Abstract
Polychlorinated biphenyls (PCBs) are toxic environmental contaminants that represent a class of 209 congeners characterized by different degree of chlorination and substitution patterns. Most of experimental studies about microbial degradation of PCBs have been conducted on PCB mixtures, even though evidence accumulated in bacteria and other organisms shows that exposure to different congeners may have different biological effects. Microcosm experiments were conducted using aerobic agitated soil slurries individually exposed to PCB congeners with different degrees of chlorination: PCB-3, 15, 28, and 77, and the commercial mixture Aroclor 1242. After four weeks of incubation, PCBs were analyzed by gas chromatography/mass spectrometry (GC/MS) showing different transformation extents: With the exception of PCB-15 that was not significantly transformed (7%), biodegradation rates decreased with the degree of chlorination, from 75% for PCB-3 to 22% for PCB-77 and Aroclor 1242. The bacterial abundance, as measured by colony counting and 16S rDNA quantification by real-time PCR, was lower (of about 40%) in soil microcosms exposed to the higher-chlorinated congeners, PCB-28, PCB-77, and Aroclor 1242, as compared to non-exposed soils and soils exposed to the lower-chlorinated congeners, PCB-3 and PCB-15. The relative abundance of different taxonomic groups, as determined by real-time PCR, revealed an increase of β-Proteobacteria and Actinobacteria in all microcosms exposed to PCBs, as compared with non-exposed soil. In addition, exposure to PCB-77 and Aroclor 1242 resulted in a higher abundance of α-Proteobacteria and Acidobacteria. Globally, these results suggest that exposure to PCBs (and especially to higher-chlorinated congeners and Aroclor 1242) selected bacterial groups involving most known PCB degraders, i.e., β-Proteobacteria and Acidobacteria. The quantification of biphenyl dioxygenase (BPH) genes - involved in the aerobic degradation of PCBs - using real-time PCR showed that exposure to all PCB congeners and Aroclor 1242 resulted in a marked increase of two out of the four BPH genes tested, similarly suggesting the selection of PCB-degrading bacteria. This paper showed that exposure to different PCB congeners leads to different structures of the soil bacterial community and BPH genes expression patterns.
Introduction
Polychlorinated biphenyls (PCBs) are toxic environmental contaminants that were manufactured widely during a half century and used for a variety of industrial applications, such as lubricants, dielectric fluids, and plasticizers. It is usually estimated that over 1.5 million tons of PCBs were produced worldwide (Pieper and Seeger, 2008). Because of their toxicity and persistence in the environment, production and usage of PCBs were banned in the U.S since 1979 (Field and Sierra-Alvarez, 2008). However, because of their relative volatility and high chemical stability, PCBs have been largely dispersed and are today detected everywhere, in virtually every compartment of the ecosystem, including air, water, soil, sediments, and living organisms. PCBs are highly lipophilic and their bioaccumulation along the food chain makes low environmental concentrations a threat for the wildlife and human health (Borja et al., 2005; Furukawa and Fujihara, 2008). Although the acute toxicity for adult humans is rather low, chronic exposure to PCBs induce neurobehavioral and immunological disorders in children. According to the Department of Health and Human Services (DHHS), the U.S. Environmental Protection Agency (EPA), and the International Agency for Research on Cancer (IARC), PCBs are today suspected to be carcinogenic in animals and humans (Borja et al., 2005; Pieper and Seeger, 2008).
Unlike many other organic compounds, the chemical stability of PCBs allows them to resist microbial biodegradation and persist over long periods of time in soil and sediments (Furukawa and Fujihara, 2008). PCB molecules consist of a biphenyl core with 1 to 10 chlorine atoms attached, resulting in 209 different congeners characterized by different degree of chlorination and substitution patterns. PCBs in the environment are generally found in complex mixtures originating from commercial formulations, such as Aroclor (Monsanto, USA) (Borja et al., 2005). Soils often contain a higher proportion of higher-chlorinated congeners, although air is generally dominated by lesser-chlorinated fractions (Hornbuckle et al., 2004).
Although they are classified as persistent pollutants, biodegradation of PCBs has been well documented (Abraham et al., 2002; Ohtsubo et al., 2004; Borja et al., 2005; Vasilyeva and Strijakova, 2007; Field and Sierra-Alvarez, 2008; Pieper and Seeger, 2008). Two major microbial metabolic routes are known: Anaerobic and aerobic degradation of PCBs, depending of the degree of chlorination of the PCB congener, the redox conditions, and the type of microorganism involved (Abraham et al., 2002; Borja et al., 2005). Generally, PCB congeners with four or more chlorine atoms undergo microbial anaerobic reductive dechlorination, an energy yielding process, where PCBs serve as electron acceptor for the oxidation of organic carbon. Chlorine atoms are preferentially removed from the meta and para positions on the biphenyl structure, leaving lesser-chlorinated ortho-substituted congeners (Olson et al., 2003). Microorganisms that reductively dechlorinate PCBs are widespread within PCB contaminated sediments and involve a variety of species (Tiedje et al., 1987; Abraham et al., 2002; Cho et al., 2003). Lower-chlorinated PCBs congeners, possibly resulting from anaerobic dechlorination, undergo co-metabolic aerobic oxidation mediated by dioxygenase enzymes, resulting in ring opening and potentially complete mineralization of the molecule (Kohler et al., 1988; Furukawa et al., 2004; Vasilyeva and Strijakova, 2007).
The number of chlorine atoms per molecule and the placement of chlorine atoms are important factors for the aerobic biodegradation via oxidative enzymes (Ohtsubo et al., 2004; Furukawa and Fujihara, 2008). Adjacent unchlorinated atoms allow for hydroxylation of the ring and formation of arene oxide intermediates, facilitating the aromatic ring cleavage (Broja et al., 2004). Generally, PCB congeners with three or fewer chlorine atoms per molecule are easily degraded, and the ones with five or more are quite recalcitrant (requiring reductive dechlorination prior to aerobic mineralization) (Field and Sierra-Alvarez, 2008). Aerobic biodegradation of PCBs typically involves two cluster of genes, the first one responsible for the transformation into chlorobenzoates (upper pathway), and the second, for the further mineralization of chorobenzoates (lower pathway) (Furukawa and Fujihara, 2008). PCBs are typically first oxidized by biphenyl dioxygenase genes (BPH) to produce cis-dihydrodiol intermediates that undergo ortho- or meta-ring cleavage to form chlorobenzoates (Borja et al., 2005). However, even lower-chlorinated congeners in soil can be slow to degrade due to catabolite repression by other readily available substrates (Callahan et al., 1979).
Although studies conducted with single PCB congeners suggest different susceptibility to degradation and metabolic routes depending on the chlorination degree and substitution pattern (Borja et al., 2005), there is currently very little information about the effect of different congeners on the soil bacterial community and the induction of genes involved in PCB degradation. Most of studies about PCB-degrading microbial communities have been conducted on PCB mixtures (Nogales et al., 1999; 2001; Leigh, 2006; Aguirre de Cárcer et al., 2007), even though evidence accumulated in other organisms shows that exposure to different congeners induces different biological effects and responses (Safe, 1993; Giesy and Kannan, 1998).
The objective of this research was to investigate the effect of different PCB congeners on the soil bacterial community structure and the abundance of genes involved in the aerobic metabolism of PCBs. Microcosm experiments were conducted using aerobic agitated soil slurries individually exposed to specific PCB congeners with different degrees of chlorination: PCB-3, 15, 28, and 77, and the commercial mixture Aroclor 1242 (Figure 1). After four weeks of incubation, the extend of PCB transformation was determined by gas chromatography/mass spectrometry (GC/MS) and the microbial abundance in soil exposed to different PCBs was measured by cell counting and real-time polymerase chain reaction (PCR) amplification of ribosomal DNA (16S rDNA). The abundance of specific bacterial groups and BPH genes were analyzed by real-time PCR using taxonomic group-specific primers and BPH-specific primers, respectively.
Figure 1.
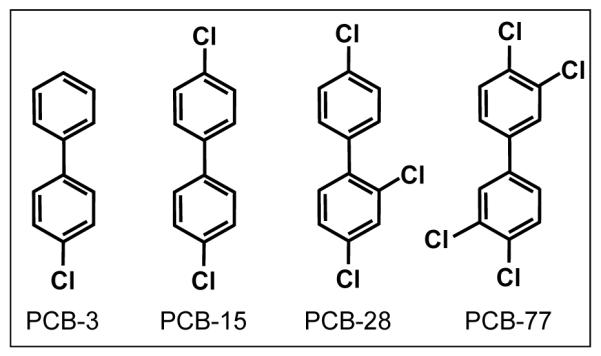
Structure of PCBs used in soil microcosm experiments. The PCB names used refer to the UIPAC numbers: PCB-3: 4-monochlorinated biphenyl, PCB-15: 4,4′-dichlorinated biphenyl, PCB-28: 2,4,4′-trichlorinated biphenyl, and PCB-77: 3,4,3′,4′-tetrachlorinated biphenyl.
Material and Methods
Soil samples
A synthetic soil medium was prepared by mixing the general-purpose peat-based medium, PRO-MIX BX (Premier Horticulture, Quakertown , PA) with all-purpose sand (Quikrete, Atlanta, GA) in the proportion of 40:60 by weight. PRO-MIX BX growing medium consisted in Canadian sphagnum peat moss (75 - 85% by volume), horticultural-grade perlite, vermiculite, and dolomitic and calcitic limestone. The medium was characterized as follows: pH: 5.5 - 6.5, electrical conductivity: 1.3 - 2.0 mmhos cm-1, air porosity: 17 - 22% by volume, and bulk density: 0.13 - 0.16 g cm-3.
Microcosm experiments
Soil slurries were prepared by mixing synthetic soil aliquots with sterile deionized water in a ratio 1:2 w/v. Forty milliliters of slurry were then introduced into pre-sterilized 250-mL penicillin bottles. Slurry samples were spiked with individual PCB congeners and Aroclor 1242 (formulated as 5 mg mL-1 acetone stock solution) to a final concentration of 60 μg mL-1. Flasks were sealed with Teflon-lined rubbers stoppers. Non-exposed control flasks consisted in similar soil slurries spiked with an equivalent volume of acetone. Control sterile PCB-exposed flasks contained soil slurry autoclaved for 1 hour at 121 °C, 1 atm and spiked with 60 μg mL-1 of PCB. Flasks were then incubated under shaking at 125 rpm, in the dark, at room temperature for 4 weeks. In order to maintain aerobic conditions, the headspace of the flasks was flushed every three days with air for 3 min under a chemical hood. After four weeks of incubation, aliquots of slurries were collected for bacterial counts, PCB analyses, bacterial community structure characterization, and quantification of BPH genes. For viable bacteria counting, samples were stored at 4 °C for less than 6 hours until processing. For PCB analysis and DNA extraction, samples were stored for longer periods of time at −80 °C. Three experimental replicates were prepared for each treatment. Bacterial number determination and DNA amplification were performed on individual replicates and results were presented as mean ± standard deviation of the replicates. For PCB analyses, equal aliquots of three replicates were combined; extraction and analysis were performed on combined samples. The statistical significance of the results was determined by conducting t-test on the values recorded in PCB-exposed samples (i.e., bacterial numbers or levels of amplification of specific markers) as compared with values recorded in non-exposed controls. Statistical analyses was performed using the function TTEST of Microsoft Excel 2003. Differences between PCB-exposed samples and non-exposed controls were considered statistically significant for p-values < 0.05.
Bacterial enumeration
To determine the bacterial concentration in slurries exposed to different PCB congeners, 1-mL aliquots were collected, weighted, and diluted 10 times in sterile phosphate buffer solution (PBS) in 15-mL centrifugation tubes. Tubes were agitated horizontally at 200 rpm for 1 hour (Ogram, 1998). Suspensions were then centrifuged under refrigeration at 1,000 rpm for 2 min to remove soil particles. Serial dilutions were made with sterile PBS and plated on two different media: Luria-Bertani (LB) agar and soil LB-supplemented agar prepared as follows: 50 g of soil were blended with 250 mL PBS and centrifuged at 4,000 rpm for 10 min to remove solid particles; the supernatant was mixed with 8 g LB agar and 9 g granulated agar, watered to 1,000 mL, and autoclaved for 15 min at 121°C, 1 atm. Plates were incubated for 2 days in the dark at 28 °C before colony forming unit (CFU) counting. Only plates containing between 100 and 300 CFU were considered and at least 3 plates were counted per sample (Gerhardt et al., 1993). Alternatively and because typically, less than 1% of soil bacteria are cultivable (Baker et al., 2003), the cell biomass was also estimated based on the quantification of bacterial ribosomal DNA (16S rDNA) using real-time PCR.
DNA extraction
Total DNA was extracted from 500 to 750 mg of pre-weighted soil slurry using UltraClean™ Soil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA) following the manufacturer’s recommendations. Purified DNA was eluted from the silica membrane using two times 50 μL of elution buffer. DNA extracts were quantified by the O.D.260 using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). The quality of the DNA was assessed by the ratio O.D.260/O.D.280 (all DNA samples showed a ratio higher than 1.9). When necessary, DNA was concentrated by ethanol precipitation using standard protocols (Ausubel et al., 1999). Purified DNA was stored at −80 °C.
Bacterial community structure
The effect of different PCB congeners on the soil bacterial community structure was assessed using real-time (quantitative) PCR following a method described by Blackwood et al. (2005) and Fierer et al. (2005). Total DNA extracted from soil slurries was used as a template for the PCR-amplification of ribosomal DNA using taxonomic group-specific primers (Table 1). PCR reactions were carried out on an ABI Prism® 7300 Sequence Detection System using SYBR® Green PCR Master Mix with the passive reference ROX™ (Applied Biosystems, Foster City, CA). Cycling conditions were as prescribed by the manufacturer: Initial activation/denaturation at 95 °C for 10 min; 40 cycles: denaturation at 95 °C for 15 sec and annealing-extension at 60 °C for 1 min. The dissociation protocol was run from 60 to 95 °C. Results were calculated according to the manufacturer’s recommendations using the relative quantification method: In order to compare the relative abundance of different bacterial groups, the amplification levels of group-specific genes were first normalized by the amplification level of 16S rDNA (universal marker), used as an internal standard (Fierer et al., 2005). Then, the relative abundance of each bacterial group was calculated as the ratio between the normalized amplification levels for each group-specific genes in PCB-exposed samples and the corresponding normalized amplification levels in non-exposed controls. Results were computed with ABI Prism 7300 software v1.3.1.
Table 1.
Primers used for group-specific bacterial identification and biphenyl dioxygenase quantification
| Target group | Sequence (5′-3′) | Primer | Reference |
|---|---|---|---|
| Universal (F) | ACT CCT ACG GGA GGC AGC AG | Eub338 | Lane (1991) |
| Universal (R) | ATT ACC GCG GCT GCT GG | Eub518 | Muyzer et al. (1993) |
| α-Proteobacteria (R) | TCT ACG RAT TTC ACC YCT AC | Alf685 | Lane (1991) |
| β-Proteobacteria (R) | TCA CTG CTA CAC GYG | Bet680 | Overmann et al. (1999) |
| Actinobacteria (F) | CGC GGC CTA TCA GCT TGT TG | Actino235 | Stach et al. (2003) |
| Firmicutes (F) | GCA GTA GGG AAT CTT CCG | Lgc353 | Meier et al. (1999) |
| Bacteroidetes (F) | GTA CTG AGA CAC GGA CCA | Cfb319 | Manz et al. (1996) |
| Acidobacteria (F) | GAT CCT GGC TCA GAA TC | Acid31 | Barns et al. (1999) |
| BPH D.1.B (F) | GGA CGT GAT GCT CGA (C/T)CG C | BPH1-F | Baldwin et al. (2003) |
| BPH D.1.B (R) | TGT T(C/G)G G(C/T)A CGT T(A/C)A GGC CCA T | BPH1-R | |
| BPH D.1.C (F) | GAC GCC CGC CCC TAT ATG GA | BPH2-F | |
| BPH D.1.C (R) | AGC CGA CGT TGC CAG GAA AAT | BPH2-R | |
| BPH D.2.A (F) | CCG GGA GAA CGG CAG GAT C | BPH3-F | |
| BPH D.2.B (F) | AAG GCC GGC GAC TTC ATG AC | BPH4-F | |
| BPH D.2.A/B (R) | TGC TCC GCT GCG AAC TTC C | BPH3/4-R | |
| 16S rDNA (F) | GAA TTG ACG GGG GCC CGC ACA AG | E916F | Baker et al. (2003) |
| 16S rDNA (R) | AGG GTT GCG CTC GTT G | E1115R |
Quantification of biphenyl dioxygenase genes
Quantification of BPH genes in slurries exposed to different PCB congeners was performed by real-time PCR using gene-specific primers, as described by Baldwin et al. (2003). Bacterial ribosomal DNA (16S rDNA) primers, used as an internal standard, were designed according to Baker et al. (2003) (Table 1). PCR reactions were carried out as previously described on an ABI Prism® 7300 Sequence Detection System using SYBR® Green PCR Master Mix (Applied Biosystems), except for the cycling conditions that were as in Baldwin et al. (2003): Initial activation/denaturation at 95 °C for 10 min; 40 cycles: denaturation at 95 °C for 15 sec, annealing at 57 – 63 °C for 30 sec, and extension at 87 °C for 1 min. The dissociation protocol was run from 60 to 95 °C. Results were calculated using the relative quantification method: The amplification levels of BPH genes were normalized by the amplification level of 16S rDNA, used as an internal standard. Results were expressed by reference to the amplification levels recorded in non-exposed soil controls.
PCB analyses
For PCB analysis, 1-mL aliquots of microcosm slurries were aseptically removed using a cut-off pipet tip, while maintaining the flask under agitation, and directly transferred into pre-weighted 8-mL glass vials sealed with Teflon-lined screw caps (aliquots were collected as four 250-μL fractions combined in one tube). Aliquots were weighted and then extracted using a procedure described in Bedard et al. (2005): PCBs were extracted with 5 mL of anhydrous diethyl ether (Acros Organics, Morris Plains, NJ) by vigorous horizontal shaking at 250 rpm on a platform shaker for 16 h. Extraction efficiency was controlled by the extraction of standards prepared by spiking sterile slurries with 0, 10, 25, and 100 μg mL-1 of PCB. Standards curves constructed after extraction and analysis showed linear trends with R2 > 0.99. Second extraction of 100 μg mL-1 standards resulted in 1 – 5% of PCB recovery. PCB analyses were as described elsewhere (Hu et al., 2008): PCB extracts were diluted by 10 folds using hexane and spiked with 2,2′,3,4,4′,5,6,6′-octachlorobiphenyl (PCB-204) as an internal standard before analysis. Samples were analyzed for a suite of 209 congeners using gas chromatography/mass spectrometry/mass spectrometry (GC/MS/MS) based on a modified EPA method 1668A. The quantification of PCB homologues was performed using an Agilent 6890N gas chromatograph equipped with an Agilent 7683 series autosampler (Agilent, Santa Clara, CA) and coupled to a Waters Micromass Quattro Micro GC mass spectrometer (Milford, MA, USA). Operational conditions were: Electron impact (EI) positive mode at 70 eV, multiple reaction monitoring (MRM), and trap current 200 μA. Two μL of sample were injected in splitless mode in a Supelco SPB™-Octyl fused silica capillary column (30 m × 250 μm i.d. × 0.25 μm film thickness; Supelco, St Louis, MO) with helium as carrier gas at a constant flow rate of 0.8 mL min-1. The collision gas was carrier-grade argon. The oven temperature profile was 75° C, held constant for 5 min, then increased to 150 °C at 15 °C per min, held for 1 min, then increased to 280° C at 2.5° C per min, and held for 3 minutes.
Results and Discussion
Microbial numbers and community structure
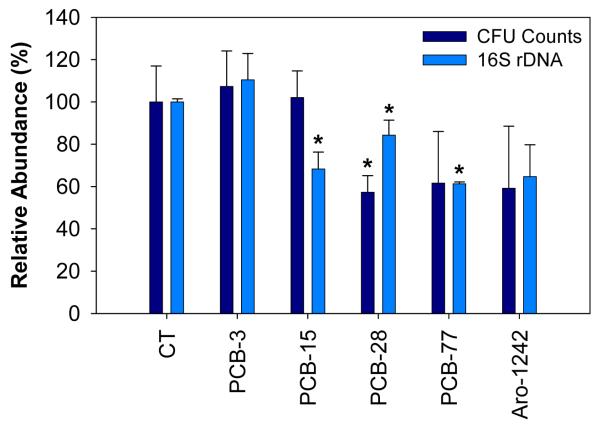
Incubation of soil slurries for 4 weeks in agitation was conducted to investigate the effect of different PCB congeners on both the bacterial abundance and community structure (distribution of bacterial groups). Total bacterial numbers were estimated by CFU counting and quantification of bacterial ribosomal DNA (16S rDNA). Figure 2 shows the relative bacterial numbers in microcosms exposed to different PCB congeners. We observed that exposure to higher-chlorinated congeners: PCB-28, PCB-77, and Aroclor 1242, resulted in lower bacterial numbers (as measured by colony counting and quantification of bacterial ribosomal DNA). The observed reduction of bacterial abundance was statistically significant (p-value < 0.05) in samples exposed to PCB-15 (p = 0.002) , PCB-28 (p = 0.019), and PCB-77 (p = 0.0) as measured by 16S rDNA, and only in the sample exposed to PCB-28 (p = 0.017) as measured by CFU counting. This may be explained by a higher toxicity of higher-chlorinated PCBs for some bacterial groups. Indeed, PCB-77 is expected to be the most toxic, both because of a higher number of chlorine atoms and chlorination at 3,4-position (Borja et al., 2005). On the other hand, as a mixture of PCBs, Aroclor 1242 may contain more toxic congeners. Toxicity of PCBs for bacterial cells has been well documented: Cámara et al. (2004) reported a significant decrease of the viability of E. coli cells exposed to 2 mM of PCB-1, 5, and 23 (377, 446, and 515 μg mL-1). However, unlike our observations, the authors noticed a slight decrease of toxicity with increased chlorine substitution. The relationship between bacterial toxicity and degree of chlorination is highly variable according to the source: For instance, using the bioluminescent bacteria Photobacterium phosphoreum, Chu, et al. (1997) observed a high variability of the toxicity of PCB congeners from dichlorinated to hexachlorinated biphenyls without clear correlation with the degree of chlorination or co-planarity of the molecule. The situation is likely to be further complicated because PCBs can be transformed by bacteria into potentially more toxic metabolites, such as dihydrodiols, dihydroxybiphenyls, chlorobenzoates, and chlorocatechols (Cámara et al., 2004; Parnell et al., 2006; Vasilyeva and Strijakova, 2007). In addition, in the presence of high concentration (as used in our experiments), the hydrophobicity of PCBs can cause membrane degradation leading to cell lysis (Ohtsubo et al., 2004; Vasilyeva and Strijakova, 2007).
Figure 2.
Bacterial abundances in soil microcosm cultures after 4 weeks of incubation in the presence of different PCB congeners and the mixture Aroclor 1242, as determined by CFU counting and real-time PCR quantification of 16S rDNA. Relative abundances were calculated by reference with non-exposed control microcosms (CT). The error bars represent standard deviations between experimental triplicates. Bacterial abundance in PCB-exposed samples showing statistically significant differences with the non-exposed controls were marked with a star (*).
Real-time or quantitative PCR has recently emerged as a popular tool to quantified DNA and it has been used for estimating the abundance of microbial groups or specific metabolic genes, or for gene expression analysis when coupled with a reverse-transcription step. In this paper, we used real-time PCB both to estimate the relative abundance of important bacterial taxonomic groups and BPH genes in soil microcosms exposed to different PCB congeners and the commercial mixture Aroclor 1242.
Except for PCB-28, a generally good correlation was obtained between estimation of bacterial biomass based on CFU counting and amplification of 16S rDNA. Notoriously, only a small fraction of bacteria in the environment is cultivable (< 1%), which could lead to significant differences between results obtained using cultivation-based and cultivation-independent methods (Abraham et al., 2002; Baker et al., 2003). However, for large and diverse bacterial communities, such as found in soil, one can expect to observe similar trends using both methods. On the other hand, estimations based on 16S rDNA do not allow to distinguish between living and dead bacteria. Even though DNA in dead cells is expected to be biodegraded, this limitation can lead to overestimation of active bacteria.
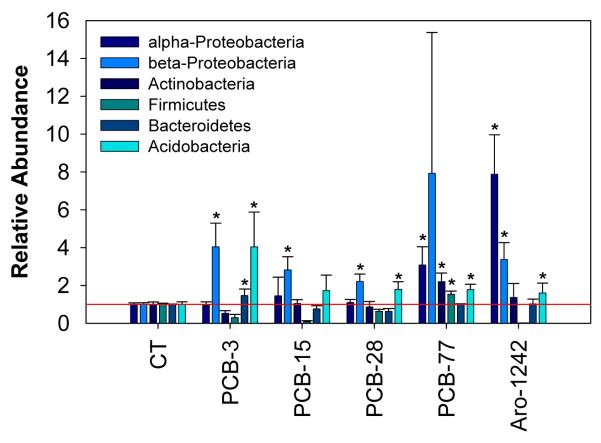
The bacterial community structure in soil microcosms exposed to different PCB congeners and Aroclor 1242 was characterized by the real-time PCR amplification of dominant bacterial groups following an approach described by Blackwood et al. (2005) and Fierer et al. (2005). Figure 3 shows the relative abundance of each bacterial group as calculated by the ratio between the copy number of different group-specific markers and the universal marker. The relative abundance (or fractional copy number) constitutes a more reliable measurement of bacterial group representation, since the efficiency of PCR amplification is expected to vary across DNA samples (Fierer et al., 2005). Although an important variability was observed between experimental replicates, an emergent pattern can be identified in the different microcosms: Exposure to individual congeners and Aroclor 1242 resulted in higher numbers of β-Proteobacteria (2.2 to 7.9 times higher) and Acidobacteria (1.6 to 4.0 times higher). The increase of relative abundance of β-Proteobacteria was statistically significant in samples exposed to PCB-3 (p = 0.007), PCB-15 (p = 0.004), PCB-28 (p = 0.002), and Aroclor 1242 (p = 0.0). The increase of relative abundance of Acidobacteria was statistically significant in samples exposed to PCB-3 (p = 0.021), PCB-28 (p = 0.012), PCB-77 (p = 0.002) and Aroclor 1242 (p = 0.028). In addition, a higher proportion of α-Proteobacteria was detected in microcosms exposed to PCB-77 and Aroclor 1242 (3.1 and 7.9 times higher with p-values = 0.02 and 0.0, respectively). Interestingly, β-Proteobacteria constitutes one of the major bacterial groups were BPH genes where described, i.e., Comamonas and Burkholderia sp., suggesting that exposure to PCBs selected for members of PCB-degraders. None of the primer sets used was specific to γ-Proteobacteria, so no information was obtained about the relative abundance of this other group of potential PCB-degraders (Pseudomonas sp.). On the other hand, exposure to PCB-77 and Aroclor-1242 selected higher fractions of Actinobacteria (2.2 times higher (p = 0.004) and 1.3 times higher (statistically non-significant), respectively). The phylum Actinobacteria comprises the genus Rhodococcus, which is another well characterized group of PCB-degraders. Our results are generally consistent with other reports about the bacterial community structure in PCB-exposed soils obtained based on 16S rDNA: Studying the phylogenetic distribution of bacterial groups and BPH genes in planted and non-planted soils, Aguirre de Cárcer et al. (2007) observed an enrichment of β-Proteobacteria, dominated by the Burkholderiales order, in PCB-contaminated soil (Lhenice, Czech Republic) by comparison with non-contaminated soil. Using RNA extracts from soil heavily contaminated with PCBs (Wittenberg, Germany), Nogales et al. (1999; 2001) identified a large majority of α-, β-, and γ-Proteobacteria (34%, 33%, and 7%), as well as Acidobacteria (14%) and Actinobacteria (6.5%), which is rather consistent with our observations using group specific primers. Especially, the authors reported a higher abundance of β-Proteobacteria than usually observed in soil (Nogales et al., 2001). In order to investigate the metabolic potential for PCB biodegradation in non-contaminated environments, Macedo et al. (2007) conducted aerobic microcosm experiments in the presence of Aroclor 1242 using bacterial inoculum from non-contaminated soils and sediments. Consistently with our findings, bacterial analyses using 16S rDNA fingerprinting revealed a predominance of α- and β-Proteobacteria in the biofilm developing on PCBs after 35 days of incubation. In similar experiments using a microbial community from a heavily PCB-contaminated soil, Tillmann et al. (2005) detected a predominance of bacteria belonging to α- and β-Proteobacteria, and, to a lesser extent, to Bacteroidetes and Firmicutes. However, it is noteworthy that the estimated relative abundances obtained using real-time PCR may not reflect the true percentages of bacterial groups analyzed because of differences in DNA extraction efficiencies, ribosomal DNA copy numbers, and the exclusion of several potentially important bacterial phyla (Fierer et al., 2005).
Figure 3.
Profiles of microorganisms in soil microcosms incubated for 4 weeks in the presence to different PCB congeners. The microbial abundance of different taxonomic groups was obtained by quantitative PCR-amplification of group-specific markers (Fierer et al., 2005). Relative abundance was calculated by normalizing the amplification level of each group by ribosomal DNA (universal marker), used as an internal standard. Results were expressed by reference with control microcosms non-exposed to PCBs (CT). The error bars represent standard deviations between experimental triplicates. Abundance of bacterial groups in PCB-exposed samples showing statistically significant increase by comparison with non-exposed controls were marked with a star (*).
PCB biodegradation
The biodegradation percentages of the different PCB congeners after 4 weeks of incubation in aerobic soil microcosms were as follow: PCB-3: 75%, PCB-15: 7%, PCB-28: 53%. PCB-77: 22%, and Aroclor 1242: 22%. Except for PCB-15 that was not significantly transformed, we observed that PCBs were efficiently biodegraded in active microcosms by comparison with autoclaved controls. Recovery of PCBs in autoclaved controls were 79% for PCB-3, 89% for PCB-15, 75% for PCB-28, 77% for PCB-77, and 103% for Aroclor 1242. Aerobic degradation rates decreased with the degree of chlorination, which is consistent with previous reports (Macedo et al., 2005; Tillmann et al., 2005; Vasilyeva and Strijakova, 2007; Furukawa and Fujihara, 2008). Indeed, the presence of more chlorine substituents reduces the electron density of the aromatic cycle(s), making more difficult the oxidative attack by dioxygenases. The rates of degradation observed are also consistent with aerobic degradation of PCBs described in the literature (Kohler et al., 1988; Barja et al., 2005; Vasilyeva and Strijakova, 2007). Biodegradation of tetra-chlorinated PCB-77, expected to proceed mainly through reductive dechlorination, might occurred in anaerobic microniches in soil particles (Macedo et al., 2005; 2007). As previously demonstrated by Abraham et al. (2005) and Macedo et al. (2007), our results show that non-contaminated soil bacteria have intrinsic potential to metabolize PCBs.
Biphenyl dioxygenase genes
Primers specific to BPHs were designed to amplified the gene coding for the large subunit (bphA1), which determines the substrate specificity (Furukawa et al., 2004; Furukawa and Fujihara, 2008). Based on the sequence similarity of the large subunit, BPHs belong to two different families of aromatic oxygenases: D.1, which includes the Gram-negative BPHs, D.1.B and D.1.C, and D.2, which includes the Gram-positive BPHs, D.2.A and D.2.B. D.1.B includes BPHs from Comamonas testosteroni B-356, Burkholderia sp. JB1, and Pseudomonas sp. KKS102; D.1.C includes BPHs from Pseudomonas sp. B4, Burkholderia sp. LB400, and P. pseudoalcaligenes KF707; D.2.A includes BPHs from Rhodococcus erythropolis TA421, R. globerulus P6, and Rhodococcus sp. M5; and D.2.B includes BPHs from Rhodococcus sp. RHA1 and R. erythropolis BD2 (Baldwin et al., 2003; Furukawa et al., 2004).
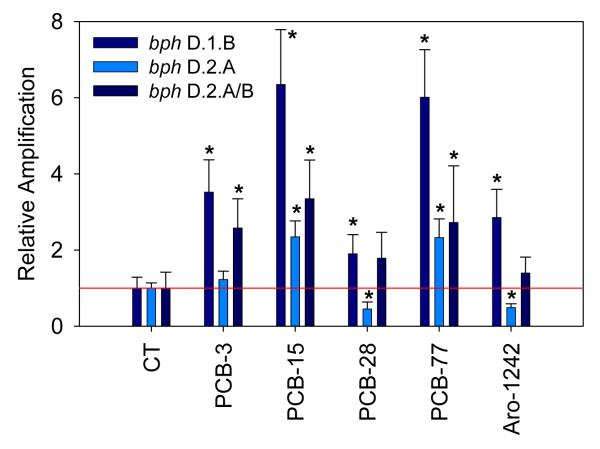
Figure 4 presents the relative abundance of bacterial BPH genes in microcosms exposed to the different PCB congeners and the mixture Aroclor 1242. Four genes were tested representing the four subfamilies of biphenyl dioxygenases: bph D.1.B (PBH-1), bph D.1.C (BPH-2), bph D.2.A. (PBH-3), and bph D.2.A/B (BPH-4) (Baldwin et al., 2003). Results are presented for three out of the four BPH genes since no significant amplification signal was obtained with bph D.1.C (PBH-2). A significant increase of two out of the three genes quantified, bph D.1.B (PBH-1) and bph D.2.A/B (BPH-4), was consistently observed in microcosms exposed to PCB congeners and Aroclor mixture: Relative abundance of bph D.1.B were 3.5 for PCB-3 (p = 0.0), 6.4 for PCB-15 (p = 0.0), 1.9 for PCB-28 (p = 0.01), 6.1 for PCB-77 (p = 0.0), and 2.9 (p = 0.002) for Aroclor 1242; relative abundance of bph D.2.A/B (BPH-4) were 2.6 for PCB-3 (p = 0.001), 3.4 for PCB-15 (p = 0.001), 1.8 for PCB-28 (statistically non-significant), 2.7 for PCB-77 (p = 0.048), and 1.4 (statistically non-significant) for Aroclor 1242. A higher amplification of bph D.2.A (BPH-3) was also detected upon exposure to PCB-15 and PCB-77: 2.3 (p = 0.002) and 2.3 (statistically non-significant), respectively. BPH-1 primers were designed to amplified specifically BPH genes of the subfamily D.1.B, which involves Comamonas testosteroni B-356 and Burkholderia sp. JB1, two well described PCB-degraders belonging to β-Proteobacteria, which were detected at higher levels in all microcosms exposed to PCBs. A similar observation can be made regarding BPH-3 and BPH-4, involving BPHs belonging to D.2.A and D.2.B subfamilies, which involve different Rhodococcus sp. Many Rhodococcus species are PCB-degraders and they belong to Actinobacteria, also detected at slightly higher levels in soil exposed to PCB-77 and Aroclor 1242. However, even though this correlation is attractive, these results do not provide evidence that the higher numbers of BPH genes observed actually belong to any of the β-Proteobacteria or Actinobacteria species detected at higher levels in the samples exposed to PCBs. No clear quantitative relationship can be observed between the relative abundance of bacterial groups and BPH genes in the microcosms exposed to different PCBs. Also it is likely that the group-specific primers used in our experiments overlap other bacterial groups or miss members of the group intended to be quantified and it is equally likely that the BPH primer sets used did not detect all BPH genes (or other genes) involved in the aerobic transformation of PCBs. Also, the relative abundance of BPH genes has been determined using 16S rDNA as an internal standard. Since bacterial species have variable copy number of 16S rDNA, the composition of the microbial community in samples exposed to different treatment may introduce a bias in the calculated relative abundance of BPH genes.
Figure 4.
Quantification of bacterial biphenyl dioxygenase (BPH) genes: D.1.B, D.2.A, and D.2.A/B in microcosm experiments after 4 weeks of incubation in the presence of different PCB congeners, as determined by real-time PCR relative quantification. Ribosomal DNA (16S rDNA) was used as an internal calibrator and the amplification levels were normalized by reference to non-exposed microcosms (CT). The error bars represent standard deviations between experimental triplicates. Abundance of BPH genes in PCB-exposed samples showing statistically significant increase by comparison with non-exposed controls were marked with a star (*).
No clear relationship was observed between the degradation extent of PCB congeners and the abundance of BPH genes, e.g., relatively high levels of the three BPH genes under study were detected after exposure to PCB-15, which was only poorly degraded; conversely, low levels of BPH genes were associated with exposure to PCB-28, which was significantly degraded. This may be explained by several factors, including different substrate specificity of BPH genes for PCB congeners, the presence of other BPH genes not captured by the primers used in this study, or the presence of other catabolic pathways for PCB degradation. Alternatively, the observed discrepancy between extent of PCB biodegradation and abundance of BPH genes could be explained by the cooperative nature of PCB degradation (Abraham et al., 2002). Tillmann et al. (2005) reported the implication of a “metabolic network” of bacteria for the biodegradation of PCBs, suggesting highly complex metabolic pathways.
Nevertheless, our observations suggest that, overall, exposure to individual PCB congeners and Aroclor 1242 resulted in higher levels of bacteria belonging to potential PCB-degraders and higher abundance of BPH genes. The regulation of BPH genes has been studied in some strains and shows different transcriptional control mechanisms by biphenyl, salicylate, and/or cleavage products of PCBs (Furukawa and Fujihara, 2008). However, in this paper, the expression of BPH genes was not specifically investigated; only BPH gene numbers were detected, reflecting the abundance of potential PCB-degraders. Higher numbers of BPH genes in microcosms exposed to PCBs is therefore better explained by a competitive advantage of PCB-degraders than by induction in the presence of specific compounds.
This paper presents evidence that, although specific patterns are difficult to discern, exposure of soil microcosms to different PCB congeners resulted in different bacterial community structures and abundance of BPH genes.
Acknowledgements
This work was supported by the National Institute of Environmental Health Sciences, NIEHS (NIH) (award ES05605), and the West Virginia University Research Corporation (PSCoR award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Cur Op Microbiol. 2002;5:246–253. doi: 10.1016/s1369-5274(02)00323-5. [DOI] [PubMed] [Google Scholar]
- Abraham WR, Wenderoth DF, Glasser W. Diversity of biphenyl degraders in a chlorobenzene polluted aquifer. Chemosphere. 2005;58:529–533. doi: 10.1016/j.chemosphere.2004.08.074. [DOI] [PubMed] [Google Scholar]
- de Cárcer D. Aguirre, Martin M, Karlson U, Rivilla R. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl Environ Microbiol. 2007;73:6224–6232. doi: 10.1128/AEM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. John Wiley and Sons; 1999. [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Meth. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Baldwin BR, Nakatsu CH, Nies L. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microbiol. 2003;69:3350–3358. doi: 10.1128/AEM.69.6.3350-3358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns SM, Takala SL, Kuske CR. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard DL, Pohl EA, Bailey JJ, Murphy A. Characterization of the PCB substrate range of microbial dechlorination process LP. Environ Sci Technol. 2005;39:6831–6838. doi: 10.1021/es050255i. [DOI] [PubMed] [Google Scholar]
- Blackwood CB, Oaks A, Buyers JS. Phylum- and class-specific PCR primers for general microbial community analysis. Appl Environ Microbiol. 2005;71:6193–6198. doi: 10.1128/AEM.71.10.6193-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja J, Taleon DM, Auresenia J, Gallardo S. Polychlorinated biphenyls and their biodegradation. Proc Biochem. 2005;40:1999–2013. [Google Scholar]
- Callahan MA, Slimak MW. Water-related environmental fate of 129 priority pollutants. U.S. Environmental Protection Agency; 1979. [Google Scholar]
- Cámara B, Herrera C, Gonzalez M, Couve E, Hofer B, Seeger M. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol. 2004;6:842–850. doi: 10.1111/j.1462-2920.2004.00630.x. [DOI] [PubMed] [Google Scholar]
- Cho YC, Sokol RC, Frohnhoefer RC, Rhee GY. Reductive dechlorination of polychlorinated biphenyls: Threshold concentration and dechlorination kinetics of individual congeners in Aroclor 1248. Environ Sci Technol. 2003;37:5651–5656. doi: 10.1021/es034600k. [DOI] [PubMed] [Google Scholar]
- Chu S, He Y, Xu X. Determination of acute toxicity of polychlorinated biphenyls to Photobacterium phosphoreum. Bull Environ Contam Toxicol. 1997;58:263–267. doi: 10.1007/s001289900329. [DOI] [PubMed] [Google Scholar]
- Field JA, Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollu. 2008;155:1–12. doi: 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fujihara H. Microbial degradation of polychlorinated biphenyls: Biochemical and molecular features. J Biosci Bioeng. 2008;105:433–449. doi: 10.1263/jbb.105.433. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Suenaga H, Goto M. Biphenyl dioxygenases: Functional versatilities and directed evolution. J Bacteriol. 2004;186:5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesy JP, Kannan K. Dioxin-like and non-dioxin-like toxic effects of polychlorinated biphenyls (PCBs): Implications for risk assessment. Crit Rev Toxicol. 1998;28:511–569. doi: 10.1080/10408449891344263. [DOI] [PubMed] [Google Scholar]
- Gerhardt P, Murray G, Wood W, Krieg N. Methods for General and Molecular Bacteriology. American Society for Microbiology; 1993. [Google Scholar]
- Hornbuckle KC, Smith GL, Miller SM, Eadie BJ, Lansing M. Magnitude and origin of PCBs resuspended in open waters of Lake Michigan. Abs Pap Am Chem Soc. 2004;228:U544–U544. [Google Scholar]
- Hu D, Martinez A, Hornbuckle KC. Discovery of non-Aroclor PCB (3, 3′-dichlorobiphenyl) in Chicago air. Environ Sci Technol. 2008 doi: 10.1021/es801823r. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler HPE, Kohlerstaub D, Focht DD. Co-Metabolism of Polychlorinated-Biphenyls - Enhanced Transformation of Aroclor 1254 by Growing Bacterial-Cells. Appl. Environ Microbiol. 1988;54:1940–1945. doi: 10.1128/aem.54.8.1940-1945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. 16s/23s rRNA sequencing. In: Goodfellow ESAM, editor. Nucleic Acid Techniques in Bacterial Systematics. John Wiley; West Sussex, UK: 1991. pp. 115–175. [Google Scholar]
- Leigh MB, Prouzova P, Mackova M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo AJ, Kuhlicke U, Neu TR, Timmis KN, Abraham WR. Three stages of a biofilm community developing at the liquid-liquid interface between polychlorinated biphenyls and water. Appl Environ Microbiol. 2005;71:7301–7309. doi: 10.1128/AEM.71.11.7301-7309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo AJ, Timmis KN, Abraham WR. Widespread capacity to metabolize polychlorinated biphenyls by diverse microbial communities in soils with no significant exposure to PCB contamination. Environ Microbiol. 2007;9:1890–1897. doi: 10.1111/j.1462-2920.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology-UK. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- Meier H, Amann R, Ludwig W, Schleifer KH. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Dewaal EC, Uitterlinden AG. Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16s ribosomal-RNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales B, Moore ERB, Abraham WR, Timmis KN. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl polluted moorland soil. Environ Microbiol. 1999;1:199–212. doi: 10.1046/j.1462-2920.1999.00024.x. [DOI] [PubMed] [Google Scholar]
- Nogales B, Moore ERB, Llobet-Brossa E, Rossello-Mora R, Amann R, Timmis KN. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl Environ Microbiol. 2001;67:1874–1884. doi: 10.1128/AEM.67.4.1874-1884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogram A. Isolation of nucleic acids from environment samples. In: Burlage RS, Atlas R, Stahl D, Geesey G, Sayler G, editors. Techniques in Microbial Ecology. Oxford University Press; New York, NY: 1998. pp. 273–288. [Google Scholar]
- Ohtsubo Y, Kudo T, Tsuda M, Nagata Y. Strategies for bioremediation of polychlorinated biphenyls. Appl Microbiol Biotechnol. 2004;65:250–258. doi: 10.1007/s00253-004-1654-y. [DOI] [PubMed] [Google Scholar]
- Olson P, Reardon K, Pilon-Smits E. Ecology of rhizosphere bioremediation. In: McCutcheon S, Schnoor J, editors. Phytoremediation. Transformation and Control of Contaminants. John Wiley; 2003. pp. 317–353. [Google Scholar]
- Overmann J, Coolen MJL, Tuschak C. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch Microbiol. 1999;172:83–94. doi: 10.1007/s002030050744. [DOI] [PubMed] [Google Scholar]
- Parnell JJ, Park J, Denef V, Tsoi T, Hashsham S, Quensen J, Tiedje JA. Coping with polychlorinated biphenyl (PCB) toxicity: Physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl Environ Microbiol. 2006;72:6607–6614. doi: 10.1128/AEM.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper DH, Seeger M. Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol. 2008;15:121–138. doi: 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- Safe S. Toxicology, structure-function relationship, and human and environmental-health impacts of polychlorinated-biphenyls - Progress and problems. Environ Health Persp. 1993;100:259–268. doi: 10.1289/ehp.93100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT. New primers for the class Actinobacteria: Application to marine and terrestrial environments. Environ Microbiol. 2003;5:828–841. doi: 10.1046/j.1462-2920.2003.00483.x. [DOI] [PubMed] [Google Scholar]
- Tiedje JM, Boyd SA. Anaerobic degradation of chlorinated aromatic hydrocarbons. Dev Ind Microbiol Ser. 1987;27:117–127. [Google Scholar]
- Tillmann S, Strompl C, Timmis KN, Abraham WR. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbial Ecol. 2005;52:207–217. doi: 10.1016/j.femsec.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Vasilyeva GK, Strijakova ER. Bioremediation of soils and sediments contaminated by polychlorinated biphenyls. Microbiology. 2007;76:639–653. [Google Scholar]