Abstract
Caveolin-1 belongs to a family of scaffolding proteins, and the P132L point mutation of this gene has been found in up to 16% of all examined breast cancers. Subsequent studies have revealed that the P132L mutation exerts a dominant negative effect through misfolding during caveolin-1 oligomerization. However, this mutation has not been validated in other series of breast cancer samples. Contradictory to the suggested tumor suppressor function, overexpression of caveolin-1 is common in various cancer types. To clarify these inconsistent results, we examined the caveolin-1 mutation in a large series of breast cancer specimens. We first used a standard direct sequencing method and found that none of the 99 breast cancers tested had this mutation. Then we developed a sensitive method for a paraffin section that could detect the mutant allele at a rate of as little as 0.1% among wild-type allele copies. Even when using this sensitive method, none of the 80 estrogen receptor–positive breast tumors had the P132L mutation. Furthermore, 270 cancers in various organs were examined, and no caveolin-1 mutations were detected. These results raise doubt about the presence of the caveolin-1 P132L mutation in breast cancer and other cancer types, and thus further studies are warranted.
Caveolae, or “little caves,” are flask-like invaginations of the plasma membrane that are coated by the oligomerized protein, caveolin.1,2,3 Caveolin-1 belongs to a highly conserved gene family and is coexpressed with caveolin-2 in the cells of various tissues, including the mesenchyme, endothelium, neuronal tissues, and some epithelial cells. The caveolin-1 gene is composed of three exons and is translated into the endoplasmic reticulum as a full-length protein of 178 amino acids (aa) (ie, the α-isoform) or as the β-isoform, which lacks the first 32 aa. Caveolin-1 plays a major role in the function of caveolae, which are involved in various cellular processes, including cholesterol homeostasis, vesicular transport, cell migration, cell cycle, and cell polarity. This molecule directly interacts with proproliferative molecules, including EGFR, ERBB2, and PI3K, through the caveolin-scaffolding domain and negatively controls signaling pathways regulating cell proliferation, differentiation, apoptosis, adhesion, and invasion.1,2,4,5 Therefore, impairment of caveolin-1 regulation would be expected to result in tumor suppressor activity.5 Indeed, the transcript and protein levels of caveolin-1 are down-regulated in cancer cell lines, and reduced levels of this protein in NIH3T3 cells leads to oncogenic transformation.6 Furthermore, in MCF7 cells, which show low levels of caveolin-1, expression of this molecule inhibits anchorage-independent growth, anoikis, and invasion.7 In addition, caveolin-1–deficient mice develop multifocal dysplastic lesions throughout the entire mammary gland.8,9
In 2001, Hayashi et al found a sporadic mutation in the caveolin-1 gene that leads to a proline-to-leucine substitution at position 132 (P132L).10 Subsequent studies have revealed that the P132L mutation has a dominant negative effect through the misfolding of caveolin-1 within the Golgi complex.11 The initial article stated that 15 of 92 primary breast tumors harbored this mutation.11 A subsequent report determined that the P132L mutation was specific to estrogen receptor (ER)-positive breast cancers; the mutation was found in six of 32 ER-positive tumors (35%) but in none of the 23 ER-negative tumors.12 However, a later study reported that the P132L mutation was not detected in any of 55 breast cancer specimens, using the same method as described in the initial article.13 Additionally, there are no articles from any other research group demonstrating the presence of this mutation in primary breast cancers. Therefore, we aimed to assess whether we could detect the mutation using a standard direct sequencing method and a novel sensitive assay in a larger series of primary breast cancer specimens.
Materials and Methods
Samples
We examined a consecutive series of 109 breast cancer specimens by RT-PCR–coupled direct sequencing. The specimens included 98 invasive ductal carcinomas, six invasive lobular carcinomas, two noninvasive ductal carcinomas, and three special subtypes of breast cancer. Among them, ten samples were excluded due to poor nucleic acid integrity; insufficient amounts of PCR product were obtained even when using the control primers for β actin, which generate a shorter PCR product. We also examined 80 ER-positive cancers, using the Cycleave PCR assay on DNA that was extracted from paraffin-embedded sections. Of the 80 cancers, 40 were selected from the samples used for RT-PCR direct sequencing analysis, while the other 40 were independent sample set selected in a concretive manner form a different period. All of the tumors were histologically reviewed by two authors (S.K. and Y.Y.), and histological classification was based on the World Health Organization classification of tumors of the breast.14 In addition, 270 cancers from various anatomical sites were analyzed for the presence or absence of the caveolin-1 mutation. The 270 cancers included 77 colorectal adenocarcinomas, 48 gastric adenocarcinomas, 100 lung cancers (80 adenocarcinomas, one each of adenosquamous carcinoma, large cell carcinoma, squamous cell carcinoma, small cell lung cancer, large cell neuroendocrine carcinoma, and carcinoid tumor), 32 ovarian tumors (11 serous carcinomas, 10 clear cell carcinomas, seven endometrioid carcinomas, and four mucinous carcinomas), and 13 pancreatic cancers (12 ductal carcinomas and an intraductal papillary mucinous tumor). For this analysis, written informed consent was obtained from each patient according to the protocol approved by the Institutional Review Board of the Aichi Cancer Center.
Immunohistochemistry
Expression status of the ER and progesterone (PR) were immunohistochemically analyzed, using antibodies for ER (6F11, Novocastra, Newcastle, UK) and PR (PR88, Biogenex, San Ramon, CA), as described previously.15 The antigens were retrieved by microwave treatment in citrate buffer, and the color was developed by 3,3′-diaminobenzidine. The expression were evaluated according to the criteria described by Harvey et al.16 HER2 status was determined using HERCEPTEST (DAKO, Copenhagen, Denmark) according to the manufacturer's instructions. The results were evaluated according to the ASCO/CAP guideline,17 including further analysis of tumors with a score of 2+ by means of fluorescent in situ hybridization (PathVysion, Vysis Inc., Downers Grove, IL).
Direct Sequencing
Using needle biopsy equipment, cancer tissue cores were obtained from the cancerous tissues. Several touch imprint slides were made, and the presence of cancer cells was confirmed with Giemsa staining, followed by total RNA extraction from the slides using an RNeasy kit (Qiagen, Valencia, CA). Briefly, the cellular contents of the slides were washed out with Buffer RLT and collected in a tube. The subsequent procedure was performed according to the manufacturer's instructions.
Caveolin-1 transcripts were amplified using the QIAGEN OneStep RT-PCR Kit (Qiagen) with primers as follows: forward, 5′-GAGGGACATCTCTACACCGTTC-3′; and reverse, 5′-ATTGGCACCAGGAAAATTAAAA-3′. The PCR products, which were purified using the QIAquick PCR Purification Kit (Qiagen), were directly sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Forester City, CA).
In addition to direct sequencing of the RT-PCR products, caveolin-1 was analyzed using DNA extracted from the paraffin-embedded sections. The tumor cell–rich area was marked on the H&E slide, and the exact same area of deparaffinized unstained sections was dissected for DNA extraction. The primers used were 5′-CCACCTTCACTGTGACGAAATA-3′ (forward) and 5′-ATTGGCACCAGGAAAATTAAAA-3′ (reverse). Subsequently, the PCR products were sequenced with a procedure similar to the direct sequencing of RT-PCR products. Mutations in the TP53 and CDH1 genes were also examined as a control using RT-PCR–coupled direct sequencing, similar to the procedure used for caveolin-1. TP53 sequencing was performed as described previously,18,19 while CDH1 was sequenced with the primer sets as follows: CDH1-1F, 5′-AAGAGAAACAGGATGGCTGAAG-3′; CDH1-1R, 5′-GGTACCACATTCGTCACTGCTA-3′; CDH1-2F, 5′-ATCTTTGTGCCTCCTGAAAAGA-3′; CDH1-2R, 5′-GCAGCAGAATCAGAATTAGCAA-3′.
In all cases, forward and reverse sequencing reactions were analyzed separately and produced concordant results. It should be noted that duplicate sequencing reactions were not performed.
Cycleave PCR Assay
A Cycleave PCR technique for the detection of the P132L caveolin-1 mutation was used with the SmartCycler system (SC-100, Cepheid, Sunnyvale, CA) for the ER-positive breast cancer tissues. The detailed principle of this method has been previously described.20,21 This assay was performed using a Cycleave PCR core kit (TAKARA, Co., Ltd., Ohtsu, Japan), and the sequences of the primer sets and the probes were as follows: caveolin-1 PCR forward primer, 5′-TTGCTGTCTGCCCTCTTTG-3′; caveolin-1 PCR reverse primer, 5′-GACGGTGTGGACGTAGATG-3′; wild-type probe, 5′ FAM-AT(G)GTACAACTGC-Eclipse 3′; and P132L mutation probe, 5′ ROX-AT(A)GTACAACTGC-Eclipse 3′(parentheses denote RNA). The PCR primers were used to amplify caveolin-1 DNA. The probe was an RNA-DNA chimera that could hybridize to either the mutation or the wild-type sequence of the amplified gene. Once the probe hybridized to the amplified product, the RNA part of the probes was cleaved by RNase H, which specifically recognized the complete RNA-DNA hybrids (Figure 1). Cleavage resulted in two products with a fluorescer and a quencher on each side of the probe, which emit fluorescence when separated. The mutant probe was labeled with FAM, while the wild-type probe was labeled with ROX. The caveolin-1 P132L mutant DNA was generated as an artificial gene, which was provided by Custom Gene Synthesis Services (Invitrogen, Carlsbad, CA). This technology allowed us to generate specifically designed DNA of a few kilobases in length. The artificial gene and placental DNA were then used as positive and negative controls, respectively.
Figure 1.
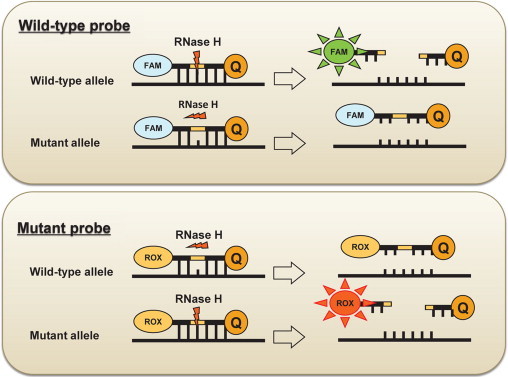
Schematic representation of the Cycleave method. The probe is an RNA-DNA chimera that hybridizes to either the mutated or the wild-type sequence of the amplified gene. Once the probe is hybridized to the amplified products, the RNA part of the probes is cleaved by RNase H, which specifically recognizes complete RNA-DNA hybrids. The cleavage led to two products with the fluorescer and quencher on each side of the probe that emitted fluorescence, when separated. In the case of a mismatch or incomplete hybridization of the RNA part of the probes and amplified products, RNase H does not affect the probe. The mutant probe was labeled with FAM, while the wild-type probe was labeled with ROX. Because the two probes were mixed and reacted simultaneously, the assay was monitored using the signal of the wild-type probe as an internal control.
Results
The P132L Caveolin-1 Mutation Was Not Detected by Direct Sequencing
We first examined the presence of caveolin-1 mutations using 109 breast cancer tissues. However, RT-PCR was unsuccessful in ten of the samples, and no products were amplified due to poor nucleic acid integrity. Adequate sequencing signals were obtained from the other 99 samples. No mutations were detected in any of the samples. Because the expression of caveolin-1 transcripts is decreased in cancer cells, we confirmed that sufficient PCR products were obtained in the individual reactions. In this series, 74 were positive for ER (Table 1). These results are contradictory to the previous studies.10,12 Two explanations for this difference are possible: 1) expression of mutated caveolin-1 might be reduced in cancer tissues, and an RNA-based assay was used; and 2) the assay used is less sensitive and might not detect low levels of the transcript.
Table 1.
Characteristics of the Breast Cancers in This Study
| Series characteristics | Consecutive series (n = 99) |
Independent consecutive series |
|
|---|---|---|---|
| ER status | Positive and negative | Positive alone | Positive alone |
| Number of samples | n = 59 | n = 40 | n = 40 |
| RT-PCR direct sequencing | Yes | Yes | |
| DNA-based direct sequencing | Yes | ||
| Cycleave PCR | Yes | Yes | |
| Age, year | |||
| Median | 55 | 56 | 56 |
| Range | 28–82 | 33–76 | 26–78 |
| Tumor size | |||
| pTis | 2 | 0 | 6 |
| pT1 (<2 cm) | 28 | 24 | 23 |
| pT2 (2–5 cm) | 21 | 9 | 8 |
| pT3/4 (>5 cm) | 8 | 7 | 3 |
| Nodal status | |||
| pN0 | 23 | 17 | 26 |
| pN1–3 | 35 | 23 | 14 |
| Estrogen receptor status | |||
| Positive | 34 | 40 | 40 |
| Negative | 25 | 0* | 0* |
| Progesterone receptor status | |||
| Positive | 26 | 29 | 30 |
| Negative | 33 | 11 | 10 |
| HER2 status | |||
| Positive | 40 | 34 | 3 |
| Negative | 19 | 6 | 37 |
| Histological grade | |||
| Low-grade | 9 | 10 | 12 |
| Intermediate | 27 | 21 | 19 |
| High-grade | 23 | 9 | 9 |
| Histology | |||
| DCIS | 2 | 0 | 6 |
| IDC | 55 | 35 | 30 |
| ILC | 0 | 4 | 0 |
| Special types | 2 | 1 | 4 |
| p53 status | |||
| Wild type | 31 | 26 | 0 |
| Mutated | 17 | 7 | 0 |
| N.A. | 11 | 7 | 40 |
| E-cadherin status | |||
| Wild type | 53 | 31 | 0 |
| Mutated | 1* | 1* | 0 |
| N.D. | 6 | 8 | 40 |
DCIS indicates duct carcinoma in situ; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; N.A., not done.
CDH1 mutation was detected in an invasive lobular carcinoma and an invasive ductal carcinoma with lobular carcinoma features.
Development of a New Technique to Detect the P132L Caveolin-1 Mutation
To investigate these two possibilities, a sensitive method that is applicable to paraffin-embedded tissues was developed. We previously reported the use of a Cycleave PCR method to detect the EGFR L858R point mutation.20,21 This method is very sensitive, and a tumor DNA prevalence of just 5% among normal tissue DNA copies is sufficient to detect the mutation. Furthermore, only a single unstained paraffin-block section of a small biopsy specimen is required for the assay. Thus, we applied this method to the caveolin-1 P132L mutation assay. As shown in Figure 2, this method can simultaneously detect wild-type and mutant signals, which can be monitored as an internal control. When we tested the mutant samples that were serially diluted with wild-type DNA, a mutant DNA prevalence of just 0.1% among wild-type DNA copies was sufficient for detection of the P132L mutation (Figure 2). In addition, the cycling threshold was linearly correlated with the log concentration of the mutant allele.
Figure 2.
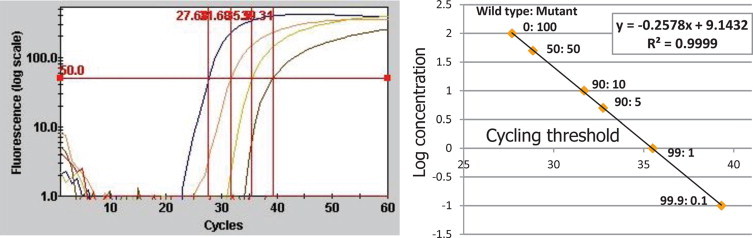
Sensitivity assay using artificial mutant DNA serially diluted with wild-type DNA. Left: Fluorescence-cycle curve. The individual lines represent dilutions of 100%, 10%, 1%, and 0.1% of mutant DNA, respectively. Right: Relationship between the log concentration and the cycling threshold. These are significantly correlated (r2 = 0.9999), suggesting that this assay is quantitative.
Screening of ER-Positive Breast Cancers with the Sensitive Cycleave PCR Method
Because the Cycleave PCR method had sufficient sensitivity, we applied this method to 80 ER-positive breast cancer tissues. Even after using DNA instead of RNA as a template, no P132L mutations were detected by this method (Tables 1 and 2). Because half of the ER-positive breast cancer tissues had also been shown to lack this mutation by RT-PCR direct sequencing, these results were not likely to be affected by the template source of the RNA or DNA. Furthermore, DNA-based direct sequencing was also performed on the 80 ER-positive breast cancers (Table 1), and the caveolin-1 mutation was not detected.
Table 2.
Caveolin-1 Mutation in Breast Cancer and Various Other Cancer Types
| Cancer type | n | Caveolin-1 mutation |
|---|---|---|
| RT-PCR direct sequencing | ||
| Breast cancer | 99 | 0 |
| DNA-based direct sequencing | ||
| Breast cancer | 40 | 0 |
| Cycleave PCR assay | ||
| Breast cancer (ER-positive) | 80 | 0 |
| Colorectal cancer | 77 | 0 |
| Gastric cancer | 48 | 0 |
| Lung cancer | 100 | 0 |
| Ovarian cancer | 32 | 0 |
| Pancreatic cancer | 13 | 0 |
Lack of Caveolin-1 P132L Mutation in Various Cancer Types
We next determined the presence or absence of the P132L mutation in a total of 270 cancers from various tissues using the Cycleave PCR method. Most of the samples (252) were adenocarcinomas, while 15 were squamous cell carcinomas, and three were neuroendocrine carcinomas of the lung. None of the samples had the caveolin-1 P132L mutation (Table 2).
Discussion
Caveolin-1 functions as a tumor suppressor in vitro and is down-regulated in sarcomas and some adenocarcinomas.4 However, many primary cancers have shown high caveolin-1 expression,22 which is associated with a negative prognostic factor for the overall and/or disease-free survival in patients with cancers of the esophagus, oral cavity, pancreas, kidney, prostate, and lung. Even in breast cancers, increased expression of caveolin-1 has been reported in some articles.23,24,25,26 Recently, Savage et al reported that 9.4% of 245 invasive breast cancers expressed caveolin-1, which was associated, in part, with gene amplification.27 Notably, most of the breast cancers with high caveolin-1 expression were categorized into the “basal-like” subtype.24,25,27,28 Because the caveolin-1 P132L mutation is restricted to ER-positive breast cancers,12 the difference in the roles that caveolin-1 plays in the molecular pathogenesis of cancer might be explained by the different cancer subtypes. Recent advances in genome-wide analysis have revealed that ER-positive breast cancers are biologically different from “basal-like” breast cancers, in terms of molecular pathogenesis.29,30,31,32,33 Thus, different roles of caveolin-1 in the pathogenesis of ER-positive and “basal-like” breast cancers might explain the discrepancy.
Here, we addressed the presence of the caveolin-1 P132L mutation in various cancers, which has not yet been reported in detail. Although some genetic alterations, such as the EGFR mutation, theTMPRSS2/ERG fusion, and the EML4/ALK fusion, are cancer-type specific, most of the cancer-associated gene mutations were detected in various cancers with different frequencies. Altered expression of caveolin-1 was reported in various cancers, as mentioned above. Therefore, it was expected that some cancers would also harbor this mutation. However, the P132L mutation was not detected in 77 colorectal cancers, 48 gastric cancers, 100 lung cancers, 32 ovarian cancers, or 13 pancreatic cancers.
In summary, we examined a total of 139 breast cancers. The caveolin-1 P132L mutation was not detected, even when using the sensitive Cycleave PCR method. Furthermore, this mutation was not detected in 270 samples from various cancer types. These results raise doubts about the presence of the caveolin-1 P132L mutation in breast cancers. Further studies on this discrepancy are warranted.
Acknowledgements
We thank Noriko Shibata, Motoko Nimura, and Akiko Yoshinari for excellent technical assistance with the molecular genetic experiments.
Footnotes
See Related Commentary on page 562
Supported in part by a Grant-in-Aid (B-21390110) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a research grant from the Princess Takamatsu Cancer Research Fund (08-24018).
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.Shatz M, Liscovitch M. Caveolin-1: a tumor-promoting role in human cancer. Int J Radiat Biol. 2008;84:177–189. doi: 10.1080/09553000701745293. [DOI] [PubMed] [Google Scholar]
- 2.Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. J Biol Chem. 2002;277:41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 3.Engelman JA, Zhang X, Galbiati F, Volonte D, Sotgia F, Pestell RG, Minetti C, Scherer PE, Okamoto T, Lisanti MP. Molecular genetics of the caveolin gene family: implications for human cancers, diabetes. Alzheimer disease, and muscular dystrophy. Am J Hum Genet. 1998;63:1578–1587. doi: 10.1086/302172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 5.Sotgia F, Rui H, Bonuccelli G, Mercier I, Pestell RG, Lisanti MP. Caveolin-1, mammary stem cells, and estrogen-dependent breast cancers. Cancer Res. 2006;66:10647–10651. doi: 10.1158/0008-5472.CAN-06-2805. [DOI] [PubMed] [Google Scholar]
- 6.Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene. 2002;21:2365–2375. doi: 10.1038/sj.onc.1205300. [DOI] [PubMed] [Google Scholar]
- 8.Williams TM, Cheung MW, Park DS, Razani B, Cohen AW, Muller WJ, Di Vizio D, Chopra NG, Pestell RG, Lisanti MP. Loss of caveolin-1 gene expression accelerates the development of dysplastic mammary lesions in tumor-prone transgenic mice. Mol Biol Cell. 2003;14:1027–1042. doi: 10.1091/mbc.E02-08-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercier I, Casimiro MC, Zhou J, Wang C, Plymire C, Bryant KG, Daumer KM, Sotgia F, Bonuccelli G, Witkiewicz AK, Lin J, Tran TH, Milliman J, Frank PG, Jasmin JF, Rui H, Pestell RG, Lisanti MP. Genetic ablation of caveolin-1 drives estrogen-hypersensitivity and the development of DCIS-like mammary lesions. Am J Pathol. 2009;174:1172–1190. doi: 10.2353/ajpath.2009.080882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- 11.Lee H, Park DS, Razani B, Russell RG, Pestell RG, Lisanti MP. Caveolin-1 mutations (P132L and null) and the pathogenesis of breast cancer: caveolin-1 (P132L) behaves in a dominant-negative manner and caveolin-1 (-/-) null mice show mammary epithelial cell hyperplasia. Am J Pathol. 2002;161:1357–1369. doi: 10.1016/S0002-9440(10)64412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Sotgia F, Vuolo MA, Li M, Yang WC, Pestell RG, Sparano JA, Lisanti MP. Caveolin-1 mutations in human breast cancer: functional association with estrogen receptor alpha-positive status. Am J Pathol. 2006;168:1998–2013. doi: 10.2353/ajpath.2006.051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ST, Lin SY, Yeh KT, Kuo SJ, Chan WL, Chu YP, Chang JG. Mutational, epigenetic and expressional analyses of caveolin-1 gene in breast cancers. Int J Mol Med. 2004;14:577–582. [PubMed] [Google Scholar]
- 14.Tavassoli FA, Devilee P. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. IARC; Lyon: 2003. [Google Scholar]
- 15.Sasaki E, Tsunoda N, Hatanaka Y, Mori N, Iwata H, Yatabe Y. Breast-specific expression of MGB1/mammaglobin: an examination of 480 tumors from various organs and clinicopathological analysis of MGB1-positive breast cancers. Mod Pathol. 2007;20:208–214. doi: 10.1038/modpathol.3800731. [DOI] [PubMed] [Google Scholar]
- 16.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 17.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, Mitsudomi T, Takahashi T. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–1688. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 19.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 20.Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T. A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn. 2006;8:335–341. doi: 10.2353/jmoldx.2006.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatabe Y, Mitsudomi T. Epidermal growth factor receptor mutations in lung cancers. Pathol Int. 2007;57:233–244. doi: 10.1111/j.1440-1827.2007.02098.x. [DOI] [PubMed] [Google Scholar]
- 22.Burgermeister E, Liscovitch M, Rocken C, Schmid RM, Ebert MP. Caveats of caveolin-1 in cancer progression. Cancer Lett. 2008;268:187–201. doi: 10.1016/j.canlet.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 23.Yang G, Truong LD, Timme TL, Ren C, Wheeler TM, Park SH, Nasu Y, Bangma CH, Kattan MW, Scardino PT, Thompson TC. Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res. 1998;4:1873–1880. [PubMed] [Google Scholar]
- 24.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia S, Dales JP, Charafe-Jauffret E, Carpentier-Meunier S, Andrac-Meyer L, Jacquemier J, Andonian C, Lavaut MN, Allasia C, Bonnier P, Charpin C. Poor prognosis in breast carcinomas correlates with increased expression of targetable CD146 and c-Met and with proteomic basal-like phenotype. Hum Pathol. 2007;38:830–841. doi: 10.1016/j.humpath.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Liedtke C, Kersting C, Burger H, Kiesel L, Wulfing P. Caveolin-1 expression in benign and malignant lesions of the breast. World J Surg Oncol. 2007;5:110–119. doi: 10.1186/1477-7819-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savage K, Lambros MB, Robertson D, Jones RL, Jones C, Mackay A, James M, Hornick JL, Pereira EM, Milanezi F, Fletcher CD, Schmitt FC, Ashworth A, Reis-Filho JS. Caveolin 1 is overexpressed and amplified in a subset of basal-like and metaplastic breast carcinomas: a morphologic, ultrastructural, immunohistochemical, and in situ hybridization analysis. Clin Cancer Res. 2007;13:90–101. doi: 10.1158/1078-0432.CCR-06-1371. [DOI] [PubMed] [Google Scholar]
- 28.Elsheikh SE, Green AR, Rakha EA, Samaka RM, Ammar AA, Powe D, Reis-Filho JS, Ellis IO. Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer. 2008;99:327–334. doi: 10.1038/sj.bjc.6604463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 30.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 31.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 32.Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C, Vatcheva R, Rayter S, Mahler-Araujo B, Fulford LG, Hungermann D, Mackay A, Grigoriadis A, Fenwick K, Tamber N, Hardisson D, Tutt A, Palacios J, Lord CJ, Buerger H, Ashworth A, Reis-Filho JS. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 33.Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DS, Jones C, Lord CJ, Vatcheva R, Rodriguez-Pinilla SM, Palacios J, Ashworth A, Reis-Filho JS. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2010;121:575–589. doi: 10.1007/s10549-009-0501-3. [DOI] [PubMed] [Google Scholar]


