Abstract
AIM: To evaluate the diagnostic value of cancer-testis antigen (CTA) mRNA in peripheral blood samples from hepatocellular carcinoma (HCC) patients.
METHODS: Peripheral blood samples were taken from 90 patients with HCC before operation. Expression of melanoma antigen-1 (MAGE-1), synovial sarcoma X breakpoint-1 (SSX-1), and cancer-testis-associated protein of 11 kDa (CTp11) mRNA in peripheral blood mononuclear cells (PBMC) was tested by nested reverse transcripts-polymerase chain reaction (RT-PCR). Serum α-fetoprotein (AFP) in these patients was also determined.
RESULTS: The positive rate of MAGE-1, SSX-1 and CTp11 transcripts was 37.7%, 34.4%, 31.1% in PBMC samples, and 74.4%, 73.3%, 62.2% in their resected tumor samples, respectively. The positive rate for at least one of the transcripts of three CTA genes was 66.7% in PBMC samples and 91.1% in their resected tumor samples. MAGE-1, SSX-1 and/or CTp11 mRNA were not detected in the PBMC of those patients from whom the resected tumor samples were MAGE-1, SSX-1 and/or CTp11 mRNA negative, nor in the PBMC samples from 20 healthy donors and 10 cirrhotic patients. Among the 90 patients, the serum AFP in 44 patients met the general diagnostic standard (AFP > 400 μg/L) for HCC, and was negative (AFP ≤ 20 μg/L) or positive with a low concentration (20 μg/L < AFP ≤ 400 μg/L) in the other patients. The positive rate for at least one of the transcripts of three CTA genes in PBMC samples from the AFP negative or positive patients with a low concentration was 69.2% and 45.0%, respectively. Of the 90 patients, 71 (78.9%) were diagnosed as HCC by nested RT-PCR and serum AFP. Although the positive rate for at least one of the transcripts of three CTA genes in PBMC samples from 53 patients at TNM stage III or IV was obviously higher than that in PBMC samples from 37 patients at stage I or II (77.9% vs 51.4%, P = 0.010), the CTA mRNA was detected in 41.7% and 56.0% of PBMC samples from HCC patients at stages I and II, respectively.
CONCLUSION: Detecting MAGE-1, SSX-1 and CTp11 mRNA in PBMC improves the total diagnostic rate of HCC.
Keywords: Hepatocellular carcinoma, α-fetoprotein, Cancer-testis antigen, Diagnosis, Nested reverse transcripts-polymerase chain reaction
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common and lethal cancers in the world[1-3]. More than one million cases of HCC occur in the world each year[4]. Although many treatment modalities for HCC are available (including hepatic resection, liver transplantation, radiofrequency ablation, transarterial chemoembolization, etc.) at present, the prognosis of HCC patients remains dismal because it is detected at an advanced, non-resectable stage. Early diagnosis of HCC can improve the prognosis of HCC patients. So far, α-fetoprotein (AFP) is the generally accepted serological marker. Serum AFP alone contributes to the diagnosis of HCC if its level is markedly elevated (over 400 μg/L as a threshold value), which occurs in less than 50% of cases at the time of diagnosis[5-8]. Moreover, serum AFP level is negative or slightly elevated in 20%-40% patients, which can significantly reduce the sensitivity of an assay based on over-expression of AFP. The serum AFP level in patients with acute or chronic hepatitis or liver cirrhosis but without malignant disease is often elevated. Since detection of serum AFP level in blood samples appears to be nonspecific[7-10]. Therefore, the diagnostic sensitivity and specificity of AFP are unsatisfactory and questionable. It is thus necessary to select other specific methods for the diagnosis of HCC.
Transcripts of tumor-specific genes can be amplified and detected by reverse transcripts-polymerase chain reaction (RT-PCR), which is a reliable technique to detect circulating tumor cells (CTC). In 1991, Smith et al[11] first successfully adopted RT-PCR technique to assess tyrosinase messenger RNA (mRNA) as a tumor marker in detecting circulating melanoma cells. Since then, this technique has been applied to the detection of CTC in solid tumors[12-14]. Melanoma antigen-1 (MAGE-1)[15], synovial sarcoma X breakpoint-1 (SSX-1)[16] and cancer-testis-associated protein of 11 kDa (CTp11)[17] antigens have been designated as cancer-testis antigens (CTA). It has been reported that MAGE-1 and SSX-1 mRNA are expressed with a high percentage and specificity in HCC[16,18-20]. Our group has verified a relatively high and specific expression of CTp11 mRNA in HCC tissues but not in the corresponding adjacent non-HCC and cirrhosis tissues[21]. In this study, we evaluated the diagnostic significance of a highly sensitive nested RT-PCR assay for the MAGE-1, SSX-1 and CTp11mRNA in peripheral blood of HCC patients.
MATERIALS AND METHODS
Cell lines
Human HCC cell lines BEL7405 expressing MAGE-1 mRNA and LM3 expressing both SSX-1 and CTp11 mRNA, purchased from the Cell Bank, Chinese Academy of Sciences, and Liver Cancer Institute, Zhongshan Hospital, Fudan University, respectively, were grown in RPMI1640 medium with 10% fetal calf serum and served as a positive control of the assay used in this study.
Patients and tissue samples
Ninety consecutive patients (79 men and 11 women) with a mean age of 45.6 ± 2.7 years (range 18-79 years) undergoing operation for HCC, including hepatectomy (48 cases) or orthotopic liver transplantation (42 cases), at the 2nd Hospital of Peking University Health Science Centre, were enrolled in this study. Of the 83 patients with virus infection, 79 were infected with hepatitis B virus, 2 with hepatitis C virus, and 2 with both hepatitis B and C viruses. The serum AFP level was negative (≤ 20 μg/L) in 26 patients, positive with a low concentration (20 μg/L < AFP ≤ 400 μg/L) and a high concentration (> 400 μg/L) in 20 and 44 patients, respectively. According to the TNM classification of International Union Against Cancer[22], 12 cases were classified as stage I, 25 as stage II, 9 as stage III, and 44 as stage IV, respectively. HCC and its adjacent non-cancerous tissue samples (the distance to the edge of HCC tissue > 2 cm) were collected during operation. Control samples collected by surgical biopsy included 20 liver tissue samples from cirrhotic patients and 20 normal liver tissue samples from patients without liver disease. Testis tissue (kindly provided by Urological Department of the 2nd Hospital of Peking University Health Science Centre) was used as a positive control. Clinical diagnosis was confirmed by pathological examination. Each sample was immediately frozen in liquid nitrogen and stored at -80°C until extraction of total RNA. Informed consent was obtained from each patient before the study. The study protocol was approved by the Ethic Committee of Peking University.
Blood samples
Whole blood samples were taken from the 90 HCC patients on the day before operation. Control blood samples were collected form 20 healthy volunteers and 10 cirrhotic patients. Ten mL of blood from each patient was collected into a heparinzed tube and peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation[15].
Extraction of total RNA and synthesis of cDNA
Total RNA was extracted from frozen tissue specimens (50-100 mg) and freshly isolated PBMC (1 × 107 cells) using TRIZOL reagent (GIBCOL BRL) according to its manufacturer’s instructions. Total RNA (2.5 μg) was primed with an Oligo (dT)15 oligonucleotide (Promega, USA) and reverse-transcribed with Superscript II (GIBCO BRL, USA) according to their manufacturers’ instructions.
PCR amplification of CTAs
PCR amplification reaction (50 μL) contained 5 μL of cDNA, 1 μL each of 10 μmol/L outer/inner primers, 1 μL of 10 mmol/L dNTP mixture, 2.5 U Taq polymerase (GIBCO BRL, USA) in a buffer solution. Thirty-five cycles of PCR amplification of cDNA from liver tissue were performed with a pre-programmed UNOIIthermocycler (Biometra, German) under the following conditions: an initial denaturation at 94°C for 5 min, a final extension at 72°C for 8 min. The PCR products were 421 base pair (bp) (MAGE-1), 422 bp (SSX-1) and 297 bp (CTp11), respectively. Twenty-five cycles of PCR amplification of cDNA from PBMC were performed with its first round conditions identical to those of cDNA from liver tissue. For the second round of PCR, 1 μL of the first-round PCR products was used as a template in combination with 1 μL each of 10 μmol/L inner primers. After heated for 2 min at 94°C, the samples were subjected to 35 cycles of PCR amplification, followed by a final extension at 72°C for 8 min. The PCR products were 299 bp (MAGE-1), 398 bp (SSX-1) and 188 bp (CTp11), respectively. The PCR conditions and outer/inner primers for MAGE-1[15], SSX-1[16] and CTp11[17] used in this study are shown in Table 1. To verify the integrity of cDNA[18], β2-microglobin (β2-MG) (primers: forward: 5'-CTCGCGCTACTCTCTCTTTCTGG-3' and reverse: 5'-GCTTACATGTCTCGATCCCACTTAA-3', 335 bp) was amplified for 30 cycles (at 94°C, 55°C and 72°C for 45 s). For analysis, 8 μL of reaction products was run in 2% agarose gel (Promega, USA), followed by ethidium bromide staining and digital camera photographing (Korda D3.5, USA).
Table 1.
Primers and conditions used in nested reverse transcripts-polymerase chain reaction
| Primers | Primers from 5' to 3' | PCR conditions denaturation annealing extension |
| MAGE-1 outer primers | Forward primer: 5'-CGGCCGAAGGAACCTGACCCAG-3' | 94°C for 45 s |
| Reverse primer: 5'-GCTGGAACCCTCACTGGGTTGCC-3' | 65°C for 45 s | |
| 72°C for 45 s | ||
| MAGE-1 inner primers | Forward primer: 5'-ACAGAGGAGCACCAAGGAGAAG-3' | 94°C for 45 s |
| Reverse primer: 5'-AGTTGATGGTAGTGGGAAAGGC-3' | 65°C for 45 s | |
| 72°C for 45 s | ||
| SSX-1 outer primers | Forward primer: 5'-CTAAAGCATCAGAGAAGAGAAGC-3' | 94°C for 60 s |
| Reverse primer: 5'-AGATCTCTTATTAATCTTCTCAGAAA-3' | 57°C for 60 s | |
| 72°C for 60 s | ||
| SSX-1 inner primers | Forward primer: 5'-TCAGAGAAGAGAAGCAAGGCCTTT-3' | 94°C for 45 s |
| Reverse primer: 5'-TTCTCAGAAATATTTGCTTTTCC-3' | 56°C for 45 s | |
| 72°C for 45 s | ||
| CTp11 outer primers | Forward primer: 5'-CTGCCCCAGACATTGAAGAA-3' | 94°C for 45 s |
| Reverse primer: 5'-TCCATGAATTCCTCCTCCTC-3' | 57°C for 60 s | |
| 72°C for 90 s | ||
| CTp11 inner primers | Forward primer: 5'-TGTGAATCCAACGAGGTG-3' | 94°C for 45 s |
| Reverse primer: 5'-TTGATTCTGTTCTCTCGGGC-3' | 60°C for 45 s | |
| 72°C for 45 s |
PCR: Polymerase chain reaction; MAGE-1: Melanoma antigen-1; SSX-1: Synovial sarcoma X breakpoint-1; CTp11: Cancer-testis-associated protein of 11 kDa.
Sensitivity of nested RT-PCR technique
The sensitivity of our nested PCR assay was evaluated by performing the procedure on healthy volunteer blood samples mixed with a certain number of hepatoma cells. Ten-fold serial dilution experiments from 104 hepatoma cells were carried out using human hepatoma cell lines BEL7405 expressing MAGE-1 transcript and LM3 expressing both SSX-1 and CTp11 mRNA. Hepatoma cells (1 × 103, 1 × 102, 1 × 101 and 1 × 100) were added to 5 × 106 PBMC from healthy donors, then total RNA was extracted and subjected to RT-PCR amplification with primers for β2-microglobin, MAGE-1, SSX-1 and CTp11 genes.
Sequence analysis of PCR products
PCR amplification-purified cDNA was cloned into the pGEM-T easy vector (Promega) by T4 DNA ligase and amplified in Escherichia coli, JM109. Four positive colonies were selected and assessed using EcoRIdigestion of mini-prepared DNA. Putative MAGE-1, SSX-1 and CTp11 cDNA samples were sequenced with T7 sequencing primers in Sangon Co., Shanghai, China.
Statistical analysis
Statistical analysis was performed by chi-square test and Fisher’s exact test. P < 0.05 was considered statistically significant.
RESULTS
Sensitivity of nested RT-PCR technique
After two rounds of PCR amplification, MAGE-1, SSX-1 and CTp11 transcript genes could be detected in the PCR products, indicating that our assay is able to detect a hepatoma cell in 5 × 106 PBMC (Figure 1).
Figure 1.
Sensitivity of nested polymerase chain reaction assay to tumor-specific markers in hepatocellular carcinoma cell lines. Lanes 1-4: 1 × 103, 1 × 102, 1 × 101 and 1 × 100 hepatoma cells detected by nested reverse transcripts-polymerase chain reaction using melanoma antigen-1 (MAGE-1), synovial sarcoma X breakpoint-1 (SSX-1), cancer-testis-associated protein of 11 kDa (CTp11), and β2-microglobin; Lane 5: Peripheral blood mononuclear cells (PBMC) from healthy donors only; Lane 6: negative control; Lane M: Molecular marker, 100 bp DNA ladder (Gibco).
Expression of CTA genes in HCC tissue samples
Expression of MAGE-1, SSX-1 and CTp11 mRNA was detectabled in 74.4%, 73.3% and 62.2% of the 90 HCC tissue samples, respectively. No expression of these genes was detected in the corresponding adjacent non-HCC tissue samples, or in the normal and cirrhotic liver tissue samples. Eighty-two HCC tissue samples (91.1%) were positive for at least one of the transcripts of three CTA genes.
Nested RT-PCR results in HCC PBMC samples
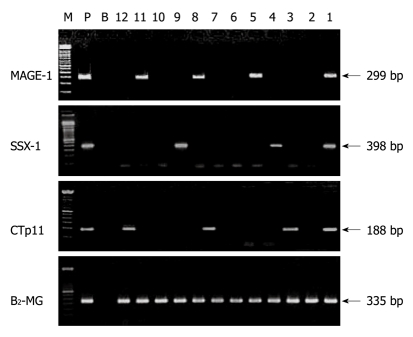
After two rounds of PCR amplification, MAGE-1, SSX-1 and CTp11 were detected in 37.7%, 34.4% and 31.1% of the PBMC samples, respectively. At least one of the three genes was expressed in 66 PBMC samples (66.7%). Of the 82 MAGEE-1, SSX-1 or CTp11mRNA positive HCC tissue samples, at least one of the transcripts of three genes was detected in PBMC from 60 patients with a positive correlation rate of 73.2%. These gene transcripts could not be detected in PBMC from patients with MAGEE-1, SSX-1 or CTp11mRNA undetectable in their liver tissue samples. MAGE-1, SSX-1 or CTp11 gene was not expressed in the 30 control PBMC samples from 20 healthy volunteers and 10 cirrhotic patients. The typical electrophoresis of nested RT-PCR products amplified from cDNA in PBMC samples from HCC patients is shown in Figure 2.
Figure 2.
Electrophoresis of second round polymerase chain reaction products amplified from cDNA of peripheral blood mononuclear cells samples. Lane M: Molecular marker, 100 bp DNA ladder (Gibco); Lane P: Positive control; Lane B: Blank control; lane Β2-MG (335 bp) as cDNA quality control; Lanes 1, 5, 8, 11: Positive melanoma antigen-1 (MAGE-1) transcript; Lanes 1, 4, 9: Positive synovial sarcoma X breakpoint-1 (SSX-1) transcript; Lanes 1, 3, 7, 12: Positive cancer-testis-associated protein of 11 kDa (CTp11) transcript; Lanes 2, 6 and 10: Negative transcript of all three CTA genes.
Sequence analysis of PCR products
Sequence analysis of PCR products verified that the nucleotide sequences of MAGE-1, SSX-1, and CTp11 cDNA fragments were identical to those in GenBank, indicating that the RT-PCR products are MAGE-1, SSX-1, and CTp11 cDNA.
CTA transcripts in PBMC and serum AFP level
The overall positive rate of AFP with a high concentration in serum and MAGE-1, SSX-1 and/or CTp11 mRNA in PBMC samples was 48.9% and 66.7%, respectively. No correlation was observed between positive AFP rate and CTA transcripts. However, the serum AFP was negative (≤ 20 μg/L) in 26 HCC patients and positive with a low concentration (20 μg/L < AFP ≤ 400 μg/L) in 20 HCC patients. Of these 46 patients, 27 (18 cases with negative AFP and 9 cases with low concentration AFP) had MAGE-1, SSX-1 and/or CTp11 mRNA transcripts detected in their PBMC samples. By contrast, of the 30 patients with negative CTA transcripts in PBMC samples, 21 had the serum AFP level higher than 400 μg/L. Totally, 78.9% of HCC patients had either the AFP level higher than 400 μg/L in serum or positive CTA transcripts in PBMC. The parameters of AFP in serum and CTA transcripts in PBMC in combination with the results of imaging studies, would enable to make a clear diagnosis of 78.9% of HCC patients, which is much higher than the test with single AFP (χ2 = 17.555, P < 0.01)
CTA transcripts in PBMC from early HCC patients
The frequency of positive MAGE-1, SSX-1 and/or CTp11 transcripts detected in PBMC from patients with HCC was 41.7% at stage I, 56.0% at stage II, 66.7% at stage III, and 79.5%% at stage IV, respectively. Of note, MAGE-1, SSX-1 and/or CTp11 mRNA was detected in PBMC from 51.4% of HCC patients at stages I and II and from 77.9% of HCC patients at stages III and IV, showing that advanced stages of HCC are correlated with the higher expression frequency of MAGE-1, SSX-1 and/or CTp11 gene mRNA (χ2 = 6.632, P = 0.010). As many as 59.5% of the HCC patients at stages I and II could be diagnosed when CTA transcripts in PBMC and serum AFP level were combined with imaging findings. However, the serum AFP level was higher than 400 μg/L in 35.1% of the HCC patients. The diagnosis rate made by combined CTAs and AFP was significantly higher than that based on single AFP (χ2 = 4.391, P = 0.036).
DISCUSSION
The integration of molecular and immunological techniques has led to the identification of a new category of tumor-specific antigens, also known as cancer-testis antigens, such as melanoma antigen (MAGE), synovial sarcoma X breakpoint (SSX), B melanoma antigen (BAGE), G melanoma antigen (GAGE), synaptonemal complex protein-1 (SCP-1), New York esophagus-1 (NY-ESO-1), and CTp11[23]. The CTAs are a distinct and unique class of differentiation antigens. Attributing genes to this gene group is based on their characteristics, including mRNA expression in normal tissues of testis, fetal ovary, and placenta, and mRNA expression in different cancers. Until now, at least 70 families of cancer-testis gene with 140 members have been attributed to this group and their expression has been studied in different types of tumors[23-26]. MAGE-1, SSX-1 and CTp11 belong to the CTA family members. The growing knowledge about CTAs indicates that the expression of CTA often shows a marked specificity for tumor cells[23,27-30]. These markers can be used to target tumor cells for early detection.
In the present study, CTAs (MAGE-1, SSX-1 and CTp11) were expressed with a high percentage and specificity in HCC. The positive rate for at least one of the transcripts of three CTA genes in HCC tissue samples was as high as 91.2%. Conversely, no expression was detected in the adjacent normal and cirrhotic liver tissue samples, or in the PBMC samples from healthy donors and cirrhotic patients. Based on the prevalent invasion of HCC cells to hepatic vessels, it is reasonable to consider that MAGE-1, SSX-1 and CTp11 are the appropriate tumor-specific markers for the detection of circulating HCC cells, which may play a complementary role in HCC diagnosis.
In this study, a sensitive and specific technique was developed, which is capable of detecting circulating HCC cells using MAGE-1, SSX-1 and CTp11 transcripts as tumor-specific markers by nested RT-PCR. Through the ten-fold serial dilution experiments with positive control cell lines, our results verified that exponential amplification of target cDNA converted from mRNA could allow to detect a single malignant cell within millions of normal blood cells and hence, to sensitively detect the metastatic tumor cells in peripheral blood. The sensitivity of this assay is within the range of other published reports[31-34], which is much more sensitive than antibody-based serology[35,36]. The positive rate of nested RT-PCR was as high as 66.7% for at least one of the transcripts of three CTA genes in the PBMC samples from HCC patients. In addition, detecting any of the transcripts of three CTA genes in PBMC samples from HCC patients would directly represent the presence of tumor cells in peripheral blood, suggesting that this method has a higher specificity than serum AFP and is thus able to improve the diagnosis of HCC. However, no expression of CTAs was detected in 8.9% of HCC tissue samples, showing that it is necessary to screen other CTAs or tumor specific antigens in these patients. If we can filter out 1-2 other markers, the diagnostic sensitivity of this method would be further improved.
Both albumin[37,38] and AFP[39-41] mRNA have been widely used as tumor markers for detecting HCC cells in circulation. However, the reliability is challenged, because albumin is abundantly expressed in normal liver cells[37] and AFP is expressed in liver cells infected with hepatitis virus or in cirrhotic liver[34]. In recent years, although an increasing number of genetic markers, such as telomerase reverse transcriptase[42], Des-g-carboxyprothrombin[43], squamous cell carcinoma antigen-immunoglobulin M complexes[44] and human cervical cancer oncogene[45], have been used in the diagnosis of early HCC, they have significant diagnostic limitations in their specific nature. In this study, MAGE-1, SSX-1 or CTp11 was not detected in PBMC from patients with their HCC tissue samples negative for these three CTA genes mRNA or in PBMC from 20 healthy donors and 10 cirrhotic patients, indicating that detecting CTA transcripts in PBMC from HCC patients has a high specificity for HCC.
So far, no methods or biomarkers demonstrate absolute superiority for early detection of HCC. It is difficult to simultaneously solve their sensitivity and specificity. Our assay by nested RT-PCR using MAGE-1, SSX-1 and CTp11 mRNA as tumor-specific makers showed a high sensitivity and specificity, indicating that it can establish the diagnosis of HCC.
Molecular biology technology contributes to the early diagnosis of HCC. However, its disadvantages are also obvious, including its cost and availability. PCR assay, a commonly used molecular biology technology, is more expensive and troublesome than serological tests, and is thus not the first choice in early detection of HCC. However, it plays a supplementary role in the diagnosis of HCC. Hopefully in the not so distant future, this technology will become increasingly popular and automatic with its cost decreased.
At present, the serum AFP level is still the gold standard for diagnosis of liver cancer. The AFP level is normal in 20%-40% of HCC patients at the time of diagnosis and usually remains low even in patients with advanced HCC[7-10]. AFP > 400 μg/L is considered diagnostic for HCC, although fewer than 50% of HCC patients may meet this standard[5-8]. With values of that magnitude, the specificity of AFP is close to 100% at the cost of its sensitivity fallen to less than 45%[6,7]. In this study, 51.1% of HCC patients were negative or positive for AFP with a low concentration. MAGE-1, SSX-1 and/or CTp11 gene was expressed in 69.2% and 45.0% of HCC patients in the two groups, respectively, suggesting that mRNA, a tumor-specific marker, can be used as an adjuvant diagnostic tool. Detecting the transcripts of three CTA genes combined with serum AFP test in PBMC from HCC patients, can improve the total diagnostic rate of HCC.
In this study, the expression of CTAs in PBMC was significantly correlated with the clinical TNM classification of HCC. Although the positive frequency of CTA mRNA in PBMC was significantly higher in HCC patients at stages III and IV than in those at stages I and II, MAGE-1, SSX-1 and/or CTp11 mRNA transcripts were detected in 41.7% of the HCC patients at stage I and in 56.0% of the HCC patients at stage II, showing that these three CTAs are reliable markers for screening hematogenous spread of early HCC cells. The combination of CTA transcripts in PBMC and serum AFP level improves early diagnosis of HCC. The classic TNM staging method[22] does not accurately reflect the actual process of HCC patients. The TNM classification criteria for HCC include tumor size, presence of portal vein invasion, and extrahepatic metastasis, etc. According to the TNM criteria, HCC at stage I (T1N0M0) or stage II (T2N0M0) should have no metastasis of tumor cells except for intra-hepatic metastasis. Assay by nested RT-PCR to detect MAGE-1, SSX-1 and/or CTp11 transcripts, the tumor-specific markers, revealed that 51.4% patients with HCC in early stages (stage I and II) have already had micrometastasis to the peripheral blood, indicating that blood dissemination of tumor cells has already occurred in the early stage of HCC when distant metastasis cannot be confirmed. Furthermore, it may be the reason why some early HCC patients still suffer from recurrence even after complete removal of the tumor. Detecting CTA transcripts in PBMC of early HCC patients can demonstrate hematogeneous dissemination of tumor cells more specifically than conventional methods, thus playing a supplementary role in the diagnosis of HCC. The traditional TNM staging criteria ignoring the presence of circulating HCC cells need to be perfected.
COMMENTS
Background
The prognosis of hepatocellular carcinoma (HCC) is poor because it is detection at an advanced, non-resectable stage. So far, α-fetoprotein (AFP) is a generally accepted serological marker. Its diagnostic accuracy is unsatisfactory and questionable. Serum AFP alone is helpful when its level is markedly elevated, occuring in less than 50% of cases at the time of diagnosis. Therefore, more sensitive and specific biomarkers are needed.
Research frontiers
The limitations of conventional AFP as a marker has led to a search for more sensitive and specific markers. In recent years, although an increasing number of genetic markers have been used in diagnosis of early HCC, they have significant diagnostic limitations in the specific nature. Cancer-testis antigens (CTAs) are frequently expressed in different types of cancer and have received considerable attention as ideal biomarkers of tumor cells.
Innovations and breakthroughs
The sensitivity and specificity of nested reverse transcripts-polymerase chain reaction assay using melanoma antigen-1, synovial sarcoma X breakpoint-1 and cancer-testis-associated protein of 11 kDa mRNA as tumor-specific multiple-makers are high and can thus be as an adjuvant diagnostic tool. This assay combined with serum AFP level may improve the diagnosis of HCC.
Applications
Detecting transcripts of CTA genes in peripheral blood mononuclear cells from HCC patients, combined with serum AFP test, improves the total diagnostic rate of HCC.
Peer review
The authors investigated the expression of some CTA genes in peripheral blood of HCC patients and showed that the positive rate for at least one of the three CTA genes was 67% in blood samples and 90% in resected tumor samples. They also showed a high specificity and sensitivity of their method for HCC. The study is very interesting and important for early diagnosis of HCC.
Footnotes
Supported by National Natural Science Foundation of China, No. 30200271
Peer reviewer: Masahiro Arai, MD, PhD, Department of Gastroenterology, Toshiba General Hospital, 6-3-22 Higashi-ooi, Shinagawa-ku, Tokyo 140-8522, Japan
S- Editor Wang JL L- Editor Wang XL E- Editor Ma WH
References
- 1.Jain S, Singhal S, Lee P, Xu R. Molecular genetics of hepatocellular neoplasia. Am J Transl Res. 2010;2:105–118. [PMC free article] [PubMed] [Google Scholar]
- 2.Paul SB, Manjunatha YC, Acharya SK. Palliative treatment in advanced hepatocellular carcinoma: has it made any difference? Trop Gastroenterol. 2009;30:125–134. [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Padma S, Martinie JB, Iannitti DA. Liver tumor ablation: percutaneous and open approaches. J Surg Oncol. 2009;100:619–634. doi: 10.1002/jso.21364. [DOI] [PubMed] [Google Scholar]
- 5.Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialecki ES, Di Bisceglie AM. Diagnosis of hepatocellular carcinoma. HPB (Oxford) 2005;7:26–34. doi: 10.1080/13651820410024049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med. 2003;139:46–50. doi: 10.7326/0003-4819-139-1-200307010-00012. [DOI] [PubMed] [Google Scholar]
- 8.Colombo M. Screening for cancer in viral hepatitis. Clin Liver Dis. 2001;5:109–122. doi: 10.1016/s1089-3261(05)70156-2. [DOI] [PubMed] [Google Scholar]
- 9.Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5:145–159. doi: 10.1016/s1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- 10.Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609–1619. doi: 10.1053/gast.2002.33411. [DOI] [PubMed] [Google Scholar]
- 11.Smith B, Selby P, Southgate J, Pittman K, Bradley C, Blair GE. Detection of melanoma cells in peripheral blood by means of reverse transcriptase and polymerase chain reaction. Lancet. 1991;338:1227–1229. doi: 10.1016/0140-6736(91)92100-g. [DOI] [PubMed] [Google Scholar]
- 12.Raj GV, Moreno JG, Gomella LG. Utilization of polymerase chain reaction technology in the detection of solid tumors. Cancer. 1998;82:1419–1442. doi: 10.1002/(sici)1097-0142(19980415)82:8<1419::aid-cncr1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Merrie AE, Yun K, McCall JL. Utilization of polymerase chain reaction technology in the detection of solid tumors. Cancer. 1999;85:248–250. doi: 10.1002/(sici)1097-0142(19990101)85:1<248::aid-cncr39>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 14.Clarke LE, Leitzel K, Smith J, Ali SM, Lipton A. Epidermal growth factor receptor mRNA in peripheral blood of patients with pancreatic, lung, and colon carcinomas detected by RT-PCR. Int J Oncol. 2003;22:425–430. [PubMed] [Google Scholar]
- 15.Mou DC, Cai SL, Peng JR, Wang Y, Chen HS, Pang XW, Leng XS, Chen WF. Evaluation of MAGE-1 and MAGE-3 as tumour-specific markers to detect blood dissemination of hepatocellular carcinoma cells. Br J Cancer. 2002;86:110–116. doi: 10.1038/sj.bjc.6600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CH, Chen GJ, Lee HS, Huang GT, Yang PM, Tsai LJ, Chen DS, Sheu JC. Expressions of cancer-testis antigens in human hepatocellular carcinomas. Cancer Lett. 2001;164:189–195. doi: 10.1016/s0304-3835(01)00379-2. [DOI] [PubMed] [Google Scholar]
- 17.Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN. CTp11, a novel member of the family of human cancer/testis antigens. Cancer Res. 1999;59:6223–6229. [PubMed] [Google Scholar]
- 18.Chen H, Cai S, Wang Y, Zhao H, Peng J, Pang X, Zhu J, Cong X, Rui J, Leng X, et al. Expression of the MAGE-1 gene in human hepatocellular carcinomas. Chin Med J (Engl) 2000;113:1112–1118. [PubMed] [Google Scholar]
- 19.Tahara K, Mori M, Sadanaga N, Sakamoto Y, Kitano S, Makuuchi M. Expression of the MAGE gene family in human hepatocellular carcinoma. Cancer. 1999;85:1234–1240. [PubMed] [Google Scholar]
- 20.Luo G, Huang S, Xie X, Stockert E, Chen YT, Kubuschok B, Pfreundschuh M. Expression of cancer-testis genes in human hepatocellular carcinomas. Cancer Immun. 2002;2:11. [PubMed] [Google Scholar]
- 21.Zhao L, Mou DC, Leng XS, Peng JR, Wang WX, Huang L, Li S, Zhu JY. Expression of cancer-testis antigens in hepatocellular carcinoma. World J Gastroenterol. 2004;10:2034–2038. doi: 10.3748/wjg.v10.i14.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson JM, Sherman M, Tavill A, Abecassis M, Chejfec G, Gramlich T. AHPBA/AJCC consensus conference on staging of hepatocellular carcinoma: consensus statement. HPB (Oxford) 2003;5:243–250. doi: 10.1080/13651820310015833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghafouri-Fard S, Modarressi MH. Cancer-testis antigens: potential targets for cancer immunotherapy. Arch Iran Med. 2009;12:395–404. [PubMed] [Google Scholar]
- 24.Ajiro M, Katagiri T, Ueda K, Nakagawa H, Fukukawa C, Lin ML, Park JH, Nishidate T, Daigo Y, Nakamura Y. Involvement of RQCD1 overexpression, a novel cancer-testis antigen, in the Akt pathway in breast cancer cells. Int J Oncol. 2009;35:673–681. [PubMed] [Google Scholar]
- 25.Lin ML, Fukukawa C, Park JH, Naito K, Kijima K, Shimo A, Ajiro M, Nishidate T, Nakamura Y, Katagiri T. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 2009;100:1443–1450. doi: 10.1111/j.1349-7006.2009.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettoni F, Filho FC, Grosso DM, Galante PA, Parmigiani RB, Geraldo MV, Henrique-Silva F, Oba-Shinjo SM, Marie SK, Soares FA, et al. Identification of FAM46D as a novel cancer/testis antigen using EST data and serological analysis. Genomics. 2009;94:153–160. doi: 10.1016/j.ygeno.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Costa FF, Le Blanc K, Brodin B. Concise review: cancer/testis antigens, stem cells, and cancer. Stem Cells. 2007;25:707–711. doi: 10.1634/stemcells.2006-0469. [DOI] [PubMed] [Google Scholar]
- 28.Parmigiani RB, Bettoni F, Vibranovski MD, Lopes MH, Martins WK, Cunha IW, Soares FA, Simpson AJ, de Souza SJ, Camargo AA. Characterization of a cancer/testis (CT) antigen gene family capable of eliciting humoral response in cancer patients. Proc Natl Acad Sci USA. 2006;103:18066–18071. doi: 10.1073/pnas.0608853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meklat F, Li Z, Wang Z, Zhang Y, Zhang J, Jewell A, Lim SH. Cancer-testis antigens in haematological malignancies. Br J Haematol. 2007;136:769–776. doi: 10.1111/j.1365-2141.2006.06484.x. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson BJ, Iseli C, Panji S, Zahn-Zabal M, Hide W, Old LJ, Simpson AJ, Jongeneel CV. Rapid evolution of cancer/testis genes on the X chromosome. BMC Genomics. 2007;8:129. doi: 10.1186/1471-2164-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumura M, Niwa Y, Kato N, Komatsu Y, Shiina S, Kawabe T, Kawase T, Toyoshima H, Ihori M, Shiratori Y. Detection of alpha-fetoprotein mRNA, an indicator of hematogenous spreading hepatocellular carcinoma, in the circulation: a possible predictor of metastatic hepatocellular carcinoma. Hepatology. 1994;20:1418–1425. doi: 10.1002/hep.1840200607. [DOI] [PubMed] [Google Scholar]
- 32.Matsumura M, Niwa Y, Hikiba Y, Okano K, Kato N, Shiina S, Shiratori Y, Omata M. Sensitive assay for detection of hepatocellular carcinoma associated gene transcription (alpha-fetoprotein mRNA) in blood. Biochem Biophys Res Commun. 1995;207:813–818. doi: 10.1006/bbrc.1995.1259. [DOI] [PubMed] [Google Scholar]
- 33.Ghossein RA, Rosai J. Polymerase chain reaction in the detection of micrometastases and circulating tumor cells. Cancer. 1996;78:10–16. doi: 10.1002/(SICI)1097-0142(19960701)78:1<10::AID-CNCR3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Komeda T, Fukuda Y, Sando T, Kita R, Furukawa M, Nishida N, Amenomori M, Nakao K. Sensitive detection of circulating hepatocellular carcinoma cells in peripheral venous blood. Cancer. 1995;75:2214–2219. doi: 10.1002/1097-0142(19950501)75:9<2214::aid-cncr2820750905>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 35.Diel IJ, Kaufmann M, Goerner R, Costa SD, Kaul S, Bastert G. Detection of tumor cells in bone marrow of patients with primary breast cancer: a prognostic factor for distant metastasis. J Clin Oncol. 1992;10:1534–1539. doi: 10.1200/JCO.1992.10.10.1534. [DOI] [PubMed] [Google Scholar]
- 36.Pelkey TJ, Frierson HF Jr, Bruns DE. Molecular and immunological detection of circulating tumor cells and micrometastases from solid tumors. Clin Chem. 1996;42:1369–1381. [PubMed] [Google Scholar]
- 37.Chou HC, Sheu JC, Huang GT, Wang JT, Chen DS. Albumin messenger RNA is not specific for circulating hepatoma cells. Gastroenterology. 1994;107:630–631. doi: 10.1016/0016-5085(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 38.Gion T, Taketomi A, Shimada M, Shirabe K, Hasegawa H, Takenaka K, Sugimachi K. Perioperative change in albumin messenger RNA levels in patients with hepatocellular carcinoma. Hepatology. 1998;28:1663–1668. doi: 10.1002/hep.510280628. [DOI] [PubMed] [Google Scholar]
- 39.Wong IH, Lau WY, Leung T, Yeo W, Johnson PJ. Hematogenous dissemination of hepatocytes and tumor cells after surgical resection of hepatocellular carcinoma: a quantitative analysis. Clin Cancer Res. 1999;5:4021–4027. [PubMed] [Google Scholar]
- 40.Witzigmann H, Geissler F, Benedix F, Thiery J, Uhlmann D, Tannapfel A, Wittekind C, Hauss J. Prospective evaluation of circulating hepatocytes by alpha-fetoprotein messenger RNA in patients with hepatocellular carcinoma. Surgery. 2002;131:34–43. doi: 10.1067/msy.2002.118954. [DOI] [PubMed] [Google Scholar]
- 41.Witzigmann H, Geissler F, Benedix F, Thiery J, Uhlmann D, Tannapfel A, Wittekind C, Hauss J. Prospective evaluation of circulating hepatocytes by alpha-fetoprotein messenger RNA in patients with hepatocellular carcinoma. Surgery. 2002;131:34–43. doi: 10.1067/msy.2002.118954. [DOI] [PubMed] [Google Scholar]
- 42.Miura N, Maruyama S, Oyama K, Horie Y, Kohno M, Noma E, Sakaguchi S, Nagashima M, Kudo M, Kishimoto Y, et al. Development of a novel assay to quantify serum human telomerase reverse transcriptase messenger RNA and its significance as a tumor marker for hepatocellular carcinoma. Oncology. 2007;72 Suppl 1:45–51. doi: 10.1159/000111706. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura S, Nouso K, Sakaguchi K, Ito YM, Ohashi Y, Kobayashi Y, Toshikuni N, Tanaka H, Miyake Y, Matsumoto E, et al. Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am J Gastroenterol. 2006;101:2038–2043. doi: 10.1111/j.1572-0241.2006.00681.x. [DOI] [PubMed] [Google Scholar]
- 44.Giannelli G, Fransvea E, Trerotoli P, Beaugrand M, Marinosci F, Lupo L, Nkontchou G, Dentico P, Antonaci S. Clinical validation of combined serological biomarkers for improved hepatocellular carcinoma diagnosis in 961 patients. Clin Chim Acta. 2007;383:147–152. doi: 10.1016/j.cca.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Yoon SK, Lim NK, Ha SA, Park YG, Choi JY, Chung KW, Sun HS, Choi MJ, Chung J, Wands JR, et al. The human cervical cancer oncogene protein is a biomarker for human hepatocellular carcinoma. Cancer Res. 2004;64:5434–5441. doi: 10.1158/0008-5472.CAN-03-3665. [DOI] [PubMed] [Google Scholar]