Abstract
Fast and reliable tests to detect mutations in human cancers are required to better define clinical samples and orient targeted therapies. KRAS mutations occur in 30–50% of colorectal cancers (CRCs) and represent a marker of clinical resistance to cetuximab therapy. In addition, the BRAF V600E is mutated in about 10% of CRCs, and the development of a specific inhibitor of mutant BRAF kinase has prompted a growing interest in BRAFV600E detection. Traditional methods, such as PCR and direct sequencing, do not detect low-level mutations in cancer, resulting in false negative diagnoses. In this study, we designed a protocol to detect mutations of KRAS and BRAFV600E in 117 sporadic CRCs based on coamplification at lower denaturation temperature PCR (COLD-PCR) and high-resolution melting (HRM). Using traditional PCR and direct sequencing, we found KRAS mutations in 47 (40%) patients and BRAFV600E in 10 (8.5%). The use of COLD-PCR in apparently wild-type samples allowed us to identify 15 newly mutated CRCs (10 for KRAS and 5 for BRAFV600E), raising the percentage of mutated CRCs to 48.7% for KRAS and to 12.8% for BRAFV600E. Therefore, COLD-PCR combined with HRM permits the correct identification of less represented mutations in CRC and better selection of patients eligible for targeted therapies, without requiring expensive and time-consuming procedures.
Cancer biomarkers play multiple roles in oncology. They can have prognostic functions by providing information on outcome and patient tractability. They can also play a predictive role by assessing the probability that patients will benefit from specific treatments. Finally, some cancer biomarkers possess pharmacodynamic criteria, measuring drug effects and orienting dose selection.1
Cancer genotyping is now often requested to tailor personalized therapies in the treatment of common cancers, like colorectal2,3 breast,4 lung,5 prostate,6 and head and neck.7 In colorectal cancer (CRC) the search for genetic markers for therapy response could help reduce the toxicity from therapy in patients who would not benefit from treatment.8 Selecting patients who are likely to take advantage of targeted treatments will ultimately result in better patient outcome and reduced costs.2
Somatic mutations in KRAS are considered a predictive marker of response to therapy in CRC, due to their association with clinical resistance to cetuximab and panitumumab, chimeric monoclonal antibodies acting as inhibitors of the epidermal growth factor receptor.9,10,11,12 These drugs improve a variety of clinically important outcomes in CRC patients who have the wild-type KRAS gene, whereas no response is observed in CRC patients with KRAS mutations.2,8,13,14 KRAS activating mutations have been reported in 30–54% of metastatic CRC patients,13,14,15 resulting in EGFR-independent activation of the mitogen-activated protein kinase pathway.
BRAF is mutated in about 10% of CRC patients, and the T>A transversion at nucleotide 1796, causing V600E, accounts for the most frequent of all BRAF variants. The high incidence of BRAFV600E mutation in cancer suggests that BRAF may also be an attractive therapeutic target16 and negative predictors of cetuximab therapy.17 The recent development of a specific inhibitor of the mutant isoform of BRAF kinase, currently in clinical trials (PLX4032),18 has increased the value of BRAFV600E identification in cancer patients who could benefit from this therapy.
The prognostic role of KRAS and BRAF in CRC has also been well investigated.19 KRAS mutations generally confer a worse prognosis in CRC patients, even if convincing evidence of the independent prognostic role of KRAS mutations is still lacking.2 Although the association of BRAF with prognostic parameters is still controversial, its pattern of mutations that mutually exclude KRAS seems to confirm its role in orienting patient therapy.20,21
Due to the cellular heterogeneity of solid cancers, the primary technical challenge encountered in the detection of somatic variants is the cellular heterogeneity in tumor biopsies. Somatic mutations can be present in low amounts within an elevated background of wild-type sequences, and more sensitive assays are therefore needed than those used for germline variants. There are many new approaches for improving sensitivity and efficiency detection of mutant alleles, such as high-resolution melting analysis,22,23,24,25 pyrosequencing,26,27 real-time PCR.28,29 Finally the use of Locked Nucleic Acid (LNA) and Peptide Nucleic Acid (PNA)30 was also proposed.
Although having higher sensitivity in variant detection, these approaches do not have an advantage for mutant alleles during the amplification protocol, which may be the only time when enrichment of variant alleles would be requested in samples with a very low percentage of mutated DNA.
Coamplification at lower denaturation temperature PCR (COLD-PCR) is a recently introduced PCR method that allows preferential amplification of minority alleles from a mixture of wild-type and mutant sequences.31The principle of this approach derived from the direct relationship between a given DNA sequence and the relative critical denaturation temperature (Tc). Sequence mismatch (heteroduplex), caused by point mutations, are responsible of an earlier denaturation step, thus by using a lower denaturation temperature during PCR, a selective amplification of mutant alleles will be performed.28,29 On this basis, the COLD-PCR application first requires definition of the optimal dissociation temperature (Td), commonly defined as critical temperature (Tc), at which the enrichment of minority alleles during PCR amplification is maximized.
COLD-PCR has been already tested in the detection of KRAS,31,32 TP53,31,33 EGFR,31 and GNAS134 mutations, and can identify mutated samples not otherwise identified by conventional PCR and cycle sequencing.
Mutation scanning with high-resolution melting analysis, HRMA, is based on the dissociation behavior of DNA when exposed to increasing temperatures. The signal modification is generated from the transition from double-to-single strand in the presence of fluorescent dyes actively intercalating double-stranded DNA.35 The HRM melting profile gives a specific sequence-related pattern that differentiates wild-type sequences from homozygote or heterozygote variants.36 As recently reported, combination of COLD-PCR and HRMA could further improves the mutation-scanning capabilities of HRM.37
In the present study we describe a COLD-PCR and HRM approach for sensitive detection of KRAS and BRAF mutations in 117 CRC patients.
Materials and Methods
Tumor Samples
Tissue samples were obtained from 117 consecutive patients with sporadic CRC (60 men and 57 women; mean age 67.5 years, range 48–89). A fragment of cancer tissue was snap-frozen in liquid nitrogen, while the remaining was processed for routine histological examination. The study was approved by the local ethical committee and an informed consent was obtained from each patient. Cancer histology and grading were defined using the World Health Organization criteria.38 CRCs were staged according to the American Joint Committee on Cancer TNM staging system.39 DNA extraction from snap-frozen tissues was performed by using EZ1 BIOROBOT and the EZ1 DNA Tissue Kit (Qiagen Inc., Germany) following to the manufacturer's protocol.
Cell Lines
DNA from SK-Mel-28 cell line, harboring the homozygous BRAFV600E mutation, and CCRF-CEM heterozygous for G12D mutation in KRAS was used as control. Mutated DNAs were diluted with MCF-7 wild-type DNA to obtain a variable percentage of mutated alleles. Even if MCF-7 cells are known to be an aneuploid cell line, we did not consider this aspect in the context of the dilution experiments a major problem. DNA extraction was performed by using QIAamp DNA Mini Kit (Qiagen, Milan, Italy).
Optimization of COLD-PCR for KRAS
PCR and COLD-PCR for KRAS codons 12 and 13 were initially performed using the same primer set already described by Zuo et al.31 During the reevaluation of the critical temperature of COLD-PCR with this primer set, we observed that the decrease in denaturation temperature (from 82.5 to 81°C) generated a double product of amplification. The first corresponded to the expected KRAS gene sequence, while the second was identified as the KRAS pseudogene (KRAS1P, GenBank NC_000006) located on chromosome 6 (data shown in Supplemental Figure 1, available at http://jmd.amjpathol.org). The two sequences have the same size (98 bp) but have a four-bases difference in the amplified region, which reduces the melting temperature (Tm) for pseudogene sequence. This fact favors its preferential amplification in comparison with the KRAS gene at a lower denaturation temperature (Td). At 81.5°C Td only pseudogene sequence was amplified. This limitation was incompatible with the definition of a correct critical temperature (Tc) for minority allele enrichment. For this reason we designed a new set of primers to amplify a fragment of 155 bp and containing a mismatch at 3′ to prevent pseudogene amplification: forward 5′-GTCACATTTTCATTATTTTTATTATAAGG-3′ and reverse 5′-TTTACCTCTATTGTTGGATCATATTC-3′.
Testing a series of Td (from 83°C to 81°C) in COLD-PCR experiments, we found that 82.5°C was the Tc for fast COLD and 82°C for full COLD formats. These temperatures provided the best mutant-sequence enrichment, with efficient amplification.
PCR and COLD-PCR for KRAS
Reactions for conventional PCR and COLD-PCR were performed in a RotorGene 6000 (Corbett Research Pty Ltd, Sidney, Australia) using 20 ng of genomic DNA in a total volume of 30 μl containing a final concentration of 1× PCR Buffer II (10 mmol/L Tris-HCl, pH 8.3; 50 mmol/L KCl) (Applied Biosystems, Milan, Italy), 2.5 mmol/L MgCl Solution, 0.2 mmol/L each dNTP, 0.5 μmol/L each primer, 0.5 μmol/L Syto9 (Invitrogen Corp., Carlsbad, CA) and 1.5 U AmpliTaq Gold DNA Polymerase (Applied Biosystems). Amplification by conventional PCR was performed with an initial hold at 95°C for 5 minutes, 45 cycles at 95°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 20 minutes.
COLD-PCR was performed using two alternative protocols: the ‘fast’ protocol, to detect Tm-reducing variants, and the ‘full’ protocol, suitable for revealing Tm-retaining and Tm-increasing alleles, as previously indicated.30 Because fast COLD-PCR guarantees higher enrichment of mutant alleles with G>T or G>A substitutions, which are the most frequent mutations in the KRAS gene, this approach was initially used to screen all samples. Then, the full protocol was used to further increase sensitivity, allowing other variant detection (ie, G>C and insertion) despite less enrichment of mutant alleles. The fast COLD-PCR protocol included an initial step at 95°C for 5 minutes followed by 20 PCR cycles at 95°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds, followed by 35 cycles at specific Tc: dissociation at 82.5°C for 3 seconds, 58°C for 30 seconds, 72° for 30 seconds, and a final extension at 72°C for 20 minutes. Full COLD-PCR cycles had an initial step at 95° for 5 minutes. The 15 cycles of PCR were performed at 95°C for 15 seconds, 60°C for 30 seconds, 72°C for 30 seconds, followed by 40 cycles at specific steps: 95° for 15 seconds, 70°C for 7 minutes, 82°C for 3 seconds, 58°C for 30 seconds, 72° for 30 seconds, with a final extension at 72°C for 20 minutes.
PCR and COLD-PCR for BRAF
Primers selected for conventional and COLD-PCR for BRAF exon 15 were forward 5′-ACAGAATTATAGAAATTAGATCTCTTACC-3′ and reverse 5′-GACAACTGTTCAAACTGATGG-3′, which amplify a 200-bp fragment. Reactions were performed using 20 ng DNA in the QuantiTect Probe PCR Master Mix (Qiagen) with 300 nmol/L of each primer and 1.5 μmol/L of Syto9 (Invitrogen) in a final volume of 20 μl. PCR was performed as follows: an initial hold at 95°C for 15 minutes, 40 cycles to 95°C for 30 seconds, 62°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 15 minutes. Because this type of mutation introduces a T>A transversion, a full COLD-PCR was directly used. Preliminarily, we tested a range of Td in COLD-PCR experiments (from 80°C to 78°C) and we found that 79.5°C was the best Tc for COLD. These temperatures provided the best mutant-sequence enrichment, with efficient amplification.
The full COLD-PCR protocol was as follows: 95°C for 15 minutes; 10 cycles of conventional PCR: 95°C for 15 seconds; 62°C for 30 seconds; 72°C for 1 minute; then 35 cycles of COLD-PCR performed at 95°C for 15 seconds, 70°C for 7 minutes, 79.5°C for 3 seconds, 60°C for 30 seconds and 72°C for 1 minute; final extension at 72°C for 15 minutes.
High-Resolution Melting Analysis and Sequencing
After conventional PCR or COLD-PCR protocols for KRAS and BRAF genes, all samples were submitted to HRM in a RotorGene 6000 (Corbett Research) with the following denaturation profile: 5 minutes at 95°C, 1 minute at 40°C and a melting profile from 74°C to 85°C using a ramping degree of 0.05. Sequencing analysis was performed in samples after purification with Qiagen PCR Purification Kit. A cycle sequencing reaction was performed with 2 μl of BigDye Terminator Ready Reaction Mix (Applied Biosystems), and the same primers used in PCR but with a concentration of 0.16 μmol/L in a final volume of 10 μl. After a second purification with a DyeEx 2.0 Spin Kit (Qiagen), samples were then analyzed with the ABI Prism 310 Genetic Analyzer (Applied Biosystems).
Results
Sensitivity of HRM COLD-PCR
To evaluate the theoretical sensitivity of our method, serial dilutions of KRAS-mutated (CCRF-CEM) and wild-type (MCF-7) DNA were prepared to obtain reconstituted samples containing 50, 25, 12.5, 6.2, 3.1, 1.5, and 0.8% of mutated alleles. These samples were submitted to conventional PCR and fast COLD-PCR protocols, respectively. The sensitivity was then evaluated by HRM analysis and sequencing. After conventional PCR, 6.2% mutated alleles were detectable in HRM differential graphs, whereas after COLD-PCR 0.8% were easily detectable, indicating an eightfold increase in sensitivity (Figure 1).
Figure 1.
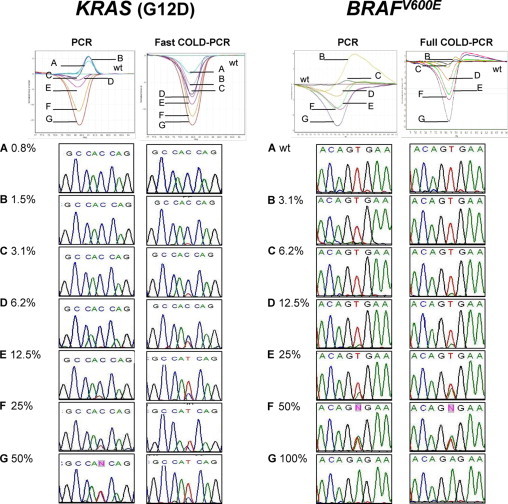
Sensitivity PCR and COLD-PCR for a KRAS mutation (G12D) and BRAFV600E. Left: Dilution tests were performed on serially diluted DNA from a KRAS-mutated cell line CCRF-CEM and wild-type DNA from MCF-7 cell line, to obtain A = 0.8%, B = 1.5%, C = 3.1%, D = 6.2%, E = 12.5%, F = 25%, G = 50%. Dilutions were submitted to conventional PCR (left) and fast COLD-PCR (right). After HRM, linearity and sensitivity were compared through differential plot analysis. Direct sequencing of corresponding reconstituted samples are also reported (reverse strand sequence). Right: Dilution tests were performed on serially diluted DNA from a BRAF-mutated cell line SK-Mel-28 and wild-type DNA from MCF-7 cell line. Mutated DNA was diluted with wild-type DNA: A = wild-type, B = 3.1%, C = 6.2%, D = 12.5%, E = 25%, F = 50%, G = 100%. Dilutions were submitted to conventional PCR (left) and full COLD-PCR (right). After HRM, linearity and sensitivity were compared through differential plot analysis. Direct sequencing of corresponding reconstituted samples are also reported.
Direct sequencing was a less sensitive technique for detecting low abundant variant sequences. The electropherograms of amplification products for KRAS obtained with conventional PCR indicated that the minimum detectable percentage was about 12.5%. Conversely, sequence analysis after COLD-PCR allowed the detection of 3.1% mutated alleles, providing a fourfold increase in the detection of this mutant (Figure 1).
The same procedure was adopted to evaluate the theoretical sensitivity of full COLD-PCR in detecting BRAFV600E in reconstituted samples. HRM was able to detect 12.5% BRAFV600E after conventional PCR. This percentage was improved by COLD-PCR that allowed to detected until 6.2% of BRAFV600E. After direct sequencing in PCR products, 25% BRAFV600E was detectable after conventional PCR and 12.5% after COLD-PCR (Figure 1).
CRC Samples
Initially, screening all of the 117 CRC samples for the research of KRAS mutations and BRAFV600E by HRMA and direct sequencing, we found that 47/117 patients (40.2%) were mutated for KRAS and 10/117 (8.5%) for BRAFV600E. The remaining 70 samples were classified as wild-type.
In a second phase, all samples (mutated and wild-type) were resubmitted to a complete screening using COLD-PCR. Firstly, adopting the fast COLD-PCR protocol to look for KRAS variants, we confirmed the presence of genetic variants in the same samples that were classified as positive after PCR and HRM. However, this approach allowed us to detect also eight new KRAS-mutated samples. In the 62 samples that were still negative, we used the full COLD-PCR protocol for KRAS mutants and were able to enrich and reclassify two CRC samples as mutated. In conclusion, a total of 10/70 (14.3%) new KRAS-mutated CRC samples were revealed by COLD-PCR protocols. Examples of conventional and COLD-PCR results are shown in Figure 2 and Supplemental Table 1 (available on http://jmd.amjpathol.org).
Figure 2.
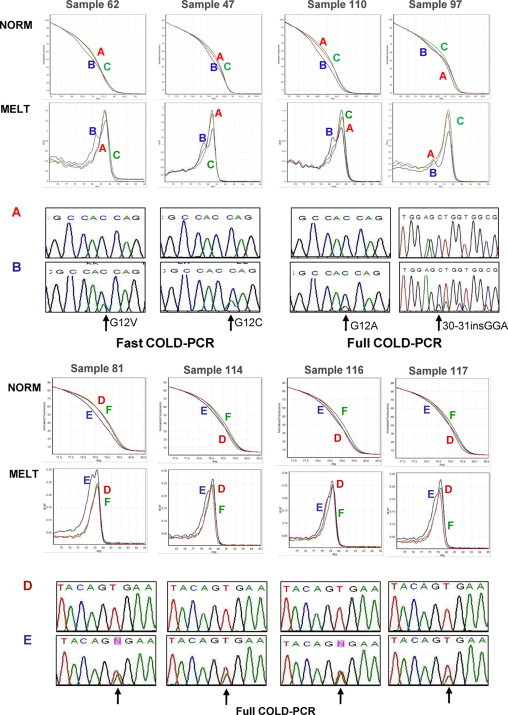
Top: Comparison between conventional PCR and COLD-PCR for KRAS mutations in CRC samples. Four examples of CRCs submitted to HRM and sequencing after either a traditional PCR protocol or COLD-PCR. Starting from the upper panels: Norm, HRM profile of a wild-type DNA control (C), unknown sample amplified by conventional PCR (A), and the same sample amplified by either fast or full COLD-PCR protocols (B); Melt, Melt analysis of the same samples of NORM panels. A double peak is evident for all samples submitted to COLD-PCR (B) but not for the same samples amplified with conventional PCR (A). A and B: Direct sequencing of the same samples, showing the appearance of KRAS mutations (indicated by the arrows) in COLD-PCR amplified samples. Bottom: Comparison between conventional PCR and COLD-PCR for BRAFV600E mutation in CRC samples. Four examples of CRCs submitted to HRM and sequencing after either a traditional PCR protocol or COLD-PCR. Starting from the upper panels: Norm, HRM profile of a wild-type DNA control (F), unknown sample amplified by conventional PCR (D), and the same sample amplified by full COLD-PCR protocol (E); Melt, melt analysis of the same samples of NORM panels. A double peak is evident for all samples submitted to COLD-PCR (E) but not for the same samples amplified with conventional PCR (D). D and E: Direct sequencing of the same samples, showing the appearance of BRAFV600E mutations (indicated by the arrows) in COLD-PCR amplified samples.
Similarly, through the full COLD-PCR protocol to look for BRAFV600E, we reconfirmed the presence of 10 mutated samples as already revealed by conventional PCR but identified five new mutated samples initially classified as negative (see examples in Figure 2).
In further five CRC samples (4 for KRAS and 1 for BRAF), HRM profiles suggested the presence of a possible genetic variant. Unfortunately, we were not able to confirm these findings after direct sequencing (data not shown). This is not surprising taking into account the differences in terms of sensitivity between HRM and sequencing (see previous paragraph). In any case these samples, in the absence of a sequencing confirmation, were classified as not mutated.
Globally, we can affirm that COLD-PCR was able to correctly identify KRAS or BRAF mutations in 15/70, or 21.5% of patients initially classified as negative for the two mutations. See Table 1 for mutations details.
Table 1.
List of KRAS and BRAFMutations Detected in Colorectal Cancers Using Conventional and COLD-PCR
| Gene | Nucleotide change | Protein mutation | Conventional PCR | COLD-PCR |
|---|---|---|---|---|
| KRAS | wt | 70 | 60 | |
| c.35G>A | G12D | 15 | 16 | |
| c.35G>T | G12V | 9 | 13 | |
| c.38G>A | G13D | 5 | 7 | |
| c.34G>T | G12C | 9 | 10 | |
| c.35G>C | G12A | 3 | 4 | |
| c.34G>A | G12S | 2 | 2 | |
| c.57G>T | L19F | 1 | 1 | |
| c.37G>A | G13C | 1 | 1 | |
| c.64C>A | Q22K | 1 | 1 | |
| c.30_31insGGA | p.10_11insG | 1 | 2 | |
| Total | 47 | 57 | ||
| BRAF | wt | 107 | 102 | |
| c.1799T>A | V600E | 10 | 15 | |
| Total | 117 | 117 |
Discussion
Some intrinsic factors can limit the detection of somatic mutations in human cancer biopsies. First, most of the mutations affecting cancer cells are heterozygous. In addition, human solid cancers have a complex cellular architecture in which both normal and pathological cells coexist. Finally, clonal heterogeneity can account for the presence of cellular clones with different mutational status. All these factors can contribute to reducing the percentage of mutated alleles in DNA extracted from tissues, favoring a dilution effect in wild-type DNA. On the other hand, direct sequencing, the reference technique for identification of sequence variants, has a defined detection limit, usually around 20% of mutated alleles. The introduction of pyrosequencing has partially modified this scenario and provides, in the best experimental conditions, as much as 10% sensitivity.26,32
To improve identification of low abundant somatic mutations in clinical samples, there are two basic strategies: increasing the sensitivity of the detection systems and enriching the proportion of mutated alleles during PCR amplification.
The first strategy is represented by the introduction of a highly sensitive screening method to identify sequence variants in PCR amplification products, evaluating their temperature-dependent melting profile through HRM. HRM is demonstrated to be a rapid closed-tube post-PCR technique to perform genetic screening in tumor samples. In a recent study40 we demonstrated the possibility of identifying at least 5% of mutated alleles in a background of wild-type DNA, even if higher sensitivities have been reported by other authors.22
The second strategy, indicated as COLD-PCR, enhanced the amplification of low abundant mutated alleles using a specific PCR protocol based on the identification of a critical temperature of DNA denaturation. This enrichment is designed for direct identification of somatic mutations, undetectable after conventional PCR.31,32 In a recent study, COLD-PCR was applied to the detection of KRAS variants in a heterogeneous group of unselected clinical samples, including 17 paraffin-embedded tissues from primary colorectal cancers and 10 metastases, reporting a mean 1.7-fold increase was obtained.31 Conversely, COLD-PCR has never been used to enrich BRAFV600E variants in clinical samples. Very recently the combined use of HRMA and COLD-PCR has been successfully applied to the screening and identification of TP53 mutations.33
In the present study, we focus our interest on detection of KRAS or BRAF hot spot mutations in CRCs with COLD-PCR and HRM. Our results on reconstituted samples confirm that COLD-PCR is a technique able to enhance sensitivity of screening of mutations of both genes. The increase was particularly evident after HRM analysis, with eightfold higher sensitivity for KRAS and fourfold for BRAF. The difference in the increment between the two genes is probably due to the different protocol used to test sensitivity: fast-COLD for KRAS and full for BRAF. When the results of COLD-PCR were confirmed with sequencing, the increased sensitivity was reduced by the innate limitation of this technique.
The increased sensitivity of COLD-PCR for KRAS and BRAF mutations was also tested in a group of 117 CRCs and compared with results obtained with a standard PCR-HRM protocol.39 By COLD-PCR we confirmed the presence of mutations in all samples initially classified as mutated using conventional PCR-HRM, and we also identified a significant subset (five BRAF and 10 KRAS of 117) of CRCs initially classified as negative for both genes. This means that COLD-PCR is able to improve the sensitivity of HRM-PCR, a technique that was already considered particularly sensitive in mutation screening, as proved by the relatively high percentage of mutated samples we initially found (40.2% for KRAS and 8.5% for BRAF).
It is also important to remark that in a small subset of 5 CRC samples, HRM evidenced an abnormal profile compatible with the presence of a mutant in the sequence not confirmed by direct sequence analysis. Due to cited difference of sensitivity of the two techniques (from 5% of HRM to the 20% of sequencing), we can postulate that few samples with a very low-level mutation can fall, after COLD-PCR, in that window detectable with HRM but not with sequencing. Further research in this matter is in progress in our laboratory.
Taking our population study as a model, we can conclude that by combining COLD-PCR and HRM, we were able to detect KRAS and BRAF mutations in about 25% of patients initially classified as wild-type CRCs and, according to our strategy, the percentage of patients with either KRAS or BRAF mutations passes from 48.7% to 61.5%. To understand the real value of this 12.8% increase of samples correctly classified for KRAS and BRAF mutations we must consider that there were 150,000 newly diagnosed colorectal cancer patients in the United States in 2009 (http://seer.cancer.gov/csr/1975_2006/). This means that, in theory, about 19,000 of these patients could be correctly classified for KRAS or BRAF mutation, using HRM COLD-PCR.
In conclusion, the correct identification of less-represented mutations in CRC can be significantly improved with COLD-PCR combined with HRM, without requiring expensive and time-consuming procedures and while maintaining a closed-tube approach. HRM COLD-PCR has the potential to improve the routine search for mutations in cancer tissues. It is important to remark that today the real significance of low-level KRAS and BRAF mutations remains to be clinically proven, mainly for their predictive role in response to targeted therapy.
Footnotes
Supported in part by a grant from Italian Ministry of Health.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Web Extra Material
Effects of improper primer design for KRAS mutation detection by COLD-PCR.Upper panel: Aligned sequences of KRAS gene and pseudogene KRAS1P. Codons 12 and 13 of KRAS are indicated. Primers used for amplification are yellow highlighted.Lower panels: Examples of Tc effects on sequencing of one mutated and one wild type CRC samples. At 81.5°C Td only pseudogene sequence was amplified in mutated sample, whereas in wild type CRC pseudogene sequence appears at 82.5°C.For this reason, to improve PCR selectivity, a new primer pair was designed with a higher homology with KRAS gene sequence and with a 3′ mismatch in both forward and reverse primers.
Comparison of detection limits for KRAS and BRAF mutations with PCR and COLD-PCR
References
- 1.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 2.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 3.Chau I, Cunningham D. Treatment in advanced colorectal cancer: what, when and how? Br J Cancer. 2009;100:1704–1719. doi: 10.1038/sj.bjc.6605061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oakman C, Bessi S, Zafarana E, Galardi F, Biganzoli L, Di Leo A. Recent advances in systemic therapy: new diagnostics and biological predictors of outcome in early breast cancer. Breast Cancer Res. 2009;11:205–216. doi: 10.1186/bcr2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.John T, Liu G, Tsao MS. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S14–S23. doi: 10.1038/onc.2009.197. [DOI] [PubMed] [Google Scholar]
- 6.Febbo PG. Genomic approaches to outcome prediction in prostate cancer. Cancer. 2009;115:3046–3057. doi: 10.1002/cncr.24350. [DOI] [PubMed] [Google Scholar]
- 7.Pai SI, Westra WH. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 9.Raymond E, Faivre S, Armand JP. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs. 2000;60:15–23. doi: 10.2165/00003495-200060001-00002. [DOI] [PubMed] [Google Scholar]
- 10.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 11.Galizia G, Lieto E, De Vita F, Orditura M, Castellano P, Troiani T, Imperatore V, Ciardiello F. Cetuximab, a chimeric human mouse anti-epidermal growth factor receptor monoclonal antibody, in the treatment of human colorectal cancer. Oncogene. 2007;26:3654–3660. doi: 10.1038/sj.onc.1210381. [DOI] [PubMed] [Google Scholar]
- 12.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 13.Mittmann N, Au HJ, Tu D, O'Callaghan CJ, Isogai PK, Karapetis CS, Zalcberg JR, Evans WK, Moore MJ, Siddiqui J, Findlay B, Colwell B, Simes J, Gibbs P, Links M, Tebbutt NC, Jonker DJ, Working Group on Economic Analysis of the National Cancer Institute of Canada Clinical Trials Group Australasian Gastrointestinal Interest Group Prospective cost-effectiveness analysis of cetuximab in metastatic colorectal cancer: evaluation of National Cancer Institute of Canada Clinical Trials Group CO 17 Trial. J Natl Cancer Inst. 2009;101:1182–1192. doi: 10.1093/jnci/djp232. [DOI] [PubMed] [Google Scholar]
- 14.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 16.Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, Ogilvie L, Hedley D, Martin J, Marshall CJ, Springer CJ, Marais R. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 17.Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaherty K, Puzanov I, Sosman J, Kim K, Ribas A, McArthur G, Lee RJ, Grippo JF, Nolop K, Chapman P. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27:15s. (abstr 9000) [Google Scholar]
- 19.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS Trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 20.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meriggi F, Di Biasi B, Abeni C, Zaniboni A. Anti-Egfr Therapy in Colorectal Cancer: how to Choose the Right Patient. Curr Drug Targets. 2009;10:1033–1040. doi: 10.2174/138945009789577891. [DOI] [PubMed] [Google Scholar]
- 22.Tindall EA, Petersen DC, Woodbridge P, Schipany K, Hayes VM. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum Mutat. 2009;30:876–883. doi: 10.1002/humu.20919. [DOI] [PubMed] [Google Scholar]
- 23.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 24.Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 25.Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans. 2009;37:433–437. doi: 10.1042/BST0370433. [DOI] [PubMed] [Google Scholar]
- 26.Ronaghi M, Shokralla S, Gharizadeh B. Pyrosequencing for discovery and analysis of DNA sequence variations. Pharmacogenomics. 2007;8:1437–1441. doi: 10.2217/14622416.8.10.1437. [DOI] [PubMed] [Google Scholar]
- 27.Langaee T, Ronaghi M. Genetic variation analyses by Pyrosequencing. Mutat Res. 2005;573:96–102. doi: 10.1016/j.mrfmmm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 28.Morlan J, Baker J, Sinicropi D. Mutation detection by real-time PCR: a simple, robust and highly selective method. PLoS One. 2009;4:e4584. doi: 10.1371/journal.pone.0004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tol J, Dijkstra JR, Vink-Börger ME, Nagtegaal ID, Punt CJ, van Krieken JH, Ligtenberg MJ. High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orum H. PCR clamping. Curr Issues Mol Biol. 2000;2:27–30. Review. [PubMed] [Google Scholar]
- 31.Zuo Z, Chen SS, Chandra PK, Galbincea JM, Soape M, Doan S, Barkoh BA, Koeppen H, Medeiros LJ, Luthra R. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22:1023–1031. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Milbury CA, Li C, Makrigiorgos GM. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum Mutat. 2009;30:1583–1590. doi: 10.1002/humu.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaney D, Diss TC, Presneau N, Hing S, Berisha F, Idowu BD, O'Donnell P, Skinner JA, Tirabosco R, Flanagan AM. GNAS1 mutations occur more commonly than previously thought in intramuscular myxoma. Mod Pathol. 2009;22:718–724. doi: 10.1038/modpathol.2009.32. [DOI] [PubMed] [Google Scholar]
- 35.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 36.Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- 37.Milbury CA, Li J, Makrigiorgos GM. COLD-PCR-enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin Chem. 2009;55:2130–2143. doi: 10.1373/clinchem.2009.131029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jass JR, Sobin LH. Histological Typing of Intestinal Tumors. 2nd Ed. Springer-Verlag; Berlin, Germany: 1989. International Histological Classification of Tumors. [Google Scholar]
- 39.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer ed 4 staging. J Natl Cancer Inst. 2004;6:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 40.Simi L, Pratesi N, Vignoli M, Sestini R, Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M, Orlando C. High-resolution melting analysis for rapid detection of KRAS. BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–253. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of improper primer design for KRAS mutation detection by COLD-PCR.Upper panel: Aligned sequences of KRAS gene and pseudogene KRAS1P. Codons 12 and 13 of KRAS are indicated. Primers used for amplification are yellow highlighted.Lower panels: Examples of Tc effects on sequencing of one mutated and one wild type CRC samples. At 81.5°C Td only pseudogene sequence was amplified in mutated sample, whereas in wild type CRC pseudogene sequence appears at 82.5°C.For this reason, to improve PCR selectivity, a new primer pair was designed with a higher homology with KRAS gene sequence and with a 3′ mismatch in both forward and reverse primers.
Comparison of detection limits for KRAS and BRAF mutations with PCR and COLD-PCR


