort et al., 1998). More recent studies have shown that Foxa2 is expressed in SHF-derived pharyngeal mesoderm where it directly regulates Tbx1 (Hu et al., 2004; Maeda et al., 2006; Yamagishi et al., 2003). Thus, it remains a possibility that FoxA2 regulates Isl1 expression in the SHF by binding to the FOX sites in the Isl1-F2 enhancer at an early time during cardiac mesoderm specification, and indeed, our EMSA results showed that FoxA2 bound specifically to each of the three FOX sites in Isl1-F2.
Foxf1 is expressed broadly in the embryonic and extraembryonic mesoderm, beginning at gastrulation (Kalinichenko et al., 2003; Mahlapuu et al., 2001; Peterson et al., 1997). Eventually, Foxf1 expression becomes more restricted to the lateral mesoderm and then to septum transversum mesenchyme and the visceral mesoderm, which ultimately gives rise to mesenchymal cells of gut-derived organs (Kalinichenko et al., 2003; Mahlapuu et al., 2001; Peterson et al., 1997). These expression data suggest that if FoxF1 regulates Isl1-F2, it is likely to do so early in mesodermal development, when its broad expression encompasses the Isl1-F2-lacZ expression pattern. However, FoxF1 is unlikely to control enhancer activity via direct binding in the splanchnic and pharyngeal mesoderm at later times in development.
Interestingly, Foxh1 null mice fail to form AHF-derived structures, including the outflow tract and right ventricle, but normally form inflow and atrial regions (von Both et al., 2004). Notably, loss of Foxh1 did not affect Isl1 expression in the splanchnic and pharyngeal mesoderm (von Both et al., 2004). Consistent with this earlier report, we did not observe any significant binding of FoxH1 to any of candidate FOX sites in the Isl1-F2 (data not shown). Together, these results suggest that Isl1 does not function downstream of Foxh1 in SHF development.
Although it is not clear exactly which Forkhead transcription factor(s) activate Isl1-F2, it is attractive to speculate that one or more Forkhead proteins may function in the SHF in a manner analogous to FoxA2 function in the endoderm (Cirillo et al., 2002; Cirillo and Zaret, 1999). In that lineage, FoxA2 functions in cooperation with the zinc finger transcription factor GATA4 by binding directly to the albumin gene to open chromatin and allow for transcriptional activation (Cirillo et al., 2002; Cirillo and Zaret, 1999). Interestingly, the Isl1-F2 enhancer has a deeply conserved GATA binding site, which exhibited robust binding to GATA4 in EMSA (data not shown). Thus, it will be interesting to test whether a Forkhead factor functions in cooperation with GATA4 upstream of Isl1 in the SHF in a manner analogous to FoxA2-GATA4 in the endoderm.
Isl1 SHF enhancer-directed transgenes as tools to mark Isl1+ progenitor cells
Isl1-expressing cardiac progenitors contribute to three major cardiac lineages, including cardiomyocytes, smooth muscle, and endothelial cells (Laugwitz et al., 2005; Moretti et al., 2006). These multipotent progenitor cells, which are prevalent in the early embryo, persist into adulthood and are found sparsely within the adult myocardium (Laugwitz et al., 2005; Moretti et al., 2006). Although postnatal Isl1+ cardiac progenitors are not prevalent, they can be isolated from embryos or from adult myocardium and expanded ex vivo (Laugwitz et al., 2005; Moretti et al., 2006), suggesting that these cells may be useful for therapeutic applications. Likewise, human or mouse ES cells can be cultured under conditions that will promote the formation of these multipotent Isl1+ progenitor cells (Laugwitz et al., 2005; Moretti et al., 2006; Qyang et al., 2007). Importantly, however, Isl1 expression alone is not sufficient to identify ES cells or iPS cells with cardiac progenitor potential, since Isl1 expression is associated with many lineages other than those derived from cardiogenic progenitors, including neuronal lineages, oral epithelium, inner ear, thalamus, pituitary cells, pancreatic β cells and distal tubular cells of the kidney (Ahlgren et al., 1997; Dong et al., 1991; Mitsiadis et al., 2003; Nakagawa and O’Leary, 2001; Pfaff et al., 1996; Radde-Gallwitz et al., 2004).
It has been necessary to use a combination of markers, including Flk1, Isl1, and Nkx2-5, to accurately identify progenitor cells that have cardiovascular potential (Moretti et al., 2006). Alternatively, transgenes, under the control of modular transcriptional enhancers, with expression restricted to the SHF or the AHF could be used to mark these multipotent cardiovascular progenitors. Indeed, a GFP transgene under the control of regulatory elements from the Mef2c gene accurately marks Isl1+ progenitor cells with cardiovascular potential (Qyang et al., 2007). However, the Mef2c-AHF enhancer is a direct transcriptional target of Isl1 (Dodou et al., 2004), and is, therefore, downstream in the hierarchy controlling the development of these cells. In this regard, the Isl1-F2-lacZ transgene described here may be a useful reagent for marking multipotent cardiovascular progenitors by combining the early expression of endogenous Isl1 with expression that is restricted to a subdomain of the endogenous Isl1 expression pattern. It will also be interesting to determine what role, if any, Forkhead proteins play in the differentiation of Isl1+ cells to cardiac, smooth muscle, and endothelial lineages.
Supplementary Material
ACKNOWLEDGEMENTS
The authors appreciate helpful comments from Sylvia Evans (UCSD). J. K. was supported by a postdoctoral fellowship from the American Heart Association, Western States Affiliate. This work was supported by NIH grants P01 HL089707, R01 HL64658, and R01 AR52130 to B.L.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–285. doi: 10.1016/j.ydbio.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Dodou E, Heidt AB, De Val SJ, Jaehnig EJ, Greene SB, Olson EN, Black BL. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol Cell Biol. 2004;24:3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Bishopric NH. Evolution of the heart from bacteria to man. Ann N Y Acad Sci. 2005;1047:13–29. doi: 10.1196/annals.1341.002. [DOI] [PubMed] [Google Scholar]
- Black BL. Transcriptional pathways in second heart field development. Semin Cell Dev Biol. 2007;18:67–76. doi: 10.1016/j.semcdb.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Dong J, Asa SL, Drucker DJ. Islet cell and extrapancreatic expression of the LIM domain homeobox gene isl-1. Mol Endocrinol. 1991;5:1633–1641. doi: 10.1210/mend-5-11-1633. [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–3025. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131:5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns. 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18:210–216. doi: 10.1016/s0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–425. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- Lin L, Bu L, Cai CL, Zhang X, Evans S. Isl1 is upstream of sonic hedgehog in a pathway required for cardiac morphogenesis. Dev Biol. 2006;295:756–63. doi: 10.1016/j.ydbio.2006.03.053. [DOI] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol. 2008;323:98–104. doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda J, Yamagishi H, McAnally J, Yamagishi C, Srivastava D. Tbx1 is regulated by forkhead proteins in the secondary heart field. Dev Dyn. 2006;235:701–710. doi: 10.1002/dvdy.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Angeli I, James C, Lendahl U, Sharpe PT. Role of Islet1 in the patterning of murine dentition. Development. 2003;130:4451–4460. doi: 10.1242/dev.00631. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent Embryonic Isl1(+) Progenitor Cells Lead to Cardiac, Smooth Muscle, and Endothelial Cell Diversification. Cell. 2006;127:1151–65. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, O’Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan E, Monovich A, Tirosh-Finkel L, Harrelson Z, Rousso T, Rinon A, Harel I, Evans SM, Tzahor E. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135:647–657. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou KN, Izumiya Y, Walsh K. Forkhead transcription factors and cardiovascular biology. Circ Res. 2008;102:16–31. doi: 10.1161/CIRCRESAHA.107.164186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech Dev. 1997;69:53–69. doi: 10.1016/s0925-4773(97)00153-6. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. Embo J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–421. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. HNF-3 beta as a regulator of floor plate development. Cell. 1994;76:103–115. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 2006;296:421–36. doi: 10.1016/j.ydbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Voon DC, Subrata LS, Baltic S, Leu MP, Whiteway JM, Wong A, Knight SA, Christiansen FT, Daly JM. Use of mRNA- and protein-destabilizing elements to develop a highly responsive reporter system. Nucleic Acids Res. 2005;33:e27. doi: 10.1093/nar/gni030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Winnier GE, Kume T, Deng K, Rogers R, Bundy J, Raines C, Walter MA, Hogan BL, Conway SJ. Roles for the winged helix transcription factors MF1 and MFH1 in cardiovascular development revealed by nonallelic noncomplementation of null alleles. Dev Biol. 1999;213:418–431. doi: 10.1006/dbio.1999.9382. [DOI] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers EN, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Rivera-Feliciano J, Pu WT. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun. 2008;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.