Abstract
The TFAP2C transcription factor is involved in mammary development, differentiation and oncogenesis. Previous studies established a role for TFAP2C in the regulation of ESR1 (ERα) and ERBB2 (Her2) in breast carcinomas. However, the role of TFAP2C in different breast cancer phenotypes has not been examined in detail. To develop a more complete characterization of TFAP2C target genes, ChIP-seq with anti-TFAP2C antibody and expression arrays with TFAP2C knock down were analyzed in MCF-7 breast carcinoma cells. Genomic sequences common to the ChIP-seq data set defined the consensus sequence for TFAP2C chromatin binding as the nine base sequence SCCTSRGGS (S=G/C, R=A/G), which closely matches the previously defined optimal in vitro binding site. Comparing expression arrays before and after knock down of TFAP2C with ChIP-seq data demonstrated a conservative estimate that 8% of genes altered by TFAP2C expression are primary target genes and includes genes that are both induced and repressed by TFAP2C. A set of 447 primary target genes of TFAP2C was identified, which included ESR1 (ERα), FREM2, RET, FOXA1, WWOX, GREB1, MYC and members of the retinoic acid response pathway. The identification of ESR1, WWOX, GREB1 and FOXA1 as primary targets confirmed the role of TFAP2C in hormone response. TFAP2C plays a critical role in gene regulation in hormone responsive breast cancer and its target genes are different than for the Her2 breast cancer phenotype.
Introduction
TFAP2C is a member of the retinoic acid-inducible, developmentally regulated family of AP-2 factors that include five members—TFAP2A (AP-2α), TFAP2B (AP-2β), TFAP2C (AP-2γ), TFAP2D (AP-2δ) and TFAP2E (AP-2ε) (Williams et al., 1988; Moser et al., 1995; Bosher et al., 1996; Zhao et al., 2001; Feng and Williams 2003). AP-2 factors bind to GC-rich regulatory elements in the promoters of target genes through a helix-loop-helix motif in the DNA binding domain, which, except for TFAP2D, is highly homologous in all the AP-2 family members (Eckert et al., 2005). The original report of the AP-2 binding site identified the sequence GCCNNNGGC for the consensus sequence (Williams and Tjian 1991), but later studies suggested greater variability for the end nucleotides and evidence for sequence preferences of the internal triad (McPherson and Weigel 1999). Although binding studies based on in vitro analysis have been valuable, detailed analysis of the sequence involved in chromatin binding by endogenously expressed protein in vivo has not been reported.
AP-2 factors play a critical role in the development and differentiation of ectodermal structures. The AP-2 factors are expressed early in differentiation of the ectoderm and specify cell fates within the epidermis and neural crest (Hoffman et al., 2007; Li and Cornell 2007). Within the human mammary gland, TFAP2A is expressed by 14 weeks gestation and by 22 weeks its expression becomes restricted to the luminal epithelial cells (Friedrichs et al., 2007). TFAP2C is widely expressed within secondary outgrowths in the human mammary gland by 19 weeks gestation, whereas later in development and in the adult mammary gland TFAP2C expression is restricted to myoepithelial cells (Friedrichs et al., 2005; 2007). Overexpression of TFAP2A or TFAP2C in mouse mammary epithelial cells using a MMTV-driven transgene induced both hyperproliferation and apoptosis resulting in lactation failure with an overall hypoplasia of the alveolar mammary epithelium during pregnancy (Jager et al., 2003; Zhang et al., 2003). In mammary tumor models, both TFAP2A and TFAP2C have been shown to be important to cell proliferation, establishment of colonies in soft agar, cell migration and xenograft outgrowth (Orso et al., 2008). However, overexpression of TFAP2C in a MMTV/neu transgenic mouse model resulted in a reduction in the number of tumors and a prolongation of latency (Jager et al., 2005).
From a clinical standpoint, breast cancer phenotypes are commonly divided into four categories based on the expression pattern of the steroid hormone receptors estrogen receptor-alpha (ERS1/ ERα) and progesterone receptor (PGR) and the cell surface receptor ERBB2/HER2/NEU (Sorlie et al., 2001). In addition, the expression patterns of ESR1 and ERBB2 are predictive of the clinical response to anti-hormonal therapy, such as Tamoxifen, and Herceptin, respectively. Furthermore, the various breast cancer phenotypes share common biologic properties and related patterns of gene expression. Interestingly, AP-2 factors have been implicated in the regulation of both ESR1 and ERBB2 expression. In cell line models, TFAP2C binds to the ESR1 promoter and regulates expression from the cloned promoter (deConinck et al., 1995; McPherson et al., 1997; Schuur et al., 2001). In addition, knock down of TFAP2C, but not TFAP2A, significantly reduced ESR1 expression and estrogen response (Woodfield et al., 2007). In contrast, expression patterns in primary breast cancers have demonstrated a correlation between ESR1 and TFAP2A expression (Gee et al., 1999; Turner et al., 1998). On the other hand, several investigators have reported that expression of TFAP2C correlates with reduced survival and Tamoxifen resistance, suggesting a role for TFAP2C in the estrogen signaling pathway (Guler et al., 2007; Gee et al., 2009).
Several studies have demonstrated a role for AP-2 factors in the regulation of ERBB2 expression. AP-2 sites were identified in the promoter of the ERBB2 (HER2) gene and TFAP2A and TFAP2C appear to be capable of inducing expression of the cloned ERBB2 promoter (Bosher et al., 1996; Begon et al., 2005; Delacroix et al., 2005; Yang et al., 2006). In MDA-MB-453 cells, TFAP2C was shown to bind to the ERBB2 promoter and knock down of TFAP2C significantly reduced ERBB2 expression (Ailan et al., 2009). In BT474 breast carcinoma cells, both TFAP2A and TFAP2C were shown to participate in the regulation of ERBB2 expression (Allouche et al., 2008). Furthermore, a correlation has been established between TFAP2A expression and the expression of ERBB2 in primary breast cancers (Turner et al., 1998; Pellikainen et al., 2004; Allouche et al., 2008). In another apparent paradox, high expression of TFAP2C was correlated with the triple negative basal breast cancer phenotype (ESR1-negative/PGR-negative/ERBB2-negative). If AP-2 factors are involved in regulation of ESR1 and ERBB2, it is not clear why high levels of AP-2 expression fail to correlate with ESR1 or ERBB2 expression in the basal phenotype.
One possibility that might explain apparent inconsistencies in AP-2 expression is that examining patterns of AP-2 expression has failed to consider alterations in the functional activity of the factors. For example, the interaction between TFAP2C and other co-factors may re-direct AP-2 to different target genes (McPherson et al., 2002; Eckert et al., 2005). Similarly, functional interactions with co-activators or co-repressors may alter the ability of bound AP-2 to alter transcription. Furthermore, epigenetic modification of certain target genes may make the promoter inaccessible to AP-2 factors, thereby essentially altering the patterns of expression controlled by AP-2 in different breast phenotypes (Woodfield et al., 2009). To better define the functional specificity of AP-2 and identify additional primary target genes, we utilized chromatin immunoprecipitation with direct sequencing of precipitated fragments (ChIP-seq) to characterize the genomic interactions of TFAP2C in hormone responsive MCF-7 cells.
Materials and Methods
Cell Lines
The MCF-7 cell line was obtained from American Type Culture Collection (ATCC) and maintained as previously described (Weigel and deConinck 1993).
Small interfering RNA transfections
On Target Plus siRNA for TFAP2C (J-005238-07), TFAP2A (WOOGE-000001) and Non-targeting siRNA (D-001210-01-05) were obtained from Dharmacon (Lafayette, CO) and used according to the manufacturer as previously described (Woodfield et al., 2007).
Expression array and RT-PCR analysis
After 72 hours following transfection with siRNA, total RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) and subjected to analysis on the Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) in conjunction with the DNA Core Facility at the University of Iowa following the GeneChip Expression Analysis Technical Manual as previously described (Woodfield et al., 2007). Expression array data included is available under the accession number GSE8640. PartekGS software (Partek Inc, St. Louis, MO) implementing the robust microarray analysis algorithm was used to generate gene expression values. For RT-PCR analysis for single genes, total RNA was analyzed by real-time RT-PCR and Western blots were performed to confirm knock-down as previously described (Woodfield et al., 2007). TaqMan primers and detection probes for specific genes were as follows: FOXA1 Assay Hs00270129_m1, WWOX Assay Hs03044790_m1, FREM2 Assay Hs00872621_m1 (Applied Biosystems, Carlsbad, CA).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP analysis for specific genes was performed as previously described (Woodfield et al., 2009) using real-time PCR for detection of immunoprecipitated chromatin for genes of interest as follows: FOXA1-S1 (FOXA1f 5-CAGATGACAAGGGGAGAGGA, FOXA1r 5-AAAAAGCCCCACTTTTGCTT); FOXA1-NCS1 (FOXA1NCS1f 5-CTGCCCATACAGAGTGCTCA, FOXA1NCS1r 5-CAGTTGCTTTCCCTCTCTGG); WWOX-S1 (WWOXf 5- GAGAGGAAGGCGGTGAAAGT, WWOXr 5-CTGGCCCGAAACTGGATG); WWOX-NCS1 (WWOX-NCSf 5-TCTGGTGGTTCAGGGAAGAC, WWOX-NCSr 5-CTGGCATGGGTTCTTACGTT); ESR1-S1(ESR-S1f 5-TTTGCAGATGTTAATACATTTCAGC, ESR-S1r 5-CCCACAAAGGTTTAGCCAGT); FREM2-S1 (FREM2f 5-ACTGCGAAGAGAACAGGAACGTCT, FREM2r 5-CTTTGCAACCGAGGTTCACAGAGT, FREM2p 5- FAMGCATTTCTCACGCATTCCAAA). For FREM2 the specific probe used for detection is shown. For FOXA1, ESR1 and WWOX amplifications, sybr-green dye was used for detection without a specific probe. All PCR reactions were also analyzed by gel electrophresis confirming size of amplicon.
Chromatin immunoprecipitation with direct sequencing
Preparation of DNA for chromatin immunoprecipitation with direct sequencing (ChIP-Seq) experiments were carried out as previously described with minor modifications (Woodfield et al., 2009). One hundred million cells were cross-linked for 10 min at 37°C using 0.7% formaldehyde, 0.125 M glycine. Cells were washed twice with PBS, re-suspended in lysis buffer [50 mmol/L HEPES (pH 8.0), 85 mmol/L KCl, 0.5% IPEGAL CA-630 + Roche complete protease inhibitors (Indianapolis, IN)] and incubated for 5 minutes. Cells were collected by centrifugation and cell pellets were frozen in liquid nitrogen and stored at minus 80° C. Cell pellets were thawed in lysis buffer, collected by centrifugation and cell nuclei were re-suspended in RIPA buffer [1×PBS, 1% IPEGAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS + Roche Complete protease inhibitors]. Chromatin was sonicated using conditions determined empirically for MCF-7 cells to achieve an optimal fragment length between 400 to 100 bp. After sonication, samples were centrifuged at 20,000 × g for 10 min at 4°C. The supernatant containing cross-linked DNA/histones was diluted with IP dilution buffer to 2 mg/ml [0.01% SDS, 1.1% Triton-X 100, 1.2 mmol/L EDTA, 16.7 mmol/L Tris-Cl, pH 8.1, 167 mmol/L NaCl] plus protease inhibitors]. Half the sample was immunoprecipitated with 10 μg of TFAP2C monoclonal antibody SC-12762X (Santa Cruz Biotechnology, Santa Cruz, CA), which was previously shown to be specific for TFAP2C without cross reactivity to TFAP2A, and half with control nonspecific IgG (Upstate, Waltham, MA) with the addition of Dynal sheep anti-mouse Dynabeads and allowed to recognize their antigens overnight at 4°C with rotation. Protein/antibody/DNA complexes were collected magnetically followed by washing and elution. Protein/DNA cross-links were reversed using 200 mmol/L NaCl at 65°C overnight. DNA was treated with Proteinase K and RNase A and was recovered with Qia Quick PCR Kit (Qiagen) according to manufacturer's suggested protocol. Input chromatin was processed identically as the IP chromatin samples. Purified DNA was quantified by using a NanoDrop ND-1000 (NanoDrop, Wilmington, DE).
Chromatin-immunoprecipitated DNA samples were prepared for and sequenced using an Illumina Genome Analyzer GAII at the Iowa State University DNA Facility according to the instructions of the manufacturer (Illumina, San Diego, CA). ChIP-Seq libraries were prepared by repairing the DNA fragment ends, adding an ‘A’ to the 3′ end of the repaired fragments, and ligating adapters using Illumina's ChIP-Seq sample prep kit. Libraries were enriched by 15 cycles of PCR amplification and size selected (200-300bp) on a 2% agarose gel. Libraries were quantified and the sizes checked on a Bioanalyzer 2100 using the DNA 1000 and high sensitivity DNA chips. For the libraries reported, the IgG library was 208 bp and the TFAP2C library was 227 bp. Libraries were clustered on the flow cell using the v.2 cluster generation kit (Illumina). The flowcell was loaded onto a Genome Analyzer II and subjected to single sequencing using v.3 36 cycle sequencing kit (Illumina). Image analysis, base calling and alignment were performed using Pipeline v.1.3 software. Sequences were aligned to the humanHG18 genome using the program ELAND.
Analysis of ChIP-Seq Data
Illumina reads were aligned using default Eland alignments (Iowa State University, DNA Facility) and imported into Partek GS v 6.4 (Partek Inc St. Louis, MO) using the standard Chip-seq workflow except that peaks were allowed to extended 300 bp rather than the default 100 bp, which optimized known TFAP2C binding locations. Peaks that overlapped between the two sets (IgG and TFAP2C) where considered likely to be false positive and the data was filtered using two criteria. A ratio between the number of reads in the TFAP2C experiment over the number of reads in the IgG control sample was calculated and any peak that had a ratio ≤ 25 was excluded. Also any peak that had more than 10 reads in the peak from IgG control was excluded and furthermore any peak that had a peak height < 100 in TFAP2C was excluded. Another filter was an in-house script, which looked at the peak boundaries and assessed its location relative to the gene location, using information from the UCSC database. Peaks were included as being unambiguously associated with a known gene if the peak was ≤ 5000 bp upstream from a gene (5′ flanking region) or contained within the 5′ UTR, or in one of the first four introns with exons excluded. This resulted in 1384 peaks. These genes were put into Ingenuity and Go Elite (Salomonis et al., 2007) to identify common themes, interactions or ontologies. DNA sequences from the peaks were also used in the Partek motif prediction tool to generate hidden markov model predictions of site conservation within the peaks listed. In order to assess the reliablilty of the Partek peak predictions, two other peak prediction programs, Findpeaks (v4.0.12) (http://sourceforge.net/apps/mediawiki/vancouvershortr/index.php) and cisGenome (v 1.2) (Ji et al., 2008), were both used (also with default settings) to predict peaks from the same source data. An in-house script was used to find overlap either complete (peaks overlap at both ends +/- 100 bp) or middle (Partek peak contained within the ends of the other prediction software +/-100 bp. cisGenome was also used to estimate a False Discovery Rate (FDR), a feature Partek does not include, to estimate how confident the peak predictions were at the thresholds chosen. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE21234 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21234).
Results
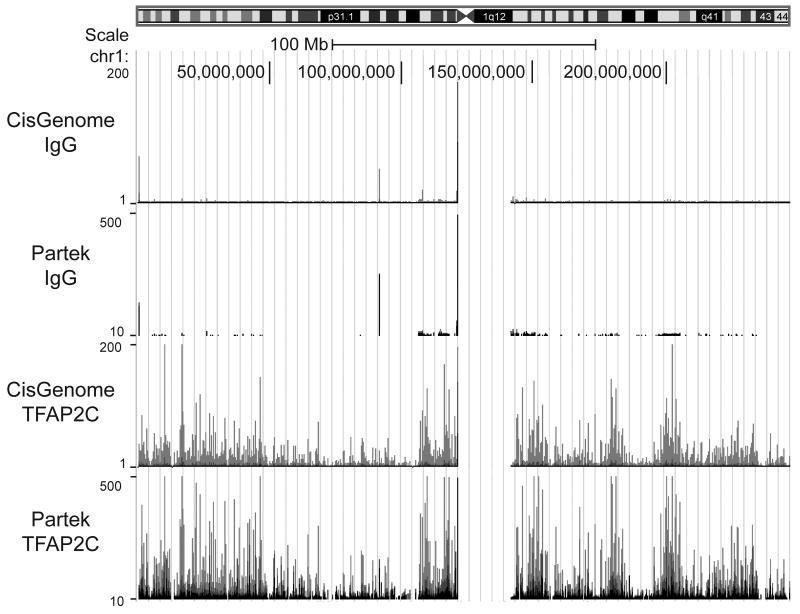
Previous efforts to identify genes that are primary transcriptional targets for AP-2 factors have utilized expression arrays from which additional experiments were performed based on possible AP-2 regulatory and promoter regions. This process may fail to identify important AP-2 target genes, may overlook important regulatory elements within genes regulated by AP-2 factors or may incorrectly identify certain genes as primary targets of AP-2 transcriptional control. Furthermore, this process is labor-intensive and lacks a mechanism to scan the genome for the most interesting AP-2 regulatory regions. To better define the scope of genes that are transcriptionally regulated by AP-2, ChIP-seq was employed to identify regions of the genome bound by endogenously expressed TFAP2C in the MCF-7 hormone responsive breast carcinoma cell line. It has previously been shown that MCF-7 cells express TFAP2C predominantly and also express significantly less amounts of TFAP2A (Woodfield et al., 2007). ChIP-seq generated 11,932,494 reads from the library prepared from immunoprecipitations with anti-TFAP2C and 8,183,070 reads were unambiguously mapped to the genome. A total of 16,048,336 reads were generated from the library prepared from IgG immunoprecipitations and 8,136,292 reads were mapped. The sequence length was 35 bp. The sequence data from ChIP-seq was analyzed to define peaks representing chromosomal regions with significant TFAP2C binding using Partek, CisGenome and FindPeaks software programs. As an additional control, a parallel set of immunoprecipitations were performed using a non-specific IgG antibody. The use of IgG as a negative control has some advantages in eliminating peaks that may be due to non-specific chromatin precipitation (Park, 2009). Figure 1 illustrates an example of IgG and TFAP2C peaks defined by CisGenome and Partek for chromosome 1. Data from ChIP-seq performed with non-specific IgG demonstrate prominent non-specific peaks located in the region of the telomeres, centromere and occasionally at other sites within the chromosome. The pattern of peak recognition is highly similar comparing the two software programs. Using CisGenome a false-discovery rate (FDR) was calculated. For peaks with 10 reads, the FDR was 0.000342 and for peaks with at least 16 reads, the FDR was less than 0.000001. For TFAP2C there were 27,759 peaks defined by cisGenome with 16 reads or more scattered across the entire MCF-7 genome or an average of 1,200 TFAP2C binding sites per chromosome.
Figure 1. Visual Display of Peaks from Chromosome 1.
Diagram of chromosome 1 with scale showing 100 Mb size marker (along top) with Genome Browser display of peaks (beneath) derived from CisGenome (panels one and three) or Partek (panels two and four) for data from ChIP-seq derived from immunoprecipitations performed with either IgG (panels one and two) or anti-TFAP2C (panels three and four). Vertical axis for each panel is approximately equal to number of sequence reads at each location. Horizontal axis is relative location along chromosome 1. Note similarity of output derived from CisGenome or Partek. The ChIP-seq dataset is accessible through GEO Series accession number GSE21234.
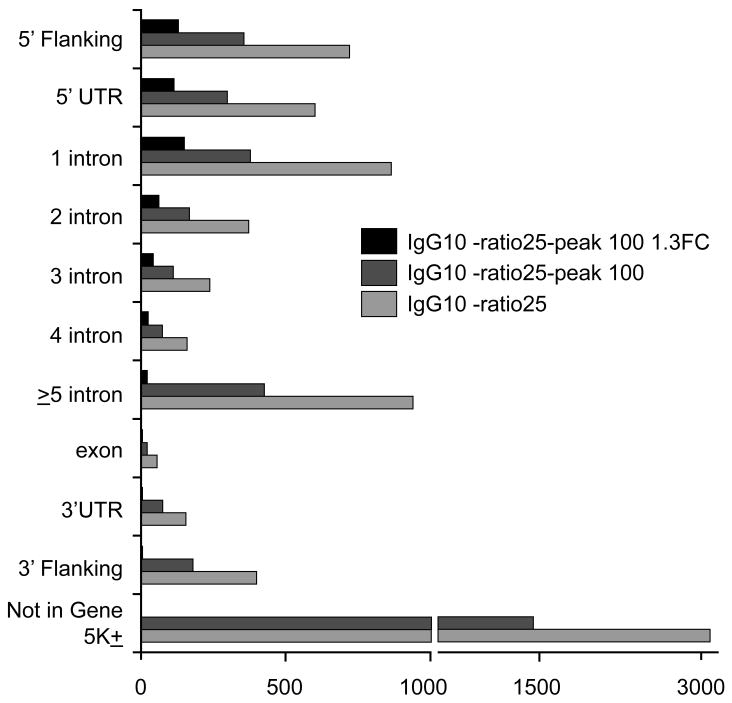
In an effort to focus on the most prominent peaks or potentially those with the most biologic relevance, the TFAP2C binding peaks were analyzed using Partek under increasingly stringent criteria. Restricting the analysis to TFAP2C peaks with less than 10 reads from IgG immunoprecipitations and a minimum of 25-fold enrichment in the ChIP performed with TFAP2C antibody compared to non-specific IgG reduced the analysis to a set of 7,643 peaks, with 3141 of these peaks greater than 5 kbp from a defined gene. The data were further restricted to include only those peaks of greater than 100 relative peak height (or reads) reducing the list to 3552 peaks. By comparison cisGenome at a criterion of 50 reads identified 6336 peaks and all but 42 of the 3552 peaks defined by Partek were contained within this set of 6336 peaks. Using FindPeaks software with the criterion of 50 reads, a total of 10,540 peaks were identified and all 3552 peaks from Partek were included in this set. Within the set of 3552 peaks defined using Partek with the stated criteria, 2,087 peaks occurred within 5 kbp of one of 1202 genes. Figure 2 summarizes the number of peaks identified with the different criteria. As seen in Figure 2, the majority of the interactions between TFAP2C and target genes occurred in the 5′ region of the gene, which included the region 5 kbp upstream of the cap site (5′ Flanking), the 5′ UTR or within the first several introns.
Figure 2. Distribution of ChIP-Seq Peaks Relative to Gene Location.
Number of peaks identified from ChIP-Seq analysis based on analysis using Partek with varying criteria. IgG10-ratio25: number of peaks from criteria examining <10 reads in the IgG control and ratio between number of reads in the TFAP2C experiment over reads in the IgG control ≥ 25. IgG10-ratio25-peak100: uses the additional criteria of filter with TFAP2C ‘peak’ output ≥ 100. IgG10-ratio25-peak100 1.3FC: adds the additional criterion based on finding a 1.3 fold change in expression after knock down of TFAP2C expression. All data is presented relative to gene location as shown. The criterion of “Not in gene” is based on peaks farther then 5 kbp from a defined gene.
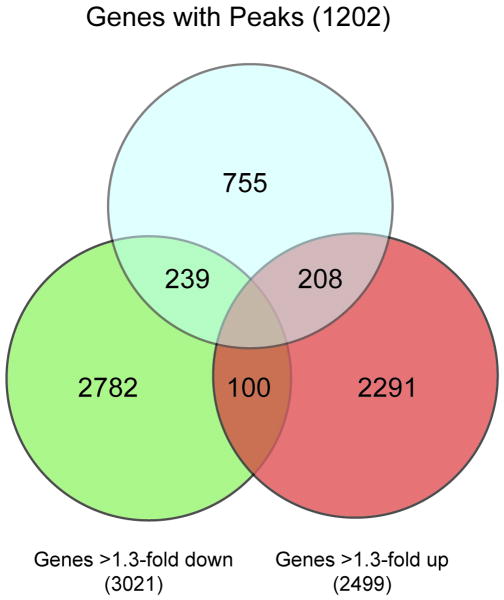
The data from ChIP-seq were compared to expression array data from MCF-7 cells in which TFAP2C was knocked down using siRNA compared to mock (non-targeting siRNA) transfected cells. Since expression arrays tend to under estimate the alterations of gene expression, a cut-off of 1.3-fold relative change in expression was used to restrict the analysis to genes that had evidence of altered expression with TFAP2C knock down (Figures 2 and 3). Expression array analysis identified 5520 genes whose relative expression was altered by 1.3-fold or greater with knock down of TFAP2C. An additional 100 genes had ambiguous results largely due to genes with multiple probes on the array with discordant expression results. Within this group of 5520 genes, 2499 (45%) increased and 3021 (55%) decreased with TFAP2C knock down. There were 1384 TFAP2C binding peaks occurring in 1202 genes identified, which met the defined criteria (IgG reads <10, ratio TFAP2C to IgG peak >25, TFAP2C peak height >100, occurring within 5′ Flanking, 5′ UTR or first four introns of a gene). Details of the set of 1384 peaks can be found within Supplemental File 1. There were 447 (447/1202, 37%) genes associated with peaks that also demonstrated altered expression of 1.3-fold or more. An additional 89 genes with defined peaks were not represented on the Affymetrix arrays and could not be assessed for gene expression changes. The 447 target genes encompassed 527 defined binding peaks with several genes having more than one AP-2 regulatory region identified. These data suggest that only 8% (447/5520) of the genes with altered expression following TFAP2C knock down are primary targets for AP-2 transcriptional regulation. The data also indicate that TFAP2C can act to either induce or repress gene expression through direct promoter interactions.
Figure 3. Summary of TFAP2C Gene Peaks and Expression Array.
Venn diagram summarizes the number of genes with identified binding peaks for TFAP2C binding to regions of defined genes compared to expression array data demonstrating increase or decrease of greater than 1.3-fold change with knock-down of TFAP2C using siRNA. A total of 447 target genes were identified with binding peaks from anti-TFAP2C ChIP with peaks defined by Partek from ChIP-seq data set and greater than 1.3 fold changes with TFAP2C knock down. There were 100 genes that demonstrated greater than 1.3-fold change in gene expression but were not associated with a peak in ChIP-seq, but whose expression pattern could not be unambiguously assigned to either the increase or decrease category. The ambiguity was due to genes with multiple probes on the expression array with discordant expression findings. A total of 3021 genes decreased by 1.3-fold or more and 2499 genes increased by 1.3-fold or more following knock-down of with siRNA directed against TFAP2C.
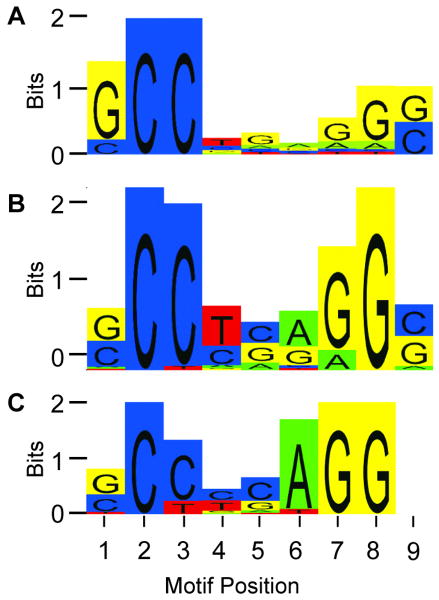
Previous studies defined a consensus binding site for TFAP2C utilizing an in vitro PCR-assisted selection process and gel shift competition (McPherson and Weigel 1999). The optimal consensus sequence for binding of TFAP2C was previously identified as the nine base pair sequence GCCTGAGGG. As shown in Figure 4A, the consensus site defined by in vitro selection was notable for an overwhelming preference for a G in position 1, a C in positions 2 and 3, a strong preference for a G in positions 7 and 8 and either a G or C in position 9. A consensus element was sought based on the chromosomal peaks defined by ChIP-seq analysis. Searching for a sequence motif, using hidden markov modeling within the set of 527 peaks derived from 447 TFAP2C target genes that demonstrated >1.3-fold expression change, generated the consensus site shown in Figure 4B. This motif was the only site reliably identified within the sequences from the set of binding peaks described. The consensus site derived from in vivo ChIP-seq analysis of TFAP2C has many of the same elements of the 9-base consensus site previously reported with some interesting differences. First, the consensus site derived from ChIP-seq data was remarkably similar to the optimal consensus site (GCCTGAGGG), which was previous defined from in vitro selection. There was a clear preference for a C in positions 2 and 3 and a G in position 8. Positions 1 and 9 demonstrated an approximately equal preference for either C or G. Positions 6 and 7 had either a G or A with a slight preference for A at position 6 and a G at position 7. Position 4 was most often a T and position 5 a C or G. The remarkable similarity to the 9-base optimal site previously defined by in vitro selection indicates that the ChIP-seq technique is likely providing meaningful chromatin binding data. An analysis based on 1384 peaks that were included regardless of transcriptional effect on gene expression with TFAP2C knock-down resulted in the consensus element shown in Figure 4C, which has many of the same features as the motif defined from regions in close proximity to known genes.
Figure 4. Sequence Logo of TFAP2C Consensus Motifs.
The motif of the TFAP2C consensus binding site indicates a 9-bp consensus element. Vertical axis (Bits) indicates the information content of the base frequency at that position. Positions that are perfectly conserved contain 2 bits of information, those with two possible nucleotides with 50% frequency equal 1 bit and if all four bases occur equally, the value is zero, since no base preference is demonstrated (D'Haeseleer 2006). The horizontal axis refers to consensus site position (1 through 9). A. The sequence logo motif of the TFAP2C consensus binding site derived from in vitro site selection as previously reported (McPherson and Weigel 1999). B. The consensus logo motif derived using Partek analysis of ChIP-seq data from the 527 peaks derived from 447 TFAP2C target genes that demonstrated >1.3-fold expression change. C. Motif logo derived from ChIP-seq data from 1384 peaks that were included regardless of transcriptional effect on gene with TFAP2C knock-down.
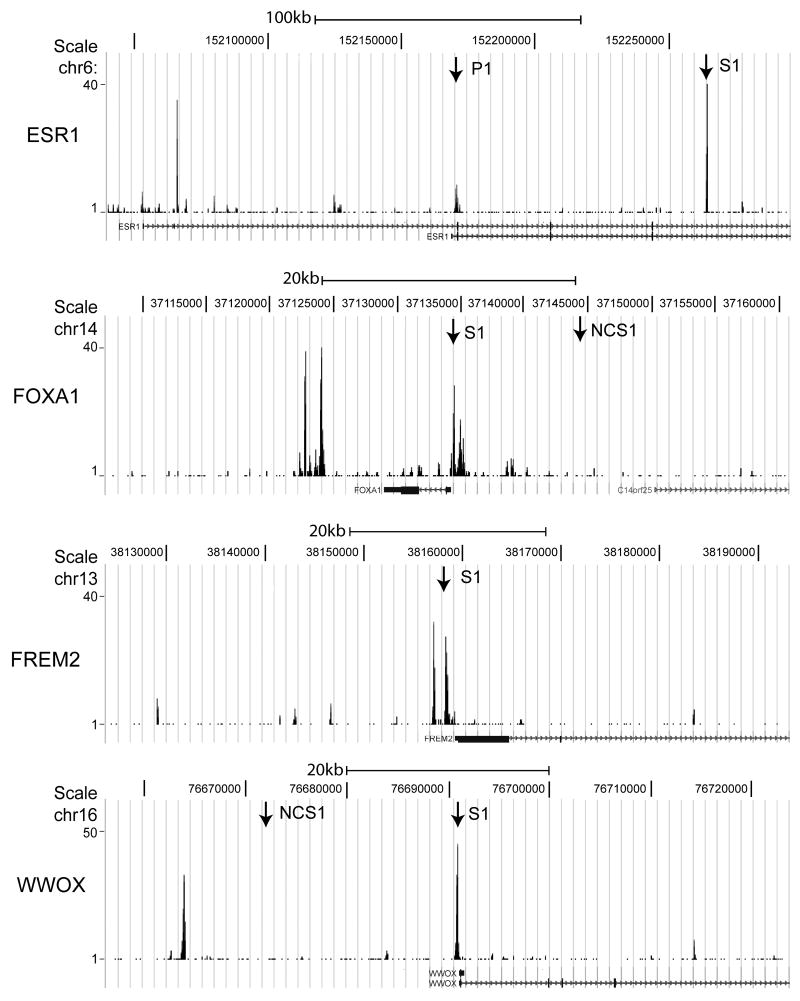
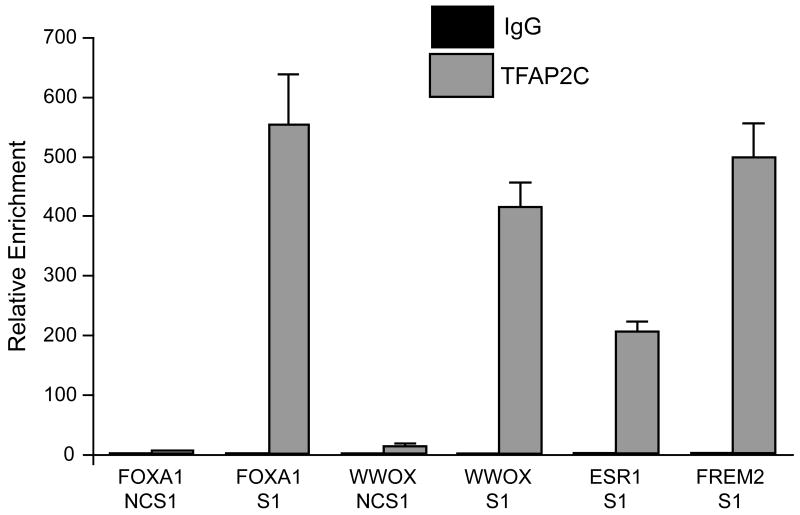
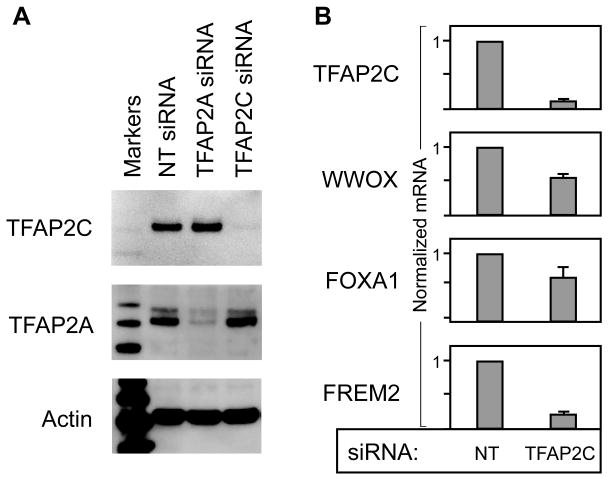
To confirm the findings from ChIP-seq, the TFAP2C binding regions of several genes were examined in more detail. Figure 5 diagrams the TFAP2C peaks in the genes for ESR1, FOXA1, FREM2 and WWOX. ChIP-seq localized a TFAP2C binding peak in the ESR1 gene within the 80 bp region of chromosome 6 from 152066108 to 152066188, which was associated with the upstream transcriptional start sites we previously characterized as the H-exons (Thompson et al., 1997). A second major TFAP2C peak of 75 bp in length was identified in the third intron of the ESR1 gene on chromosome 6 from 152264110 to 152264185 (marked as S1 in top panel, Figure 5). A third, less prominent peak was evident at the transcriptional start site for the main ESR1 cap site located on chromosome 6 at 152170670 (marked as P1 in top panel, Figure 5), which corresponds to the AP-2 regulatory region previously identified (deConinck et al., 1995; Schuur et al., 2001). Major TFAP2C peaks were found near the main transcriptional start sites for FOXA1, FREM2 and WWOX, as shown in Figure 5 (all designated as S1). In the case of FOXA1, two prominent TFAP2C peaks can also been seen approximately 5 kbp downstream of the gene. Primer pairs were generated at each of the main TFAP2C peaks for ESR1, FOXA1, FREM2 and WWOX as well as sites near the genes for FOXA1 and WWOX that were not associated with TFAP2C binding (referred to as negative control sites, NCS). Conventional real-time PCR analysis of ChIP comparing immunoprecipitations with anti-TFAP2C and IgG was performed at each site as shown in Figure 6. Amplification of 200 to 550-fold was found at each site for ChIP performed with anti-TFAP2C compared to IgG. By comparison, minimal differences between anti-TFAP2C and IgG were noted at the NCS in the FOXA1 and WWOX genes. Previous published quantitative ChIP analysis performed at the P1 site in the ESR1 gene demonstrated an intermediated amplification over IgG of approximately 35-fold enrichment (Woodfield et al., 2009). Data from expression arrays were similarly confirmed by analyzing RNA from MCF-7 cells transfected with siRNA directed against TFAP2C or NT siRNA. Western blot confirmed appropriate knock-down of AP-2 protein (Figure 7A). We had previously shown significant reduction in ESR1 mRNA expression after knock-down of TFAP2C in MCF-7 cells (Woodfield et al., 2007). RT-PCR of RNA comparing NT and TFAP2C siRNA confirmed knock-down of TFAP2C mRNA and associated reduction in the expression of WWOX, FOXA1 and FREM2 (Figure 7B).
Figure 5. Graphical Display of TFAP2C Binding Peaks for Selected Genes.
Shown is the graphical display from Genome Browser for the ChIP-seq data performed with anti-TFAP2C antibody for four TFAP2C target genes. First panel: approximately 250 kbp region of the ESR1 gene chromosomal region showing peaks representing TFAP2C binding. Arrows denote regions chosen for detailed analysis corresponding to binding peaks (ESR1 S1) and site of previous TFAP2C binding site identified (ESR1 P1). Second panel: Approximately 50 kbp chromosomal region of the FOXA1 gene showing region with significant TFAP2C binding (FOXA1 S1) and location chosen as negative control site (FOXA1 NCS1). Third panel: Approximately 60 kbp chromosomal region for the FREM2 gene showing location of the TFAP2C binding peaks near the transcriptional start site (FREM2 S1). Fourth panel: Approximately 60 kbp of the WWOX gene showing location of the TFAP2C binding peak near the transcriptional start site (WWOX S1) and the location of a negative control site (WWOX NCS1).
Figure 6. Conventional ChIP Analysis for Selected TFAP2C Binding Sites.
Conventional ChIP analysis was performed using primer pairs in the regions defined by TFAP2C binding locations displayed in Figure 5. Real-time qPCR was used to amplify chromatin derived from immunoprecipitations with either IgG or anti-TFAP2C antibody as indicated. (Note: the data is normalized to IgG and bar graph with value of 1 is not well visualized). Amplification with each primer pair was normalized to input chromatin and values subsequently normalized to values derived from IgG ChIP with data reported as relative enrichment. Data demonstrate significant relative enrichment at each TFAP2C binding site identified by ChIP-seq (FOXA1 S1, WWOX S1, ESR1 S1 and FREM2 S1) and minimal binding for negative control sites (FOXA1 NCS1, WWOX NCS1).
Figure 7. Confirmation of Gene Expression at Selected Genes.
A. Western blot of protein prepared from MCF-7 cells 72 hours after transfection with the siRNA indicated; NT siRNA: Non-targeting siRNA, TFAP2A siRNA or TFAP2C siRNA. Each blot is probed with antibody indicated and demonstrates knock-down of either TFAP2C or TFAP2A appropriately with transfection of related siRNA with approximately equal protein loading indicated by Actin blot. B. Real-time RT-PCR of mRNA expression for the genes indicated, which demonstrates successful knock-down of TFAP2C and corresponding decrease in expression of WWOX, FOXA1 and FREM2 with TFAP2C siRNA compared to transfection with NT siRNA. Data reported as normalized mRNA expression for the gene indicated.
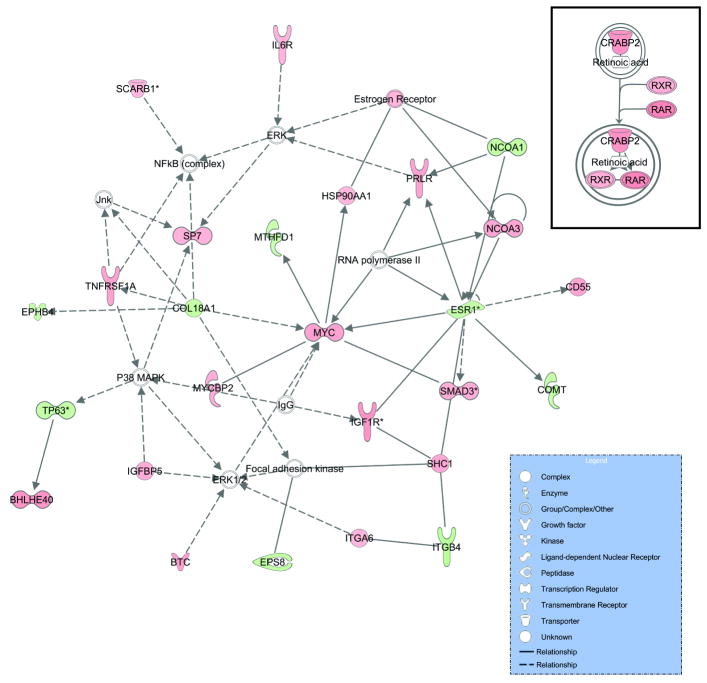
A number of primary target genes suggest important biologic function for TFAP2C in breast cancer including the regulation of ESR1, FREM2, RET, FOXA1, WWOX, GREB1, MYC and several members of the retinoic acid response pathway. Several genes involved in hormone response were found to be TFAP2C primary target genes in MCF-7 cells including ESR1, WWOX, FOXA1, and GREB1. A summary of the primary TFAP2C target genes is presented in Table 1. The list is presented according to GO Category and genes with diverse functions were listed in the first category shown and not repeated to avoid duplication of the list. Based on the set of 447 primary target genes, relationships between the genes and network associations were sought using Ingenuity Systems Analysis. The main pathway identified from the list of primary TFAP2C target genes identified a major role for TFAP2C in regulation of cell growth and proliferation (Fig. 8). In addition to ESR1 and a role in hormone response, MYC and several genes in the MYC pathway were also identified as TFAP2C target genes. The genes involved in proliferation also included the ERK kinase network. TFAP2C repressed the expression of several genes associated with retinoic acid signaling (Fig. 8 inset). The network involving RARA and RXRA included CRABP2, which was previously shown to be regulated by AP-2 transcription factors (McPherson et al., 2007).
Table 1. Summary of TFAP2C Target Genes.
| Go term | Repressed by TFAP2C | Induced by TFAP2C |
|---|---|---|
| Intracellular Signaling | AZI2,CALCR,RAB31,RASL12,RHOD,RREB1,SPSB1 | C9orf86,CORO2A,NCOA1,NPR3,PLCB4,RAB26, RAB8B,RCAN3,RHOBTB2 |
| Developmental Process | ANKRD11,AP3D1,BCAN,BCOR,CRABP2, CUX1,ELF3,ETV7,FLNB,FNDC3B,GTF2IRD1, IFRD1,IL4R,KLF3,MYC,NAB1,OSGIN1, POGZ,POU2F3,RUNX1,S100A9,SP7,ST8SIA1,SVIL,TXNRD1,ZFPM1 | CALML5,CNNM4,DAG1,FAM5B,FGF12,FOXA1, FREM2,KIAA1217,MKX,NACC2,OLFM1,PCP4, PCSK2,RPL22,SESN1,SOX3,SPDEF,SUV39H1, TPD52,UPK2 |
| Cell Projection Organization | BBS4,CELSR2,CNP,IGSF9,KALRN,MBP, MYCBP2,RXRA,SEMA3B | EFHD1,EFNA5,ITGB4,PTPRF |
| Cell Surface | CEP290,FURIN,HSPA5,IL6R,IRAK2,PRLR, RARA,SLC3A2,STX4 | C20orf3,CLEC7A,EPHB4,FGFR2,KAL1,KCNMA1, NOTCH2 |
| Enzyme Regulator | ADAP1,ASAP3,CST6,ERRFI1,MYO9A, PLEKHG3,SRGAP1 | CABLES1,CKS2,DOCK10,PKIB,RAB11FIP3, SERPINI1,SET |
| Regulation of Cell Growth | IL20,NCOA3 | ASAP1,DEPDC6,ESR1,MCF2L,RGL1,RGS12, TBC1D8,TCF7L1,WWOX |
| Adherens Junction | FERMT2,ITGA6,JUP,KEAP1,LMO7,LPP, NEDD9,VASP,ZYX | CDH3,CTNND2,DNAJC15,SDC1 |
| Cell Migration | ACTN4,NR2F2,PLXNA2,PPAP2B | CLIC4,COL18A1,COL5A1,NTN1,OVOL2,PODXL,RET, SEMA3F,VAV2 |
| Phospholipid Binding | DAPP1,PLD1,SCARB1,SHC1,SNX5,SYTL2, ZCCHC14 | ANXA4,CPNE3,DGKA,HIP1,HS1BP3,OSBPL1A, TIAM1 |
| Phosphate Metabolism | ADCY9,DUSP8 | ADCY5,PTPRS |
| Receptor Signaling | FRS2,IGF1R,IGFBP5,MYO1E,PRKCA,RGS6,SMAD3,SMURF1,TRIO | EPHA6,EPS8,FGFR4,GAB1,LTBP2,PTPRG, SMAD6 |
| Extrinsic to Membrane | BEGAIN,CLINT1,EPB41L4B,EPS15L1, RAB11FIP4,SELENBP1,ZNRF1 | CERK,DOK7,GGA2,GNA14,JAK1,KSR2,LIN7A, MAGI3,N4BP3,PPP1R14C |
| Apoptosis | BCL3,BTC,CBX4,DEDD2,EYA2,MEF2D, RYBP,SHB,SHISA5,TNFRSF1A,TRAF3 | KLF11,LY86,RABEP1,TNFAIP8,TNFRSF19 |
| Protein Domain Specific Binding | BAIAP2L1,DAPK3,FUT8,HSP90AA1,LNX1, PPP5C,RAPGEF1,SQSTM1,STRN4,SYNJ2 | CDC25C,PMEPA1,SH3BGR,SH3BP5,SKI, SLC22A4,TBC1D10A,TP63 |
| Kinase Activity | C9orf98,CAMK2D,CSNK1E,KSR1,MARK2, MAST4,MKNK2,NAGK,PAK2,PRKAG2, TAOK3 | ARSG,CAMK1D,CDKL3,DCAKD,LIMK2,NME7, NRK,PBK,PTK7 |
| Sulfur Metabolism | GSR,MTRR | CHST3,COMT,HS6ST1,IDH1,MTHFD1,SULF1, SULF2,UST |
| Unknown | ABCC3,ATXN7L1,BCL9L,BHLHE40,BRWD1, BTBD16,C10orf18,C14orf43,C1orf86, C22orf30,C2orf18,C3orf59,C6orf141,C7orf43, C9orf44,CAPN8,CD55,CD58,CDC34,CLDN4, CYS1,DCHS1,DGAT2,EHBP1,FAM102B, FAM129B,FAM160A2,FAM169B,FAM19A5, FUBP1,GABPB1,GFOD1,GMEB2,GPR126, GPRC5A,GRAMD1A,GREB1,H2AFY,HAPLN2, HIVEP3,HSH2D,IGFN1,INPP4A,ISG20, KIAA0284,LMNA,LOC154822,LPIN1,LYPD6B, NCOA7,NSUN6,P4HA2,PALM3,RDH13,REPIN1,RIF1,RNF25,RNF43,S100A8,SEC24D, SLC24A3,SLC29A3,SOLH,SPOP,SPTBN2, SYT12,TPM4,TPPP,TRIM56,UBE2H,ZFHX3, ZKSCAN1,ZNF444,ZNF462,ZNF750,ZNFX1 | ALG10,AMMECR1,AMZ1,APITD1,ARMC1,ATP2A3, BTBD11,C10orf58,C12orf76,C13orf23, C14orf132,C1orf21,C1orf26,C2CD2,C6orf125,7orf41, C9orf3,CALHM2,CANX,CBY3,CCNG2,HPT1,CMBL, CPNE4,CSTF3,CTDSPL,DIP2C,SCR8,EIF2C2,EML5, EMP2,ENSA,ERGIC1,FAM107B,FAM171B,FAM174B, FAM54A,FAM65C,BP1,FCHSD2,FMO4,FRMD6,FXYD3,GALNT6,FOD2,GPRC5B,GRAMD3,HS6ST3,IDH2,IGSF3, IGSF5,INF2,KCNK2,KCNMB1,KCTD10,KCTD3, KIAA0247,KIAA0922,KIAA1324,KIAA1549,KIF23,KMO, LMBR1,LOC100128822,LYRM1,MACROD2,MANBA, MCC,MCM6,MFSD6,MICALL2,MOCS1,MOSC1,MPRIP, NEURL1B,NR3C1,OCIAD2,PCYT2,PDE4D,PLEKHA6, PLEKHF2,PLS1,POLE2,PQLC3,PRIM2,PROM2,PSCA, RFTN1,RNASET2,SDK2,SESTD1,SIDT1,SLC12A4, SLC35A1,SLC38A1,SLC6A6,SLC7A8,SPNS2,STARD10,STOM,TACC1,TDRKH,TMEM141,TMEM168,TMEM40, TRIM9, TSC22D3,TSPAN1,TSPAN15,TSPAN5,TSPAN9, TTC39A,USP13, ZNF718 |
Figure 8. Pathway Associations of TFAP2C Target Genes.
The 447 TFAP2C target genes were analyzed using Ingenuity Systems to determine associations between genes for the purpose of defining physiologic pathways regulated by TFAP2C. The main pathway (with the greatest number of genes) is shown and was characterized as involved in cell growth and proliferation. Prominent genes noted in this network include MYC, ESR1 and ERK. Red tone indicates genes repressed by TFAP2C and green indicates genes induced by TFAP2C. Inset: Retinoic acid signaling pathway was also identified as a prominent network involved with TFAP2C target genes, RXR, RAR and CRABP2.
Discussion
Over the last several years, numerous studies have described associations between the expression of TFAP2C and important clinical findings in breast cancer (Pellikainen and Kosma 2007). However, most studies have examined expression of TFAP2C, which may not correlate with functional activity of the protein. For example, epigenetic modifications in the regulatory regions of certain TFAP2C target genes can alter the binding capability and hence the transcriptional activity of TFAP2C at certain promoters (Woodfield et al., 2009). It is also possible that the interaction between TFAP2C and other factors may alter the ability for TFAP2C to bind to or activate specific promoters (Eckert et al., 2005). ChIP-seq is a new technique that allows an examination of transcription factor binding to the entire genome without limiting the examination to a set of known target genes. Herein we have described the genomic interactions of TFAP2C in hormone responsive MCF-7 cells. Our findings corroborate earlier studies of TFAP2C activity and extend those findings by defining primary targets versus genes that are regulated by TFAP2C activity though secondary or tertiary effects. The findings also identified the precise location for genomic interactions of TFAP2C.
One goal of the current analysis was to define the DNA consensus binding site as determined by in vivo chromatin interactions between the human genome and endogenously expressed TFAP2C. Most notably, the consensus site from the ChIP-seq data, SCCTSRGGS (S=G/C, R=A/G), is consistent with the optimal binding site, GCCTGAGGG, which was determined by in vitro PCR-assisted binding site selection (McPherson and Weigel 1999). The finding that the consensus site from the ChIP-seq data closely matched the previously defined optimal binding site for TFAP2C provided one level of evidence supporting the technical success of the ChIP-seq experiments. However, it should also be recognized that the criteria used to define the optimal binding site in the in vitro experiments were partly based on gel-shift competitions, which established the relative strength of binding. The criteria used to filter the ChIP-seq data may have similarly selected the strongest interactions. Hence, there may be physiologically important interactions between TFAP2C and certain target genes that may have been excluded by the selection criteria.
It was surprising to find that only 8% of genes with significant alterations in expression with knock down of TFAP2C were defined as primary target genes. This finding implies that over 90% of the changes in gene expression were due to secondary or tertiary effects. Indirect mechanisms of gene regulation include the regulation of transcription factors (such as RARA, RXRA and ZFHX3) that regulate other genes and could account for a significant alteration of gene expression by TFAP2C without the factor directly regulating the expression of genes with altered expression. It is also possible that TFAP2C alters patterns of expression by influencing the processing, splicing or stability of certain mRNAs. It should be noted that the selection criteria used to define binding peaks focused attention on the strongest interactions but may have overlooked physiologically significant interactions between TFAP2C and AP-2 regulatory regions. There are likely to be several physical and functional reasons for ChIP-seq to identify certain locations over another and may relate to the stability of the interaction, the number of AP-2 proteins bound or the accessibility of the antibody to the AP-2 epitope. As more data is acquired using ChIP-seq, it may be found that additional chromosomal regions of interaction are significant and the number of primary target genes might increase. Furthermore, the criterion that a binding peak be located within 5 kbp of a defined gene was used to allow a degree of certainty about the gene regulated by a given site. It is possible that TFAP2C binding located at greater distances could account for regulation of genes that were not included in the current list. The criteria used in the current analysis were chosen to be conservative to assure that the genes identified were indeed primary targets. As noted in Figure 3, there were also a significant number of binding peaks not associated with significant alterations in gene expression with TFAP2C knock down. Several possibilities might explain this finding. First, knock down of TFAP2C by siRNA is not absolute and it is possible that certain genes are sensitive to low levels of residual expression after knock down. Second, genes that did not demonstrate an alteration with TFAP2C knock down may also be transcriptional targets for TFAP2A. Hence, eliminating TFAP2C alone might not be sufficient to alter expression. It will be interesting to determine how many of the genes in this category are also found to be targeted by TFAP2A. Finally, the role of TFAP2C might be to modulate the level of expression of target genes and our inclusion criteria of 1.3 may exclude genes whose effect by TFAP2C is more subtle. As noted above, the criteria chosen for the analysis were conservative to allow for a relative level of certainty to identify primary TFAP2C target genes.
The ChIP-seq data provide important mechanistic insight into gene regulation that had not previously been possible. Regulation of the ESR1 gene is one example of the power of the ChIP-seq technique. Previous studies identified the ESR1 gene as a primary target of TFAP2C (deConinck et al., 1995; Schuur et al., 2001; Woodfield et al., 2007). Indeed, ChIP-seq identified TFAP2C binding at the main transcriptional start site of the ESR1 gene, however, this was clearly not the main binding location as determined by strength of the interaction. Prominent interactions of TFAP2C were also identified just downstream of exon Hb and within the third intron of the main ESR1 transcript. Neither of these sites had previously been suspected and would not likely have been identified using conventional techniques to scan the entire ESR1 regulatory region, which spans over 200 kbp. Similarly, TFAP2C binding sites were identified 5 kbp downstream of the FOXA1 transcribed region. Currently there is no definitive method to determine how one or another binding region relates to regulation of gene transcription. For example, it is possible that the relatively weak TFAP2C binding at the main ESR1 transcriptional start site plays a more influential role in transcription than the very prominent site in the third intron 93 kbp downstream of the main transcriptional start site. Alternatively, some sites might be activating while others may actually repress transcription. Despite these limitations, knowing the precise location of TFAP2C binding will allow a more in-depth analysis of specific regulatory regions.
An examination of the TFAP2C target genes indicated several pathways regulated in Luminal A breast cancer. The data confirmed involvement of the estrogen response pathway and the finding was consistent with previous results examining expression patterns controlled by TFAP2C in MCF-7 cells, which concluded that TFAP2C regulated the expression of multiple pathways related to hormone response (Woodfield et al., 2007). Other primary target genes of TFAP2C that were identified in the current study included WWOX, GREB1 and FOXA1, which play a critical role in hormone responsive breast cancer. WWOX and TFAP2C expression are associated with hormone resistance, although a direct correlation between WWOX expression and TFAP2C protein expression was not evident in patterns of gene expression in primary breast carcinomas (Guler et al., 2009). GREB1 is an estrogen-regulated gene that is important to hormone response in breast cancer. (Ghosh et al., 2000; Rae et al., 2005) FOXA1 is necessary for ESR1 (ERα) to interact with regulatory regions and knock down of FOXA1 blocks ESR1 (ERα) chromatin binding and gene expression (Carroll et al., 2005). Furthermore, FOXA1 is associated with ESR1 expression and correlates with an improved prognosis in hormone responsive breast cancer (Thorat et al., 2008; Albergaria et al., 2009). These associations demonstrate an expanded role for TFAP2C in hormone response in breast cancer. TFAP2C was also found to regulate pathways related to retinoic acid response and MYC regulation. These networks play important roles in regulating growth and differentiation in breast cancer and further define the critical role of TFAP2C in breast cancer biology. The findings related to MYC is particularly interesting in light of the recent finding that TFAP2A also binds to the MYC promoter to repress its transcription (Yu et al., 2009).
One recent study examined the genomic binding of TFAP2C in MDA-MB-453 breast carcinoma cells and reported four target genes—ERBB2, CDH2, HPSE and IGSF11 (Ailan et al., 2009). Interestingly, none of these genes were identified as TFAP2C targets by either the current ChIP-seq analysis or expression array data in MCF-7 cells. On the other hand, IGSF5 and another cadherin family member (CDH3) were identified as primary TFAP2C target genes in the current study. This apparent discrepancy is informative and suggests that the genes transcriptionally regulated by TFAP2C are dependent upon the specific breast cancer phenotype examined. Furthermore, the data support a model where TFAP2C binds to different regions of the genome in various cell phenotypes. The MCF-7 cell line is a model for the Luminal A (ESR1-positive/ERBB2-negative) breast cancer phenotype, whereas the expression pattern in MDA-MB-453 is more consistent with the Her2 phenotype (ESR1-negative/ERBB2-positive). The role of AP-2 in different breast cancer phenotypes will likely have important clinical relevance. As demonstrated by Pellikainen et al. (2004), patients with an ERBB2-negative/AP-2-positive expression pattern have a better prognosis than patients with an ERBB2-positive/AP-2-positive pattern. Given the role of AP-2 in regulation of ERBB2, this suggests that AP-2 factors have different activities in different breast cancer phenotypes. Hence, it is likely that TFAP2C will have different roles in the various histologic phenotypes of breast cancer and furthermore, TFAP2C transcriptional activity is responsible for the clinically relevant breast cancer biology. Determining the molecular basis for altered TFAP2C activity in the various breast cancer phenotypes will provide important insight into the clinical phenotypic differences in breast carcinomas.
Supplementary Material
Acknowledgments
We acknowledge Michael D. Baker, PhD, from the DNA Sequencing and Synthesis Facility, Iowa State University for technical assistance in performing ChIP-seq library construction and sequencing.
Supported by: The National Institutes of Health grant R01CA109294 (PI: R.J. Weigel) and the Kristen Olewine Milke Breast Cancer Research Fund.
References
- Ailan H, Xiangwen X, Daolong R, Lu G, Xiaofeng D, Xi Q, Xingwang H, Rushi L, Jian Z, Shuanglin X. Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC Cancer. 2009;9:279. doi: 10.1186/1471-2407-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, Lunet N, Schmitt F. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem. 2005;280:24428–34. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene. 1996;13:1701–7. [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- D'Haeseleer P. What are DNA sequence motifs? Nat Biotechnol. 2006;24:423–5. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- deConinck EC, McPherson LA, Weigel RJ. Transcriptional regulation of estrogen receptor in breast carcinomas. Mol Cell Biol. 1995;15:2191–6. doi: 10.1128/mcb.15.4.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix L, Begon D, Chatel G, Jackers P, Winkler R. Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol. 2005;24:582–94. doi: 10.1089/dna.2005.24.582. [DOI] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Williams T. Cloning and characterization of the mouse AP-2 epsilon gene: a novel family member expressed in the developing olfactory bulb. Mol Cell Neurosci. 2003;24:460–75. doi: 10.1016/s1044-7431(03)00209-4. [DOI] [PubMed] [Google Scholar]
- Friedrichs N, Jager R, Paggen E, Rudlowski C, Merkelbach-Bruse S, Schorle H, Buettner R. Distinct spatial expression patterns of AP-2alpha and AP-2gamma in non-neoplastic human breast and breast cancer. Mod Pathol. 2005;18:431–8. doi: 10.1038/modpathol.3800292. [DOI] [PubMed] [Google Scholar]
- Friedrichs N, Steiner S, Buettner R, Knoepfle G. Immunohistochemical expression patterns of AP2alpha and AP2gamma in the developing fetal human breast. Histopathology. 2007;51:814–23. doi: 10.1111/j.1365-2559.2007.02887.x. [DOI] [PubMed] [Google Scholar]
- Gee JM, Eloranta JJ, Ibbitt JC, Robertson JF, Ellis IO, Williams T, Nicholson RI, Hurst HC. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol. 2009;217:32–41. doi: 10.1002/path.2430. [DOI] [PubMed] [Google Scholar]
- Gee JM, Robertson JF, Ellis IO, Nicholson RI, Hurst HC. Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J Pathol. 1999;189:514–20. doi: 10.1002/(SICI)1096-9896(199912)189:4<514::AID-PATH463>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone- responsive breast cancer. Cancer Res. 2000;60:6367–75. [PubMed] [Google Scholar]
- Guler G, Huebner K, Himmetoglu C, Jimenez RE, Costinean S, Volinia S, Pilarski RT, Hayran M, Shapiro CL. Fragile histidine triad protein, WW domain-containing oxidoreductase protein Wwox, and activator protein 2gamma expression levels correlate with basal phenotype in breast cancer. Cancer. 2009;115:899–908. doi: 10.1002/cncr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler G, Iliopoulos D, Guler N, Himmetoglu C, Hayran M, Huebner K. Wwox and Ap2gamma expression levels predict tamoxifen response. Clin Cancer Res. 2007;13:6115–21. doi: 10.1158/1078-0432.CCR-07-1282. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–91. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Jager R, Friedrichs N, Heim I, Buttner R, Schorle H. Dual role of AP-2gamma in ErbB-2-induced mammary tumorigenesis. Breast Cancer Res Treat. 2005;90:273–80. doi: 10.1007/s10549-004-4815-x. [DOI] [PubMed] [Google Scholar]
- Jager R, Werling U, Rimpf S, Jacob A, Schorle H. Transcription factor AP-2gamma stimulates proliferation and apoptosis and impairs differentiation in a transgenic model. Mol Cancer Res. 2003;1:921–9. [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol. 2007;304:338–54. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Baichwal VR, Weigel RJ. Identification of ERF-1 as a member of the AP2 transcription factor family. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4342–7. doi: 10.1073/pnas.94.9.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Loktev AV, Weigel RJ. Tumor Suppressor Activity of AP2alpha Mediated through a Direct Interaction with p53. J Biol Chem. 2002;277:45028–33. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- McPherson LA, Weigel RJ. AP2alpha and AP2gamma: a comparison of binding site specificity and transactivation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–9. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Woodfield GW, Weigel RJ. AP2 transcription factors regulate expression of CRABPII in hormone responsive breast carcinoma. J Surg Res. 2007;138:71–8. doi: 10.1016/j.jss.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Cloning and characterization of a second AP-2 transcription factor: AP-2 beta. Development. 1995;121:2779–88. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- Orso F, Penna E, Cimino D, Astanina E, Maione F, Valdembri D, Giraudo E, Serini G, Sismondi P, De Bortoli M, Taverna D. AP-2alpha and AP-2gamma regulate tumor progression via specific genetic programs. FASEB J. 2008;22:2702–14. doi: 10.1096/fj.08-106492. [DOI] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikainen J, Naukkarinen A, Ropponen K, Rummukainen J, Kataja V, Kellokoski J, Eskelinen M, Kosma VM. Expression of HER2 and its association with AP-2 in breast cancer. Eur J Cancer. 2004;40:1485–95. doi: 10.1016/j.ejca.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Pellikainen JM, Kosma VM. Activator protein-2 in carcinogenesis with a special reference to breast cancer-a mini review. Int J Cancer. 2007;120:2061–7. doi: 10.1002/ijc.22648. [DOI] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–9. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur ER, McPherson LA, Yang GP, Weigel RJ. Genomic structure of the promoters of the human estrogen receptor-alpha gene demonstrate changes in chromatin structure induced by ap2gamma. J Biol Chem. 2001;276:15519–26. doi: 10.1074/jbc.M009001200. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, McPherson LA, Carmeci C, deConinck EC, Weigel RJ. Identification of two estrogen receptor transcripts with novel 5′ exons isolated from a MCF7 cDNA library. J Steroid Biochem Mol Biol. 1997;62:143–53. doi: 10.1016/s0960-0760(97)00029-0. [DOI] [PubMed] [Google Scholar]
- Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol. 2008;61:327–32. doi: 10.1136/jcp.2007.052431. [DOI] [PubMed] [Google Scholar]
- Turner BC, Zhang J, Gumbs AA, Maher MG, Kaplan L, Carter D, Glazer PM, Hurst HC, Haffty BG, Williams T. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res. 1998;58:5466–72. [PubMed] [Google Scholar]
- Weigel RJ, deConinck EC. Transcriptional control of estrogen receptor in estrogen receptor-negative breast carcinoma. Cancer Res. 1993;53:3472–4. [PubMed] [Google Scholar]
- Williams T, Admon A, Luscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–69. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–82. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- Woodfield GW, Hitchler MJ, Chen Y, Domann FE, Weigel RJ. Interaction of TFAP2C with the estrogen receptor-alpha promoter is controlled by chromatin structure. Clin Cancer Res. 2009;15:3672–9. doi: 10.1158/1078-0432.CCR-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfield GW, Horan AD, Chen Y, Weigel RJ. TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res. 2007;67:8439–43. doi: 10.1158/0008-5472.CAN-07-2293. [DOI] [PubMed] [Google Scholar]
- Yang JW, Lee EY, Kang KW. ErbB2 overexpression in p53-inactivated mammary epithelial cells. FEBS Lett. 2006;580:6501–8. doi: 10.1016/j.febslet.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Yu L, Hitchler MJ, Sun W, Sarsour EH, Goswami PC, Domann FE. TFAP2A inhibits c-MYC induced oxidative stress and apoptosis in HaCaT human keratinocytes. J Oncol. 2009 doi: 10.1155/2009/780874. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Brewer S, Huang J, Williams T. Overexpression of transcription factor AP-2alpha suppresses mammary gland growth and morphogenesis. Dev Biol. 2003;256:127–45. doi: 10.1016/s0012-1606(02)00119-7. [DOI] [PubMed] [Google Scholar]
- Zhao F, Satoda M, Licht JD, Hayashizaki Y, Gelb BD. Cloning and characterization of a novel mouse AP-2 transcription factor, AP-2delta, with unique DNA binding and transactivation properties. J Biol Chem. 2001;276:40755–60. doi: 10.1074/jbc.M106284200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.