Abstract
The phospholipid fatty acid biomarkers 18:1ω9, 18:2ω6,9 and 18:3ω3,6,9 are commonly used as fungal biomarkers in soils. They have, however, also been found to occur in plant tissues, such as roots. Thus, the use of these PLFAs as fungal biomarkers in sieved soil, which may still contain small remains of roots, has been questioned. We used data from a recent beech tree girdling experiment to calculate the contribution of roots to these biomarkers and were able to demonstrate that not more than 0.61% of 18:1ω9 and 18:2ω6,9 in sieved soil samples originated from roots (but 4% of 18:3ω3,6,9). Additionally, the abundance of the biomarker 18:2ω6,9 in the soil was found to be highly correlated to ectomycorrhizal root colonization, which further corroborates its fungal origin. PLFA biomarkers were substantially reduced in vital roots from girdled trees compared to roots of control trees (by up to 76%), indicating that the major part of PLFAs measured in roots may actually originate from ectomycorrhizal fungi growing inside the roots. We calculated, that even a near to 50% reduction in fine root biomass – as observed in the girdling treatment – accounted for only 0.8% of the measured decrease of 18:2ω6,9. Our results demonstrate that both 18:1ω9 and 18:2ω6,9 are suitable biomarkers for detecting fungal dynamics in soils and that especially 18:2ω6,9 is a reliable biomarker to study mycorrhizal dynamics in beech forests.
Keywords: PLFAs; Fungal biomarkers; 18:2ω6,9; 18:1ω9; Soil microbial community; Ectomycorrhizal fungi; Girdling; Beech roots
Phospholipid fatty acid (PLFA) analysis is widely used to quantitatively assess microbial community composition of soils. One of the advantages of this approach is that it covers a wide range of microbial groups, including both bacteria and fungi. The PLFAs 18:2ω6,9, 18:1ω9 and to a lesser extent also 18:3ω3,6,9 are commonly used as fungal biomarkers in soil microbial community studies (e.g. Hill et al., 2000; Högberg, 2006; Joergensen and Wichern, 2008; Leckie, 2005) but have also been found to occur in plant tissues (Laczko et al., 2004; Zelles, 1997). Soil samples used for PLFA analysis are usually sieved and/or hand-picked to remove visible roots prior to analysis. Although this pre-treatment certainly eliminates the majority of all roots, remains of fine roots in the soil samples can not be ruled out. It has therefore been argued that analysis of fungal PLFA biomarkers in soils may be biased by possible contribution of plant remains to soil samples (Joergensen and Wichern, 2008; Leckie, 2005; Olsson, 1999). This possibility of contamination with root-borne PLFAs may thus reduce the reliability of results from studies that use PLFAs as fungal biomarkers in soil. Despite this limitation, there has been, to our knowledge, no attempt to quantify the possible contamination of plant roots to PLFAs measured from soil samples so far.
In a large-scale girdling experiment, we found a strong decrease (about 50% on average between 2 to 20 months after girdling) of the biomarker 18:2ω6,9 compared to untreated controls (Kaiser et al., in press). A similar effect has also been found before by other girdling studies (Högberg et al., 2007). The obvious interpretation that the reduction of this biomarker after girdling reflects the decline of ectomycorrhizal fungi has recently been supported by DNA profiling of the soil microbial community (Yarwood et al., 2009). However, since tree girdling reduces fine root biomass in soil, some part of this reduction could also have been caused by decreasing fine root biomass if the methodology used reflects to some extent also PLFAs in roots. To address this problem we measured the PLFA content of fine roots of beech from our experiments and calculated the contribution of fine roots to the total 18:2ω6,9, 18:1ω9 and 18:3ω3,6,9 biomarkers in our soil samples.
1. PLFA measurements of forest soil and beech root samples from a girdling experiment
Soil and root samples were taken from a tree girdling experiment in an approximately 65 year old beech forest (Fagus sylvatica) near the city of Vienna (for details see: Kaiser et al., in press). The soil was a dystric cambisol (over flysh, pH 4.8) and samples were taken from the upper mineral soil (A horizon). Girdling had taken place 14.5 months before the sampling (May 9th, 2006) in three 400 m2 girdling areas. Two 5 × 5 m sampling plots were installed in the central part of each girdling area; two control plots were installed in the vicinity of each girdling area (i.e., six girdling and six control plots in total). Five soil cores (14.5 cm height, 7 cm diameter) were taken randomly from each of the girdling and control plots and pooled to give one replicate sample. Fine roots (diameter < 1 mm) were thoroughly separated from soil and from coarse roots, washed carefully and weighed. Soil samples were sieved to 2 mm and freed from all visible roots by hand picking (n = 6). Ectomycorrhizal root colonisation was measured from aliquots of the pooled fine root samples by counting ectomycorrhizal root tips in petri-dishes at 30 x magnification (n = 6). For PLFA analysis, roots were pooled to three final samples from girdling and control plots, respectively (n = 3). Fine root samples were kept frozen (−20 °C) until PLFA extraction and crushed in liquid N2 with a mortar and pestle immediately before extraction. Soil samples were processed fresh (within 24 hours of sampling). PLFAs were extracted in soil samples and in fine roots using a modified procedure as described by Frostegård and co-workers (1991). Phospholipids were converted to fatty acid methyl esters (FAME) by alkaline methanolysis. FAMEs were analyzed by gas chromatography on a DB23 column (J&W 60 m × 0.25 mm, 0.25 μm). Mixtures of bacterial FAMEs (Bacterial acid methyl ester mix, Supelco, and 37 Comp. FAME Mix, Supelco) were used as qualitative standards. Concentrations of single FAMEs were calculated using the internal standard (19:0) peak as a reference.
2. PLFA fungal biomarkers found in beech roots are predominantly of mycorrhizal origin
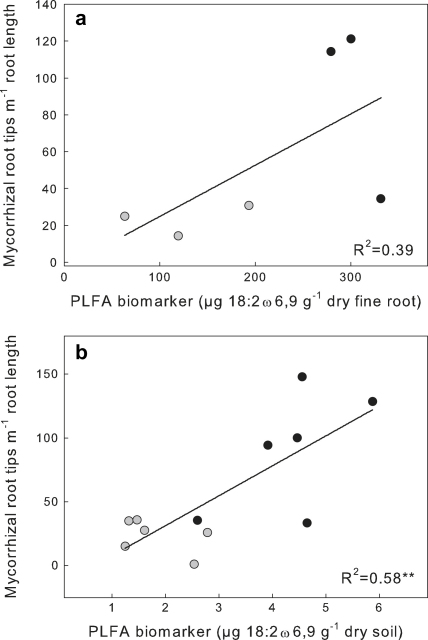
Fine roots of beech contained all of the three PLFAs 18:2ω6,9, 18:1ω9 and 18:3ω3,6,9 commonly used as fungal biomarkers (Högberg et al., 2007; Joergensen and Wichern, 2008): 18:2ω6,9 showed the highest concentration of all biomarkers, followed by 18:3ω3,6,9 and 18:1ω9 (303, 130 and 59 μg g−1 dry fine roots, respectively; Table 1). Interestingly, these biomarkers were substantially reduced in vital roots from girdled trees by as much as 56 to 76%. Since we have no reason to believe that girdling alters the PLFA composition or concentration of the membranes of vital roots, the reduced concentration of these biomarkers in roots of girdled trees most likely reflects the decrease of ectomycorrhizal fungi growing inside the roots. This is supported by a weak correlation of the concentration of these biomarkers in fine roots with the ectomycorrhizal root colonization (R2 = 0.39, p = 0.185, Fig. 1a) and suggests that the predominant part of the PLFAs measured in root tissue may in fact originate from colonization with ectomycorrhizal fungi.
Table 1.
Dominant PLFA biomarkers in fine roots of Fagus sylvatica and soils of girdled and control plots 14.5 months after girdling and estimation of the contribution of root-borne PLFAs to PLFAs measured in sieved soil samples. Values are means (1 SE). Roots, n = 3; soil, n = 6.
| Control | Girdling | Difference G − C (%) | |
|---|---|---|---|
| PLFA concentration in fine roots (μg g−1 dry matter) | |||
| 18:1ω9 | 59.02 (4.31) | 25.72 (5.80) | −56.4 |
| 18:2ω6,9 | 303.41 (15.18) | 125.25 (37.62) | −58.7 |
| 18:3ω3,6,9 | 130.24 (5.45) | 30.69 (12.23) | −76.4 |
| 18:0 | 30.09 (6.80) | 17.47 (2.51) | −41.9 |
| 20:0 | 7.64 (2.75) | 2.99 (1.52) | −60.8 |
| 16:0 | 145.98 (19.39) | 75.56 (6.96) | −48.2 |
| 17:0 | 6.05 (0.76) | 3.40 (1.81) | −43.8 |
| PLFA concentration in soils (μg g−1 dry soil)a | |||
| 18:1ω9 | 7.46 (1.45) | 7.15 (1.05) | −4.1 |
| 18:2ω6,9 | 4.34 (0.44) | 1.82 (0.27) | −58.0 |
| 18:3ω3,6,9 | 0.28 (0.05) | 0.12 (0.05) | −56.6 |
| 18:0 | 1.82 (0.38) | 1.98 (0.31) | +9.1 |
| 20:0 | 0.56 (0.22) | 0.81 (0.05) | +45.3 |
| 16:0 | 7.79 (1.79) | 6.24 (1.42) | −19.9 |
| 17:0 | 0.51 (0.06) | 0.44 (0.07) | −12.5 |
| Control |
Girdling |
Contribution of fine root biomass loss to the observed decrease in soil PLFAs (%)d | |||
|---|---|---|---|---|---|
| Root-borne PLFA (ng g−1 dry soil) | Root-borne PLFAs (%)c | Root-borne PLFA (ng g−1 dry soil) | Root-borne PLFAs (%)c | ||
| Root-borne PLFAs in soil samplesb | |||||
| 18:1ω9 | 4.97 (0.43) | 0.07 (0.01) | 1.18 (0.62) | 0.02 (0.01) | 1.25 |
| 18:2ω6,9 | 25.51 (1.68) | 0.61 (0.10) | 5.65 (3.35) | 0.31 (0.16) | 0.79 |
| 18:3ω3,6,9 | 10.95 (0.63) | 4.05 (0.55) | 1.48 (0.97) | 1.22 (0.88) | 6.01 |
| 18:0 | 2.53 (0.58) | 0.15 (0.05) | 0.90 (0.44) | 0.05 (0.03) | n.a. |
| 20:0 | 0.64 (0.23) | 0.14 (0.06) | 0.11 (0.10) | 0.01 (0.01) | n.a. |
| 16:0 | 12.30 (1.83) | 0.18 (0.05) | 3.38 (1.54) | 0.07 (0.04) | 0.57 |
| 17:0 | 0.51 (0.72) | 0.11 (0.03) | 0.13 (0.12) | 0.03 (0.02) | 0.59 |
Soil was sieved and visible roots were hand-picked prior to analysis.
Calculated contribution of root-borne PLFAs to total PLFAs measured in sieved soil samples. Calculations were based on the observed PLFA-concentrations in fine roots and on fine root biomass measurements for each plot (control plots: 1.68 ± 0.09 mg g−1 soil, girdling plots: 0.914 ± 0.27 mg g−1 soil) under the assumption that 5% of roots remained in soil samples after sieving and hand-picking (PLFA concentration in fine roots x total fine root biomass per g soil × 0.05).
Estimated contribution of root-borne PLFAs to total PLFAs measured in sieved soil samples in percent (root-borne PLFAs per g soil/total PLFAs per g soil × 100).
Estimated contribution of the root biomass decrease (∼45%) to the total observed decrease of soil-borne PLFAs in girdled plots compared to controls (difference between root-borne PLFAs per g soil in control and in girdled plots/difference between total PLFAs per g soil in control and in girdled plots × 100).
Fig. 1.
Correlations between ectomycorrhizal root tips and the amount of the PLFA biomarker 18:2ω6,9 found in beech fine roots (a) and bulk soil (b). In (a) each point represents roots from two plots (comprising five subsamples each) which were pooled to give three samples from girdled and three samples from control plots. In (b) each point represents one plot (five pooled subsamples). Black circles, control plots; grey circles, girdling plots. Significance of regressions: (a) p = 0.185 (not significant), (b) p = 0.0038 (**).
3. The contribution of roots to 18:2ω6,9 and 18:1ω9 found in soil samples is negligible
Our data allowed us to calculate the possible contribution of 18:2ω6,9, 18:1ω9 and 18:3ω3,6,9 from root tissue to soil samples. Control plots contained on average 1.68 mg fine roots g−1 soil (±0.09 mg g−1) in the upper 14.5 cm (Kaiser et al., in press). Based on a 18:2ω6,9 concentration of 303 μg g−1 fine roots, the amount of this biomarker in soil originating from fine roots would be 0.51 μg g−1 soil if all roots were present in the soil. However, since our soil samples were sieved to 2 mm and freed from all visible roots by hand picking prior to analysis, we assume that we removed at least 95% of all roots by this treatment. Thus, the amount of root-borne 18:2ω6,9 in our soil samples (contaminated with 5% of the original fine root biomass) would be 0.025 μg g−1 soil, or only 0.6% of the amount of 18:2ω6,9 measured (Table 1). The contribution of roots to the other two biomarkers would also be small, but of different magnitudes (0.07% and 4% for 18:1ω9 and 18:3ω3,6,9, respectively, Table 1). These differences are caused by the difference of the ratios at which these biomarkers occur in roots (where the ratio of 18:1ω9 to 18:2ω6,9 to 18:3ω3,6,9 is equal to 1:5:2) and soils (27:15:1, respectively). The differences of these ratios on their own, already suggest that there is no significant contribution of root-borne PLFAs to sieved soil samples, which would have required a higher similarity in the relative abundances of these markers in roots and soil.
4. Effect of a reduction in fine root biomass on fungal biomarkers found in soil
The observed decrease of fine root biomass by 45% in girdled plots at the time we took these samples would translate into a decrease of the root-borne 18:2ω6,9 biomarker in sieved soil samples by 0.020 μg g−1 soil (i.e. from 0.0255 in girdled to 0.0056 μg g−1 soil in control plots, Table 1). However, both, amount and changes of the fungal biomarker measured in our soil samples were by levels of magnitudes higher than that, i.e. 18:2ω6,9 decreased from 4.34 μg g−1 soil in control plots to 1.82 μg g−1 soil in girdled plots. Thus, a variation of 0.02 μg g−1 soil caused by the loss of fine root biomass in girdled plots would account for less than 0.8% of the observed change of 18:2ω6,9 in soil. The influence would be higher for 18:1ω9 and 18:3ω3,6,9 (1.25% and 6.0%, respectively), due to less change of these biomarkers in response to girdling.
In conclusion our results show that, although the concentrations of 18:2ω6,9 and 18:1ω9 in beech roots are high, their contribution to the total PLFA content of a sieved beech forest soil is negligible, mainly due to the high abundance of these biomarkers in the soil. If there would be a significant contribution of roots to total soil PLFAs we would expect similar ratios of these biomarkers in soils and roots, which clearly is not the case. Rather, we found a significant correlation between fungal PLFA biomarkers in the soil and ectomycorrhizal colonization of root tips, suggesting the fungal origin of these biomarkers in the soil. (R2 = 0.58, p < 0.005 Fig. 1b). PLFAs with relatively low concentrations in soils, but high concentrations in roots, such as 18:3ω3,6,9 may, however, still be biased to some extent by root-borne PLFAs. We conclude that the PLFA biomarkers 18:2ω6,9 and 18:1ω9 are well suited for reliably capturing fungal dynamics in sieved forest soils.
Acknowledgements
We thank Peter Schweiger for determination of ectomycorrhizal colonization of roots and Jörg Schnecker for help in fine root biomass determination. This work was supported by the Austrian Science Fund (FWF, Project numbers P18495-B03 and S1001-B07).
References
- Frostegård A., Tunlid A., Bååth E. Microbial biomass measured as total lipid phosphate in soils of different organic content. Journal of Microbiological Methods. 1991;14:151–163. [Google Scholar]
- Hill G.T., Mitkowski N.A., Aldrich-Wolfe L., Emele L.R., Jurkonie D.D., Ficke A., Maldonado-Ramirez S., Lynch S.T., Nelson E.B. Methods for assessing the composition and diversity of soil microbial communities. Applied Soil Ecology. 2000;15:25–36. [Google Scholar]
- Högberg M.N. Discrepancies between ergosterol and the phospholipid fatty acid 18: 2 <omega> 6,9 as biomarkers for fungi in boreal forest soils. Soil Biology & Biochemistry. 2006;38:3431–3435. [Google Scholar]
- Högberg M.N., Högberg P., Myrold D.D. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia. 2007;150:590–601. doi: 10.1007/s00442-006-0562-5. [DOI] [PubMed] [Google Scholar]
- Joergensen R.G., Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biology & Biochemistry. 2008;40:2977–2991. [Google Scholar]
- Kaiser, C., Koranda, M., Kitzler, B., Fuchslueger, L., Schnecker, J., Schweiger, P., Rasche, F., Zechmeister-Boltenstern, S., Sessitsch, A., Richter, A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytologist, in press., doi:10.1111/j.1469-8137.2010.03321.x [DOI] [PMC free article] [PubMed]
- Laczko E., Boller T., Wiemken V. Lipids in roots of Pinus sylvestris seedlings and in mycelia of Pisolithus tinctorius during ectomycorrhiza formation: changes in fatty acid and sterol composition in a beech forest soil. Plant Cell and Environment. 2004;27:27–40. [Google Scholar]
- Leckie S.E. Methods of microbial community profiling and their application to forest soils. Forest Ecology and Management. 2005;220:88–106. [Google Scholar]
- Olsson P.A. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiology Ecology. 1999;29:303–310. [Google Scholar]
- Yarwood S.A., Myrold D.D., Högberg M.N. Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiology Ecology. 2009;70:151–162. doi: 10.1111/j.1574-6941.2009.00733.x. [DOI] [PubMed] [Google Scholar]
- Zelles L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 1997;35:275–294. doi: 10.1016/s0045-6535(97)00155-0. [DOI] [PubMed] [Google Scholar]