Abstract
Somatic mutations in the PIK3CA gene have been discovered in many human cancers, and their presence correlates to therapy response. Three “hotspot” mutations within the PIK3CA gene are localized in exons 9 and 20. High-resolution melting analysis (HRMA) is a highly sensitive, robust, rapid, and cost-effective mutation analysis technique. We developed a novel methodology for the detection of hotspot mutations in exons 9 and 20 of the PIK3CA gene that is based on a combination of PCR and HRMA. The PIK3CA HRMA assay was evaluated by performing repeatability, sensitivity, and comparison with DNA sequencing studies and was further validated in 129 formalin-fixed paraffin-embedded breast tissue samples: 99 tumors, 20 noncancerous, and 10 fibroadenomas. The developed methodology was further applied in a selected group of 75 breast cancer patients who underwent Trastuzumab treatment. In sensitivity studies, the assay presented a capability to detect as low as 1% of mutated dsDNA in the presence of wtDNA for both exons. In the 99 tumor samples (validation group), 12/99 (12.1%) exon 9 mutations and 20/99 (20.2%) exon 20 mutations were found. No mutations were found in noncancerous tissues. In fibroadenomas, we report one PIK3CA mutation for the first time. In the selected group, 30/75 (40%) samples were detected as mutants. The PIK3CA HRMA assay is highly sensitive, reliable, cost-effective, and easy-to-perform, and therefore can be used as a screening test in a high-throughput pharmacodiagnostic setting.
Somatic mutations and gene amplification for the gene encoding for phosphatidylinositol 3-kinase (PI3K) p110α catalytic subunit, PIK3CA, located in chromosome 3, have been discovered in many different human cancers.1,2,3,4,5,6,7,8,9 PIK3CA mutations in exons 9 and 20 account for more than 90% of mutations reported for this gene, according to COSMIC Database (Catalogue Of Somatic Mutations In Cancer Database, v41 release; Wellcome Trust Genome Campus, Hinxton, Cambridge; Accessed May 2009; http://www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=gene&ln=PIK3CA). There are three recurrent or “hotspot” mutations within these exons: c.1624G>A(E542K), c.1633G>A(E545K) in exon 9 (helical domain), and c.3140A>G(H1047R) in exon 20 (kinase domain). These hotspot mutations have been shown to have oncogenic effects.1,10,11
PIK3CA mutations appear to have a clinical significance.12 Recent studies have shown that PIK3CA mutations can independently hamper the therapeutic response to anti-EGFR biological therapies (panitumumab or cetuximab) in metastatic colorectal cancer13 and demonstrate resistance to dietary restriction therapies.14 Moreover, several efforts are underway nowadays to target the PI3K pathway with therapeutic inhibitors.
Additionally, keeping in mind that not all patients with HER2-overexpressing metastatic breast cancer respond to Trastuzumab (Herceptin), activated PI3K signaling has been proposed to predict Trastuzumab resistance.15,16 Loss of the PTEN (Phosphatase and Tensin Homolog) protein has been suggested as a key factor for the development of resistance to this drug.15,16 However, PTEN loss alone,16,17 and in combination with phosphorylated AKT expression,17 proved inadequate to predict response to therapy. Conversely, combining PTEN loss and gain-of-function mutations of the PIK3CA gene, resistance to Trastuzumab was successfully predicted.16
The introduction of High-Resolution Melting Analysis (HRMA) technique in 200318 came with several advantages. HRMA, a closed-tube probe-free technique, with genotyping and mutation scanning capabilities, is rapid, simple, cost-effective, and nondestructive. Especially, HRMA of small amplicons (<100bp) overcomes the difficulty of classic HRMA analysis to detect mutant homozygotes, and demonstrates a higher diagnostic sensitivity in detecting homozygotes and heterozygotes,19,20,21,22 with an increase in rapidity.20 Moreover, small amplicons display a higher analytical sensitivity compared with larger amplicons.23,24 This is a major advantage when scanning for somatic mutations, where the tumor tissue is contaminated with adjacent normal tissue.
Archival tissue specimens, such as formalin-fixed paraffin-embedded (FFPE) tissues, represent a vast source of tissue genomic DNA (gDNA), easily obtained from clinical archives. However, they present difficulties in amplification due to DNA degradation, therefore demanding a reduction of PCR amplicons’ length. In addition, use of FFPE tissue decreases diagnostic specificity when used as a template for HRMA.25
Until recently, several techniques have been used for scanning and/or genotyping somatic mutations of the PIK3CA gene. These include DNA sequencing,1,2,3,5,6,8 denaturing High Performance Liquid Chromatography (dHPLC), Single Strand Conformation Polymorphism (SSCP),5 multiplex PCR amplification and primer extension,26 and Amplification Refractory Mutation System (ARMS) PCR.27 ARMS PCR has the highest analytical sensitivity reported; however, it does not allow mutation scanning capabilities and the use of Scorpion probes carries a high cost. A HRMA method for hotspot regions of the PIK3CA gene has been reported, using amplicons larger than 100bp and not applied in archival tissue specimens.28
Herein, we report a novel methodology for the detection of PIK3CA hotspot mutations using HRMA of small amplicons. By performing novel in silico primer design and extensive optimization studies we maximized analytical sensitivity and obtained very accurate results for FFPE samples, as verified by DNA sequencing. After validation using a group of 129 breast tissue samples, the method was applied on a selected group of 75 patients who underwent Trastuzumab treatment.
Materials and Methods
DNA Samples
Five gDNA samples isolated from peripheral blood (PB) of healthy volunteers served as wild-type controls. Two gDNA samples isolated from MCF-7 (c.1633G>A:E545K;heterozygous), and T47D (c.3140A>G:H1047R;heterozygous)29 breast cancer cell lines served as mutant controls. Genomic DNA was also isolated from 129 FFPE breast tissues (validation group): 99 carcinomas, 10 histologically normal samples adjacent to tumors, 10 fibroadenomas, and 10 reduction mammoplasty samples. All carcinoma patients (validation group) were diagnosed with early-stage breast cancer, and their clinical characteristics were previously described.30 Additionally, gDNA was isolated from 75 FFPE breast tissues (selected group) from patients with HER2-positive metastatic breast cancer. These patients received Trastuzumab in combination with first-line chemotherapy. All patients gave their informed consent, and the study has been approved by the Ethical and Scientific Committees of our Institutions.
From PB, gDNA was extracted using a salting-out DNA extraction protocol.31 From cell lines and FFPE samples, gDNA was extracted using the High Pure PCR Template Preparation Kit (Roche, Germany). DNA purity and quantity was determined using the Nanodrop-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Primer Design
Two sets of primers were in silico designed for each of PIK3CA exons 9 and 20, by using the PrimerPremier 5 software (Premier Biosoft International, Palo Alto, CA), and synthesized by FORTH (Greece). For each exon, one primer set was designed to span the hotspot region, with a reduced length (<100 bp), and another set to span the exons’ boundaries. Primer oligonucleotide sequences are presented in Table 1.
Table 1.
PIK3CA Oligonucleotide Sequences and Locations Used in this Study
| Exon | Primers | Primer set | Amplicon position* | Amplicon size | Amplicon Tm (°C)† | Recurrent mutations‡ | Mutation position within amplicon |
|---|---|---|---|---|---|---|---|
| 9 | F:5′-GCTCAAAGCAATTTCTACACGAGA-3′ | S9 | 180, 418, 742–180, 418, 833 | 92 bp | 81.0 | c.1624G>A | 35 |
| R:5′-TCCATTTTAGCACTTACCTGTGAC-3′ | c.1633G>A | 44 | |||||
| F:5′-ATCCAGAGGGGAAAAATATG-3′ | L9 | 180, 418, 634–180, 418, 894 | 261 bp | 82.5 | c.1624G>A | 143 | |
| R:5′-ATGCTGAGATCAGCCAAAT-3′ | c.1633G>A | 152 | |||||
| 20 | F:5′-GAGGCTTTGGAGTATTTCAT-3′ | S20 | 180, 434, 739–180, 434, 808 | 70 bp | 79.5 | c.3140A>G | 41 |
| R:5′-AATCCATTTTTGTTGTCCAG-3′ | |||||||
| F:5′-TCATTTGCTCCAAACTGACCAA-3′ | L20 | 180, 434, 534–180, 434, 885 | 352 bp | 83.5 | c.3140A>G | 246 | |
| R:5′-TGGAATCCAGAGTGAGCTTTCA-3′ |
According to GenBank accession no. NC_000003.10.
The stated Tms are according to the wild-type gDNA samples.
According to GenBank accession no. NM_006218.2.
For exon 9, set S9 was designed to amplify the region (92 bp) that includes the hotspot mutations of exon 9 (Figure 1A), while set L9 embrace the whole exon (261 bp). Both our primer sets were designed with either 1 (L9) or 2 (S9) mismatches at the 3′-end of the reverse primer, to a highly homologous (98%) genomic region (chromosome 22), to avoid false positives. For exon 20, set S20 was designed to amplify the region (70 bp) including the hotspot mutation (Figure 1B), and set L20 to amplify the whole exon 20 into the 5′-UTR (352 bp).
Figure 1.
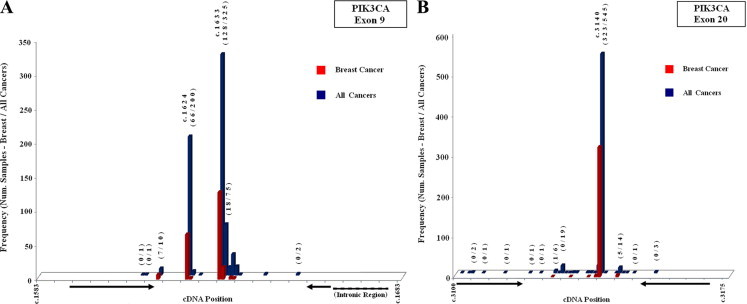
Diagrams of mutation frequencies of hotspot regions of exon 9 (A) and exon 20 (B) of the PIK3CA gene. Frequencies are reported for breast cancer (red color) and for all cancer (blue color) types. Arrows indicate primer sites of small amplicons (Sets S9 and S20). Mutation frequency data are according to COSMIC database v41 release.
Our primer sets S9 and S20 for the small amplicons were designed so that all reported somatic mutations in breast cancer, in the hotspot regions, according to the data registry at the COSMIC database, were included within the amplified region while excluded from primer sites (Figure 1). Hotspot mutations were placed as close to the center of the amplicons as possible (Figure 1 and Table 1). Amplicon melting domains were in silico tested by using the POLAND program32 (POLAND program; Institut für Physikalische Biologie, Heinrich-Heine-Universität, Düsseldorf, Germany; Accessed May 2008; http://www.biophys.uni-duesseldorf.de/local/POLAND/poland_help.html#Poland), and presented a single melting transition with the exception of the 261bp amplicon of exon 9 (Set L9), which presented a double melting transition.
PCR
Real-time PCR was performed in the LightCycler instrument (Roche Applied Science, Mannheim, Germany) using glass capillary tubes (Roche Applied Science). Extensive optimization experiments were performed to maximize PCR amplification efficiency, including PCR program parameters, Mg2+, primer, and template concentrations. The PCR reaction mix consisted of 1× PCR Buffer (Invitrogen, Carlsbad, CA), 0.4 mmol/L for each dNTP (Invitrogen), 0.05 U/μl Platinum TaqDNA Polymerase (Invitrogen), 0.15 μg/μl BSA (Sigma, Munich, Germany), 1× LCGreen Plus Dye (Idaho Technology, Salt Lake City, UT), 0.3 μmol/l primers, 25 ng gDNA and Mg2+ (2.5 mmol/L for S9 and 3.0 mmol/L for all other sets). Sterile water was used to supplement up to 10 μl.
The real-time PCR protocols began with one cycle at 94°C for 2 minutes followed by 60 cycles of the following: Set S9: 94°C for 10 s, 67°C for 20 seconds and 72°C for 20 s; Set L9: 94°C for 20 s, 61°C for 20 seconds and 72°C for 30 s; Set S20: 94°C for 10 s, 59°C for 15 seconds and 72°C for 20 s; Set L20: 94°C for 20 s, 58°C for 20 seconds and 72°C for 40 s.
After amplification, all protocols were followed by one cycle at 95°C for 1 minute and a rapid cooling to 40°C for 1 minute. Real-time fluorescence acquisition was set at 72°C (F1 channel). All PCR reactions were performed in triplicate for each sample.
Melting Analysis
After real-time PCR, melting curves were obtained using the HR-1 High-Resolution Melter (Idaho Technology). Melting with data acquisition began 10°C below and ended 6°C above the Tm of each amplicon (Table 1), rising at 0.1°C/s. Data processing included normalization, temperature shifting (2 to 5%), and fluorescence difference plots, using the accompanied software. The accompanied software did not allow processing of more than 32 samples simultaneously. In cases when more samples were needed for comparison, data were exported and curves were generated using Microsoft Excel 2003 software.
Sensitivity Study
We evaluated the sensitivity by mixing mutated gDNA from cell lines, with wild-type from PB at ratios of 50%, 30%, 20%, 10%, 5%, 2%, 1%, and 0.2%. Samples that were used for dilutions were selected to match gDNA quantity, quality and quantification cycle (Cq), to minimize PCR bias. Evaluation of sensitivity was performed for both small and large amplicons for both exons.
DNA Sequencing
All samples from both validation and selected groups that presented mutant melting transitions and ten samples presenting wild-type transitions were subjected to bidirectional DNA sequencing using the small amplicons. After HRMA, 7.5 μl of the PCR reaction mix was treated with 3 μl of ExoSAP-IT (USB). Five microliters of the treated PCR mix was used as a template for cycle sequencing, using the Big Dye Terminator v1.1 kit (Applied Biosystems, Carlsbad, CA). The reaction mix consisted of 1 μl ready reaction terminator premix, 7 μl buffer, 1 μl of 0.5 μmol/L primer (same used for PCR), and 6 μl of sterile water. The reactions were performed according to manufacturer's instructions, purified with NucleoSeq Columns (Macherey-Nagel, Düren, Germany), ran on an ABI 310 Genetic Analyzer (Applied Biosystems), and manually called.
Results
Genomic DNA from the cancer cell lines MCF-7 and T47D as well as wild-type PB samples was first used to evaluate the developed methodology. HRMA was able to readily discriminate between different wild-type controls and cell line samples in both small and large amplicons (see Supplemental Figure S1 at http://jmd.amjpathol.org). No dimers or “nonspecific” products were observed.
During optimization studies we observed a high baseline variation for FFPE samples. In an effort to reduce the wild-type baseline variation, we increased the number of PCR cycles to have FFPE samples fully amplified (see Supplemental Figure S2, A and B at http://jmd.amjpathol.org). It has been previously shown that this could be an important issue in HRMA.24,25 We also tested the amount of template used (100 ng, 50 ng, and 25 ng) in PCR reactions, using two wild-type FFPE samples presenting high and one presenting low variation. The samples that presented high variation showed a reduction of variation with template amount of 25 ng (see Supplemental Figure S2, C and D at http://jmd.amjpathol.org). By adjusting these two parameters (high number of cycles and low DNA input) a reduction of FFPE wild-type variation was apparent in a global level.
To evaluate the analytical sensitivity, dilutions of each cell line to wild-type gDNA (50–0.2%) were assessed. Fluorescence difference plots were generated, and the ability to discriminate melting transitions of the cell line dilutions from that of five wild-type samples was assessed. For exon 9, with the 92-bp amplicon we could discriminate a dilution corresponding to 2% of MCF-7 cell line, while with the 261-bp amplicon 5% could be discriminated (Figure, 2A and 2B). For exon 20, with the 70-bp amplicon we could discriminate a ratio of 1% of T47D cell line dilution, whereas with the 352-bp amplicon the detectable ratio was 10% (Figure, 2C and 2D). Melting curves were highly reproducible.
Figure 2.
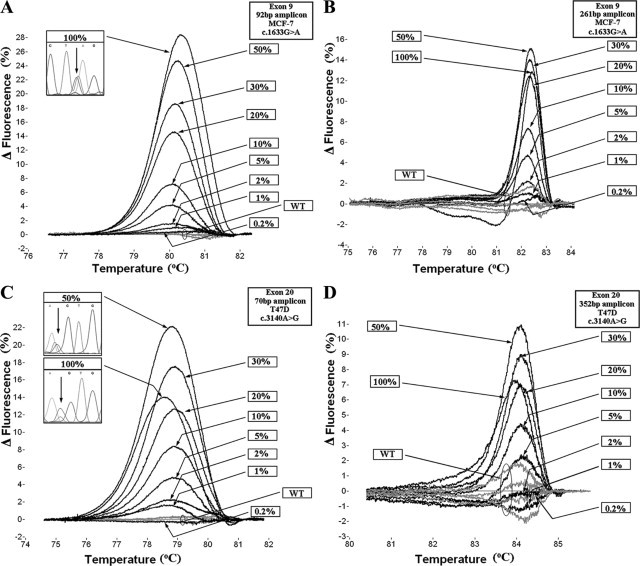
Fluorescence difference plots of sensitivity studies. The 92-bp amplicon (A) and 261-bp amplicon (B) of exon 9, and 70-bp amplicon (C) and 352-bp amplicon (D) of exon 20 of the PIK3CA gene. Figure windows present sequencing results of indicated ratios of the cell lines in the reverse direction.
In the case of exon 20, where T47D cell line was used, we observed that the 50% dilution presented a melting transition with higher differentiation than the undiluted cell line. This phenomenon was observed for both small and large amplicons (Figure, 2C and 2D). We considered this observation to be inconsistent with previous findings demonstrating maximum heteroduplex yield with fully heterozygous samples.33 Indeed, as can be seen in Figure 2C, sequencing results demonstrated that the 50% dilution of T47D simulated more authentic heterozygous sequence. We hypothesize that this may be due to preferential amplification of the mutant allele, because there are reports demonstrating gene amplification of this cell line.34
Our purpose of developing HRMA methodology for the larger amplicons was not limited to test and compare the analytical sensitivity with respect to the smaller amplicons. The objective was to detect mutations missed within these exons, not included in the small amplicon region. However, using larger amplicons, approximately 50% of the FFPE samples presented low or no PCR amplification. Therefore, only small amplicons were used for mutation scanning.
In the validation group, scanning the 20 noncancerous FFPE tissue samples no mutant melting transitions were observed. Scanning the 99 FFPE breast tumor samples, 12 samples in exon 9 and 20 in exon 20 were found to be mutated (in total: 32/99, 32%) (Figure 3). Interestingly, one FFPE fibroadenoma sample (1/10, 10%) presented a mutant transition, which was further defined by DNA sequencing as a c.3140A>G substitution (Figure 3B). All mutations detected were further defined by DNA sequencing (Figure 3; Supplemental Figure S3, A and B at http://jmd.amjpathol.org) and demonstrated in Table 2. All mutations were mutually exclusive. All samples were detected as heterozygous.
Figure 3.
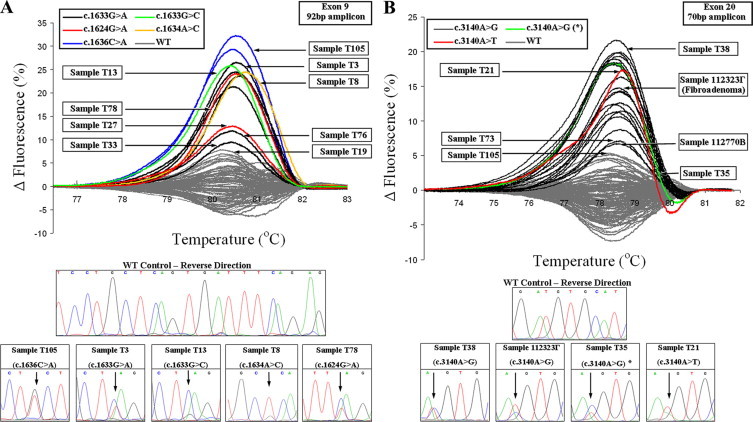
Fluorescence difference plots of HRMA, and sequencing results of the 129 formalin-fixed paraffin-embedded tissues, 92-bp amplicon for exon 9 (A) and 70-bp amplicon for exon 20 (B). Sequencing results of samples indicated by arrows can be found in the lower section of each of the figures or Supplemental Figure S3 at http://jmd.amjpathol.org. The sample indicated with an asterisk, in HRMA and sequencing, is the sample found to bear a high proportion for the c.3140A>G mutation, not compatible with an heterozygote.
Table 2.
PIK3CA Gene Variants in Breast Cancer (FFPE) Samples, as Detected by HRMA, and Defined by DNA Sequencing
| Nucleotide change | Protein change | Number of samples | Relative (%) | Total (%) |
|---|---|---|---|---|
| Validation group | ||||
| Exon 9 | ||||
| c.1633G>A | E545K | 6 | 18.8 | 6.1 |
| c.1624G>A | E542K | 2 | 6.2 | 2.0 |
| c.1636C>A | Q546K | 2 | 6.2 | 2.0 |
| c.1633G>C | E545Q | 1 | 3.1 | 1.0 |
| c.1634A>C | E545A | 1 | 3.1 | 1.0 |
| Exon 20 | ||||
| c.3140A>G* | H1047R | 19 | 59.5 | 19.2 |
| c.3140A>T | H1047L | 1 | 3.1 | 1.0 |
| Total | 32 | 100 | 32.3† | |
| Selected group | ||||
| Exon 9 | ||||
| c.1633G>A | E545K | 3 | 9.7 | 4.0 |
| c.1624G>A | E542K | 2 | 6.5 | 2.7 |
| c.1636C>G | Q546E | 1 | 3.2 | 1.3‡ |
| Exon 20 | ||||
| c.3140A>G | H1047R | 23 | 74.2 | 30.7 |
| c.3140T>G | H1047L | 1 | 3.2 | 1.3 |
| c.3136G>A | A1046T | 1 | 3.2 | 1.3‡ |
| Total | 31 | 100 | 40.0ठ|
One fibroadenoma sample, found to bear this mutation, is not included.
Of the 99 tumor samples.
The two mutations c.1636C>G and c.3136G>A were detected in the same sample, and therefore counted as a single mutated sample.
Of the 75 tumor samples.
However, one sample was found to bear a high proportion for the c.3140A>G mutation (higher the 50%). This sample presented a transition with minor differences from the rest of the samples bearing this mutation (Figure 3B, green color & marked with asterisk).
Additionally, only one wild-type FFPE sample and in only one out of the three PCR amplification experiments for exon 9 presented a transition wrongly assigned as mutant. Although this occurred in only one of the three runs, it was forwarded for DNA sequencing to be verified as wild-type. This false positive call corresponds to 1/675 PCR runs (0.15%) performed for wild-type FFPE samples.
The high baseline variation of the wild-type FFPE samples that was observed (Figure 3) has also been reported in our previous work on BRCA1 exon 20 HRM scanning.24 Some MUT samples presented a transition close to the wild-type baseline spread. The threshold for calling these transitions as MUT was attributed to the shape of the curve. In the case of exon 9 (92-bp amplicon), the broader range of the transitions with respect to the wild-type baseline assisted in calling (Figure 3A). For exon 20 (70-bp amplicon), a shift to the right of the curve peak assisted in calling these samples as MUT (Figure 3B). Because the MUT proportion varies, melting transitions from the same genotype do not present cluster together. Nevertheless, samples with the same genotype were similar or identical in shape. HRMA results were highly reproducible (see Supplemental Figure S4A at http://jmd.amjpathol.org).
When we applied the developed methodology in a high-throughput format, using a 96-well plate format LightCycler 480 (II) instrument (Roche, Germany) (data not shown), melting transitions presented almost identical profiles to those obtained by the HR-1 instrument (See Supplemental Figure S4, B and C at http://jmd.amjpathol.org).
Finally, the developed methodology was applied in a selected group of 75 breast cancer patients treated with Herceptin. When scanning these 75 FFPE tumor samples, six samples in exon 9 and 25 in exon 20 were found to be mutated. All mutations were further defined by DNA sequencing (Table 2). Interestingly, one sample presented mutant transitions in both exons. These transitions were defined by DNA sequencing (See Supplemental Figure S3C at http://jmd.amjpathol.org) as c.1636C>G and c.3136G>A substitutions and were the only mutations not encountered in the validation group. In total, 30/75 samples (40%) were detected as mutant. Only one wild-type FFPE sample in only one of the three PCR amplification experiments (exon 9) presented a transition wrongly assigned as mutant, to be verified as wild-type by sequencing.
Discussion
HRMA was introduced in 2003 as a cost-effective, probe-free, rapid, closed-tube, and simple approach for genotyping and/or mutation scanning.18 The ability of this technique to scan for mutations in samples from archival tissue specimens such as FFPE tissues is of high importance due to their ease of recovery and frequent use. However, FFPE samples present difficulties in PCR amplification and a reduction of diagnostic specificity when analyzed by HRMA.24,25
Herein, we report the development and validation of a highly sensitive and reliable HRMA mutation scanning methodology, using small amplicons, for hotspot regions of the PIK3CA gene. These mutations present a high prevalence in breast, as well as in other cancer types, and have an important clinical significance.35 We validated this methodology on 129 FFPE tissue samples and managed to minimize their disadvantages during PCR and HRMA, as shown by comparison studies with DNA sequencing. We further applied this methodology on FFPE tissue samples from 75 breast cancer patients who underwent treatment with Herceptin. All mutations in this group were also verified by DNA sequencing. However, we avoid reporting any clinical significance of these findings, because this is beyond the scope of this article.
The two small amplicons were used to scan within hotspot regions in exons 9 and 20 of the PIK3CA gene that bear the three recurrent mutations. According to COSMIC database, these three recurrent mutations represent the majority (80%, 488/609) of the reported mutated breast cancer samples of the PIK3CA gene. However, within these regions the presence of several additional mutations has been reported. Our primer sequences are in silico designed to amplify a region spanning 92% (561/609) of the reported mutated breast cancer samples (Figure 1). Moreover, in respect to mutated samples reported for all cancer types, the regions cover 84% of mutations (1305/1558).
In addition to our effort to include all frequent mutations within the small amplicons, we also tried to avoid mutations on primer sites, because this could compromise PCR amplification. Thus, regions bearing reported mutations in breast cancer were completely avoided, while regions bearing mutations in all other cancers that we were unable to exclude from primer sites presented a negligible frequency (0.008–0.024%) (Figure 1).
The high wild-type baseline variation detected in the FFPE samples tested is generally present in such samples.24,25,36 Additionally, reduction of diagnostic specificity has been reported when using HRMA of FFPE samples and is compounded by longer amplicons.25 In our validation study, only one false positive of the 129 samples occurred, and this in only one of the three runs performed for this sample. We believe that minimization of false positive calls and wild-type baseline variation may be credited to the reduction of amplicon length, and the optimization performed in terms of maximizing PCR amplification efficiency, increasing the number of PCR cycles and reducing DNA input.
Of the 2411 breast cancer samples reported in the COSMIC database that have been tested for PIK3CA mutations, 609 (25%) bear mutations. The regions covered by the two small amplicons include 23% (561/2411). However, in our study, we report a mutant frequency of 32% (32/99) of tumor samples in the validation (unselected) group. This difference was statistically significant from the expected (P < 0.05, using binomial test). This could be attributed to the higher analytical sensitivity of our methodology. Specifically, the best ever reported sensitivity for DNA sequencing has been demonstrated as being 5%,37 and as can be seen from our sequencing results (Figure 3; Supplementary Figure S3 at http://jmd.amjpathol.org), several mutant samples could easily be missed as wild-type. Our HRMA sensitivity study from the two small amplicons demonstrates an ability to detect as low as 1% of MUT alleles to total DNA, in a background of five different PB wild-type samples (Figure, 2A and 2C). On the contrary, larger amplicons have a lower analytical sensitivity (2.5% for 261-bp and 10% for 352-bp amplicon) (Figure, 2B and 2D), higher baseline variation, and increased difficulties in FFPE amplification. However, HRMA of FFPE tissue increases wild-type baseline variation, which may cause a reduction of analytical sensitivity. We estimate an analytical sensitivity of ∼2.5 to 5% when scanning FFPE samples, for the two mutations for which a sensitivity study was performed. As the estimated analytical sensitivity of HRMA is higher than this of DNA sequencing, in the case of a low MUT/wild-type proportion that cannot be verified by sequencing, digital PCR techniques can be used to confirm the results. Although digital PCR could be a labor-intense method, recent advances in microfluidics and real-time PCR allow a less laborious setting.38
In this study, we report for the first time a fibroadenoma sample mutated in the exon 20 of the PIK3CA gene (c.3140A>G), and it could be very interesting and instructive to monitor the disease outcome of this patient. Moreover, this result, along with recent findings demonstrating that PIK3CA mutations occur in preinvasive breast lesions39 and present high prevalence in papillary breast lesions,40 could have important implications with regard to therapeutic targeting of the PI3K pathway.
We also report for the first time c.1636C>A (validation group) and 3136G>A (selected group) substitutions in breast cancer. One of the tumor samples bearing a c.3140A>G mutation had a high percentage of the mutant allele (Figure 3B). This could be explained by a homozygous tumor sample contaminated with normal adjacent tissue or preferential gene amplification of the mutant allele.
In the selected group of 75 breast cancer patients who underwent Trastuzumab therapy, 40% (30/75) of the samples were detected by HRMA as mutants and were all verified by DNA sequencing. The clinical importance of these findings with respect to the connection of PIK3CA mutations resistance to Trastuzumab is to be reported in the near future by an extensive statistical analysis and in combination with results from PTEN immunohistochemistry staining.
The developed HRMA methodology has many advantages because it is rapid, demanding less than one hour for PCR, and approximately two hours for high-resolution melting per 30 samples, when using the HR-1 instrument and a heating rate of 0.1°C/s. Moreover, it can be performed in a high-throughput HRMA instrument and apart from the (96- and 384-well) plate-format HRMA instruments available (LightScanner, LightCycler 480), the recent introduction of the LightScanner32 instrument could minimize the turnaround time of this methodology, using the same (glass capillary) format.
The developed HRMA methodology represents an inexpensive approach for the detection of PIK3CA mutations because reaction volume is limited to 10 μl and does not require gene-specific probes. Samples are loaded for HRMA without postamplification processing, and because there is no destruction of the sample, we were able to forward PCR samples to sequencing. Although we were unable to undoubtedly define mutations by their melting transitions, we were able to reduce the amount of samples for sequencing down to ∼13% (33/258) for the validation and ∼21% (31/150) for the selected group.
The presence of PIK3CA mutations in many cancer types is connected with response to therapy. Furthermore, recent studies provide strong rationale for the clinical testing of combination therapies that target PI3K,41 while additional inhibitors of PI3K are expected to become available. Therefore, rapid and reliable screening for PIK3CA mutations will be essential as a marker of response to therapy. HRMA methodology based on the amplification of small amplicons is highly sensitive, reliable, cost-effective, easy-to-perform, and can be used as a screening test before DNA sequencing in a high-throughput pharmacodiagnostic setting.
Acknowledgements
We thank Prof. Eleptherios P. Diamandis (University of Toronto, Canada) for kindly providing the cancer cell lines used in this study. We would also like to thank Dr Dimitris Christou and Dr. Nikos Mavroeidis (from Bioanalytica SA) for helpful technical assistance.
Footnotes
Supported by the Special Account for Research grants (SARG) of the National and Kapodistrian University of Athens.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Current address for P.A.V.: Department of Biomolecular Medicine, SORA Division, Faculty of Medicine, Imperial College London, South Kensington, UK.
Web Extra Material
References
- 1.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7:R609–R616. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with a high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 4.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 5.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Helland A, Holm R, Kristensen GB, Børresen-Dale AL. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25:322–326. doi: 10.1002/humu.9316. [DOI] [PubMed] [Google Scholar]
- 7.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, Liu JM, Yang DM, Yang WK, Shen CY. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 8.Miyake T, Yoshino K, Enomoto T, Takata T, Ugaki H, Kim A, Fujiwara K, Miyatake T, Fujita M, Kimura T. PIK3CA gene mutations and amplifications in uterine cancers, identified by methods that avoid confounding by PIK3CA pseudogene sequences. Cancer Lett. 2008;261:120–126. doi: 10.1016/j.canlet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Okudela K, Suzuki M, Kageyama S, Bunai T, Nagura K, Igarashi H, Takamochi K, Suzuki K, Yamada T, Niwa H, Ohashi R, Ogawa H, Mori H, Kitamura H, Kaneko T, Tsuneyoshi T, Sugimura H. PIK3CA mutation and amplification in human lung cancer. Pathol Int. 2007;57:664–671. doi: 10.1111/j.1440-1827.2007.02155.x. [DOI] [PubMed] [Google Scholar]
- 10.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, Giovannucci EL, Fuchs CS. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 14.Kalaany NY, Sabatini DM. Tumors with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, Hortobagyi GN, Hung MC, Yu D. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, Bernards R. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 17.Yonemori K, Tsuta K, Shimizu C, Hatanaka Y, Hashizume K, Ono M, Kouno T, Ando M, Tamura K, Katsumata N, Hasegawa T, Kinoshita T, Fujiwara Y. Immunohistochemical expression of PTEN and phosphorylated Akt are not correlated with clinical outcome in breast cancer patients treated with trastuzumab-containing neo-adjuvant chemotherapy. Med Oncol. 2009;26:344–349. doi: 10.1007/s12032-008-9127-2. [DOI] [PubMed] [Google Scholar]
- 18.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 19.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 20.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 21.Gundry CN, Dobrowolski SF, Martin YR, Robbins TC, Nay LM, Boyd N, Coyne T, Wall MD, Wittwer CT, Teng DH. Base-pair neutral homozygotes can be discriminated by calibrated high-resolution melting of small amplicons. Nucleic Acids Res. 2008;36:3401–3408. doi: 10.1093/nar/gkn204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Idaho Technology Inc Small amplicon genotyping using internal temperature calibration and high resolution-melting. Biotechniques. 2008;44:577–578. doi: 10.2144/000112882. [DOI] [PubMed] [Google Scholar]
- 23.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295–307. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vorkas PA, Christopoulos K, Kroupis C, Lianidou ES. Mutation scanning of exon 20 of the BRCA1 gene by high-resolution melting curve analysis. Clin Biochem. 2009;43:178–185. doi: 10.1016/j.clinbiochem.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 25.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8:142–156. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst CD, Zuiverloon TC, Hafner C, Zwarthoff EC, Knowles MA. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes. 2009;2:66–72. doi: 10.1186/1756-0500-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Board RE, Thelwell NJ, Ravetto PF, Little S, Ranson M, Dive C, Hughes A, Whitcombe D. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54:757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 28.Simi L, Pratesi N, Vignoli M, Sestini R, Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M, Orlando C. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–253. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjöld B, Rutqvist LE, Skoog L, Stål O. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13:3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 30.Kioulafa M, Kaklamanis L, Stathopoulos E, Mavroudis D, Georgoulias V, Lianidou ES. Kallikrein 10 (KLK10) methylation as a novel prognostic biomarker in early breast cancer. Ann Oncol. 2009;20:1020–1025. doi: 10.1093/annonc/mdn733. [DOI] [PubMed] [Google Scholar]
- 31.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994;22:2760–2768. doi: 10.1093/nar/22.14.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palais RA, Liew MA, Wittwer CT. Quantitative heteroduplex analysis for single nucleotide polymorphism genotyping. Anal Biochem. 2005;346:167–175. doi: 10.1016/j.ab.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 35.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomoto K, Tsuta K, Takano T, Fukui T, Fukui T, Yokozawa K, Sakamoto H, Yoshida T, Maeshima AM, Shibata T, Furuta K, Ohe Y, Matsuno Y. Detection of EGFR mutations in archived cytologic specimens of non–small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006;126:608–615. doi: 10.1309/N5PQNGW2QKMX09X7. [DOI] [PubMed] [Google Scholar]
- 37.Miller CJ, Cheung M, Sharma A, Clarke L, Helm K, Mauger D, Robertson GP. Method of mutation analysis may contribute to discrepancies in reports of (V599E) BRAF mutation frequencies in melanocytic neoplasms. J Invest Dermatol. 2004;123:990–992. doi: 10.1111/j.0022-202X.2004.23468.x. [DOI] [PubMed] [Google Scholar]
- 38.Sundberg SO, Wittwer CT, Gao C, Gale BK. Spinning disk platform for microfluidic digital polymerase chain reaction. Anal Chem. 2010;82:1546–1550. doi: 10.1021/ac902398c. [DOI] [PubMed] [Google Scholar]
- 39.Troxell ML, Levine J, Beadling C, Warrick A, Dunlap J, Presnell A, Patterson J, Shukla A, Olson NR, Heinrich MC, Corless CL. High prevalence of PIK3CA/AKT pathway mutations in papillary neoplasms of the breast. Mod Pathol. 2010;23:27–37. doi: 10.1038/modpathol.2009.142. [DOI] [PubMed] [Google Scholar]
- 40.Dunlap J, Le C, Shukla A, Patterson J, Presnell A, Heinrich MC, Corless CL, Troxell ML. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120:409–418. doi: 10.1007/s10549-009-0406-1. [DOI] [PubMed] [Google Scholar]
- 41.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, García-Echeverría C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


