Abstract
Biomineralization processes such as formation of bones and teeth require controlled mineral deposition and self-assembly into hierarchical biocomposites with unique mechanical properties. Ideal biomaterials for regeneration and repair of hard tissues must be biocompatible, possess micro and macroporosity for vascular invasion, provide surface chemistry and texture that facilitate cell attachment, proliferation, differentiation of lineage specific progenitor cells, and induce deposition of calcium phosphate mineral. To expect in-vivo like cellular response several investigators have used extracellular matrix proteins as templates to recreate in-vivo microenvironment for regeneration of hard tissues. Recently, several novel methods of designing tissue repair and restoration materials using bioinspired strategies are currently being formulated. Nanoscale structured materials can be fabricated via the spontaneous organization of self-assembling proteins to construct hierarchically organized nanomaterials. The advantage of such a method is that polypeptides can be specifically designed as building blocks incorporated with molecular recognition features and spatially distributed bioactive ligands that would provide a physiological environment for cells in-vitro and in-vivo. This is a rapidly evolving area and provides a promising platform for future development of nanostructured templates for hard tissue engineering. In this review we try to highlight the importance of proteins as templates for regeneration and repair of hard tissues as well as the potential of peptide based nanomaterials for regenerative therapies.
Keywords: tissue engineering, scaffold, bone, teeth, cartilage, self-assembly, biomaterial, proteins
INTRODUCTION
Tissue engineering is the ability to engineer biological activity into synthetic or natural materials in order to repair damaged tissues or regenerate tissues de-novo. It is a multidisciplinary field involving biology, medicine and engineering and this combined consortium aims to improve the health and quality of life for millions of people. In Greek mythology the regrowth of Prometheus liver has become a classical paradigm to researchers for the possible renewal of damaged human organs.
Bone fractures, osteoarthritis, osteoporosis or bone cancer represent common and significant clinical problems. More than 6 million fractures in the U.S. annually require bone graft procedures to ensure rapid skeletal repair [1]. The American Academy of Orthopedic Surgeons also reported that in just a 4 year period, there was an 83.72% increase in the number of hip replacements performed from nearly 258,000 procedures in 2000 to 474,000 procedures in 2004 [2] However, traditional implant materials only last 10–15 years on average and implant failures originating from implant loosening, inflammation, infection, osteolysis and wear debris frequently occur [2]. Therefore, there is a need to develop cytocompatible biomaterials for bone and cartilage tissue engineering. Further, acceleration of the repair process with degradable biologically active engineered scaffolds can potentially reduce patient recovery time [3].
The replacement or restoration of osseous defects caused by fracture or trauma is a major clinical problem. Thus far, several osteoconductive and osteoinductive materials have been investigated for orthopedic and dental repair. However, the current implants suffer from 2 major drawbacks (1) The mechanical properties of the implants are far from the natural bone tissue (2) The lack of interaction between the implants and the natural tissue environment. Nature has always been a source of inspiration for technical developments but only in recent years have material scientists started to consider the hierarchical structure of natural materials as a model for the development of new types of high-performance engineering materials [4–7]. Specifically, inspiration for the design of new biomaterials for tissue regeneration has been derived from structure-function analysis on various length scales of the extracellular proteins that cells use to organize themselves into tissues [7].
Knowledge of basic biological principles for bone and teeth formation is necessary for hard tissue regeneration. Natural bone and dentin is a 2 phase porous composite material composed of a mineral phase of hydroxyapatite and a soft hydrogel reinforcing phase made of mainly type I collagen [8] (Fig. 1A&B). Another important class of proteins in the ECM of calcified tissues are the noncollagenous proteins [9]. Noncollagenous anionic proteins have been implicated to regulate the size, orientation and crystal habit of the mineral deposits [10]. This unusual combination of a hard inorganic material and an underlying collagenous network endows native bone with unique mechanical properties such as low stiffness, resistance to tensile and compressive forces, and high fracture toughness [11, 12]. Specifically, bone and dentin are biocomposites of protein and mineral with strength, hardness and fracture toughness. Understanding the functionality and complexity of biological systems has helped researchers to mimic nature’s principles and design for tissue regeneration.
Fig 1. Mineralized collagen fibrils are the basic building block for bone and dentin formation.
Fig 1A represents a generalized schematic representation of the self-assembly of the collagen fibrils subsequent nucleation and growth of hydroxyapatite within the compartment provided by the self-assembled collagen fibrils. The collagen matrix is responsible for the c-axis oriented hydroxyapatite formation. Hydroxyapatite is represented by grey blocks.
Fig 1B represents a transmission electron micrograph of mineralized collagen in bovine dentin.
The fundamental requirement for tissue engineering/regeneration strategies is the availability of three key components, namely:
Biomaterials: Function as scaffolds and provide 3-D templates. They include natural and synthetic biomaterials that are designed to direct the organization, growth and differentiation of cells in the process of forming functional tissue by providing both physical and chemical cues.
Cells: Acquiring the appropriate source of cells such as autologous cells, allogenic cells, xenogenic cells, stem cells and immunological manipulation is important. Methodologies enabling proliferation and differentiation of cells are key criteria in tissue engineering.
Biomolecules: This group comprises angiogenic factors, growth factors, differentiation factors and signaling molecules.
Traditionally, biomaterials were designed to be inert and not interact with the biology of the surrounding tissue. Current biomaterials can be classified based on their mode of application for tissue engineering: one in which biomaterials are used as carriers for introducing cells into damaged or diseased tissue, and one in which biomaterials are used to augment the function of endogenous progenitor cells.
Scaffolds serve primarily as osteoconductive vehicles, since new bone is deposited by creeping substitution from adjacent living bone [13]. In addition to osteoconductivity, scaffolds function as delivery vehicles for cytokines that can activate or recruit precursor cells from the host into osteogenic lineage, thereby demonstrating an osteoinductive response. An appropriate scaffold for tissue engineering hard tissues should accommodate several requirements, namely, an appropriate 3-D porous scaffold, which would serve as a template for initial cell attachment and subsequent tissue formation; be biodegradable and biocompatibile, have a high surface area to volume ratio with sufficient mechanical integrity, and possess the ability to provide a suitable environment for new-tissue formation that can integrate with the surrounding tissue [14, 15].
During the last decade there has been tremendous progress in designing novel nanomaterials incorporated with biologically active components and functional domains from ECM molecules for hard tissue engineering. Biomaterials are rapidly being developed to deliver regulatory signals in a precise and physiological manner in order to mimic microenvironments required for specific tissue regeneration. Protein templates are slowly replacing polymers as biomaterials for tissue engineering bone, cartilage and dentin. The main advantage is that proteins are nanoscale biological materials that can be integrated with other organic or inorganic components to form nanocomposites. The biomimetic features and the excellent physico-chemical properties of such nanomaterials play a key role in activating cellular interaction and initiating tissue regeneration. In this review, we have tried to highlight the trend in the development of naturally occurring proteins as scaffolds as well as the design of self-assembly based biomaterials that are being used as templates for hard tissue repair and regeneration.
Naturally Occurring Proteins as Biomaterials for hard tissue engineering
Natural extracellular matrix (ECM) proteins are widely used to make biomaterial scaffolds for tissue engineering and regenerative medicine. Gelatin, collagen, chitosan are excellent candidates for hard tissue engineering applications due to their biodegradability and ease of fabrication. These purified and reconstituted ECM proteins can form hydrogel matrices with unique 3-D architecture coupled with intrinsic cell signaling that guide remodeling and functional tissue regeneration [16]. Modifications in natural matrices can be performed to create desired elastic modulus conducive for tissue specific cell types. Natural polymers also permit diffusion of soluble molecules to the basal as well as apical surface. There are some drawbacks of using such recombinant proteins, namely, a limited control over the physical properties and biodegradation. Some of the common ECM proteins used in hard tissue engineering are highlighted below.
Type I Collagen
Type I collagen is the main component of natural ECM and is the most abundantly occurring protein in many connective tissues. It has been used for a long time as a scaffold for engineering cartilage and bone tissue either by itself or blended with other naturally occurring or artificial biopolymers. The ability of type I collagen to exist as monomer in acidic conditions and polymerize in vitro under physiological pH and temperature has been utilized successfully in various tissue engineering applications. This property also makes collagen attractive for use as injectible hydrogels. Collagen hydrogels provide extremely good cell adhesive properties and promote proliferation, growth and differentiation of many different cell types. However, one disadvantage of type I collagen is that the hydrogels lack load bearing ability and other mechanical properties required to successfully replace hard tissue during the initial phases of implantation. This problem has been overcome by utilization of type I collagen blended with different materials as hydrogel blends.
Type I collagen in cartilage tissue engineering
Type 1 collagen sponges comprising of 0.5 weight percentage of collagen that has been lyophilized and UV crosslinked have been shown to support the proliferation and differentiation of bovine articular chondrocytes (BACs) in vitro [17]. Addition of a small amount of hyaluronan to the sponges was shown to increase the expression of aggrecan core protein and type II collagen [18]. Upregulation of these two genes is an indication for chondrogenic differentiation. Additionally, the UV treatment also reduced the immune response to collagen when implanted in vivo. Collagen scaffold by itself has been proven to aid regeneration of meniscal cartilage in FDA approved human trials. This study showed that collagen scaffold, over a period of 36 weeks reduced pain, increased activity level of the patients and promoted regeneration in patients with damaged meniscus [19].
A blend of type I collagen scaffold and stem cells have been shown to heal white zone meniscal tears. Cultured stem cells embedded within collagen scaffolds between two layers of bovine meniscal tissue in vitro showed that the scaffold integrated well with the tissues and induced chondrogenic differentiation of the stem cells and improved the mechanical properties of the construct. However, addition of TGFβ1 a known initiator of chondrogenesis, was shown to inhibit the integration of the scaffold with the tissues [20]. On the other hand, collagen scaffolds seeded with mesenchymal cells and cultured in a bioreactor while being subjected to prolonged tensile stress and strain showed increased expression of type II collagen and aggrecan indicating the effect of prolonged tensile stress on the chondrogenic differentiation of mesenchymal cells on collagen scaffolds. This study indicates that it is possible to grow cartilage- like- tissues in vitro by simulating the physical loading conditions that the tissue experience in vivo [21]. These studies highlight the ability of type I collagen as a functional material for cartilage tissue regeneration.
Blends of type I collagen in cartilage tissue engineering
Polymeric scaffolds offer an enticing alternative to protein scaffolds primarily because of their non immunogenic nature. However, a lot of their degradation products are acidic and their ability to support cell culture cannot compare to that of ECM protein scaffolds. Recently, blends of type I collagen and PLGA polymer have been shown to be a promising alternative incorporating the ideal properties of both components. The PLGA meshes improved the mechanical aspect of the scaffold and the collagen filling provided the scaffold with bioactivity [22].
Type I Collagen in bone tissue engineering
Bone tissue engineering, unlike cartilage requires that the scaffold possess mechanical properties comparable to that of the hard tissue. Therefore, scaffolds comprising of type I collagen as the major biopolymer component often contain other materials (naturally occurring or synthetic) to boost their mechanical properties. Such additives, besides improving mechanical properties, also impart desired cues that are necessary for the differentiation of precursor cells into functional osteoblasts. The following are a few examples of type I collagen containing blends that are used as scaffolds for bone regeneration.
Hydroxyapatite coated type I collagen and blends of type I collagen
One of the many ways to augment the mechanical strength of type I collagen scaffolds is to coat the collagen fibrils with hydroxyapatite. Recombinant collagen scaffolds coated with crystalline hydroxyapatite have been shown to be osteoinductive and heal critical sized defects in animal models [23]. The recombinant human collagen alleviates concerns over the use of animal collagens as scaffolding materials and can prove to be a safer alternative. Various methods have been devised to coat type I collagen with hydroxyapatite. Recently collagen foams containing apatite nanocrystals and also blends of collagen, nanoapatite and poly lactic acid (PLA) have been developed. This scaffold has been shown to possess excellent cell attachment properties in vitro and good osteoinductive properties in vivo [23]. As the material surface regulates cell behavior, therefore, the property of type I collagen to bind morphogens has been exploited by tissue engineers. Bone morphogenetic protein 2 (BMP2) has been shown to induce differentiation of mesenchymal stem cells into osteoblast phenotype and animal models and human trials have shown that the use of type I collagen incorporated with BMP2 can be extremely useful for bone regeneration [24].
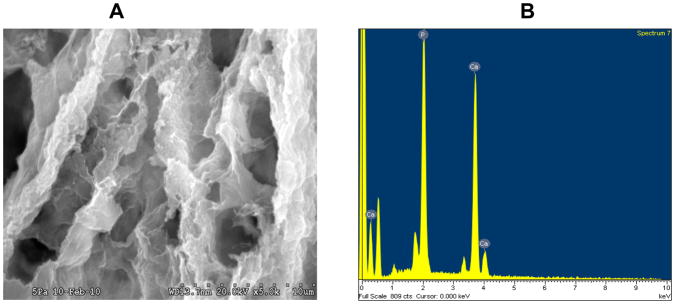
In a recent study we have developed a collagen-chitosan blend for tissue engineering applications [25]. Increase in chitosan concentration proportionately increased the mechanical strength as determined by atomic force microscopy based analysis of the hydrogels in their native hydrated state. Inerestingly, we have demonstrated that this scaffold can bind calcium ions to form hydroxyapatite coated hydrogels by virtue of chitosan’s calcium binding affinity (unpublished data) (Fig. 2A and 2B). This biomimetic scaffold exhibits great promise to regenerate bone as it possesses good mechanical properties, antimicrobial effects contributed by chitosan [26] and cell attachment properties (Fig 3B & C). In support of this concept, Zhang et al. have demonstrated osteogenic properties with electrospun collagen/HAP/chitosan nano fibers. The results from this study showed that the addition of collagen to chitosan/HAP significantly increased the cell attachment properties and osteogenic ability of the scaffolds [27].
Fig 2. SEM analysis of calcium phosphate coated collagen-chitosan scaffold.
Fig 2A represent an SEM image of collagen-chitosan (1:1) scaffold (25). The scaffold was immersed in 1M calcium chloride solution then washed with deionized water to remove non-specifically bound calcium and then immersed in 1M solution of sodium phosphate. The scaffold was then dried, processed and viewed under Hitachi S3000N variable pressure SEM.
Fig 2B represent an energy dispersive X-ray scan of the mineralized deposits on the collagen-chitosan scaffold. The Ca/P ratio was 1.7 indicative of hydroxyapatite deposition.
Figure 3. Morphology differences between cells grown on 2-D and 3-D matrices.
Fig 3A is a confocal image of rat marrow stromal cells (RMSCs) grown on tissue culture plates and stained with phalloidan conjugated to TRITC to label actin filaments.
Fig 3B represents an x-y projection of a z-stack of confocal images of RMSCs embedded within a 1:1 type I collagen-chitosan biopolymer matrix.
Note the differences in cellular maorphology between cells grown on 2D versus 3D surfaces.
Fig 3C represents a scanning electron micrograph obtained by imaging a similar sample as in 2A using a field emission scanning electron microscope. The white arrow points to the adhesion of a cellular process to the extracellular matrix.
Silk Proteins as Biomaterials
Silks are protein polymers that are synthesized and secreted by insects such as spider, silk worm and scorpion [28]. Silk worm (Bombyx mori) silk has been used for decades as a suture material. In recent times some biocompatibility issues have been reported due to the presence of serichin contaminants. However, these issues can be eliminated by using pure silk. Silk polymers offer a side chain chemistry that can be used for binding to globular proteins and growth factors providing a platform for differentiation of precursor cells. Additionally, silk fibroin offers equal cell attachment properties when compared to collagen fibers [29, 30].
Silk fibroin scaffolds in hard tissue regeneration
Silk fibroin scaffolds as hydrogels, meshes and membranes have been used without cells for guided bone regeneration [31]. Silk fibroin has been shown to be osteoconductive when osteoblasts are seeded on the scaffolds. The rate of calcium deposition and differentiation of the cells increased when RGD tripeptides were incorporated to the scaffold surface [32, 33]. Such nanoscale cell attachment motifs exhibited on the surface could aid efficient cell attachment to the scaffold via cell surface integrins. Although silk fibroins provide scaffolding, it is the cell attachment domains and other surface properties that mediate the bioactivity of the scaffold.
Another example of surface modification of the silk scaffolds is by reinforcing the fibroins with calcium phosphate. For this, poly aspartic acid residues were incorporated on to the fibroins to bind calcium and thus improve the mechanical properties [34]. Addition of BMP2 to electrospun silk scaffolds was specifically developed for bone tissue engineering. The BMP2 survived the electrospinning process and retained its bioactivity. This modified scaffold enhanced osteogenic differentiation of mesenchymal stem cells [35]. Although silk fibroin has been shown to possess excellent bioactive properties for bone regeneration, the effects of its degradation products remained largely unknown. A recent study has shown that when MG63 osteoblasts were treated with low molecular weight silk fibroin similar to the degradative products of silk fibroin, significant increase was noted in the levels of osteoblastic differentiation genes and alkaline phosphatase activity. Thus, controlled degradation of silk can by itself induce osteogenic differentiation [36]. These inherent properties of silk fibroin from Bombyx mori has made it a very attractive template for bone tissue engineering.
Silk fibroin scaffolds in cartilage tissue regeneration
Traditionally, collagen scaffolds have been used extensively as a matrix for cartilage tissue engineering, however, silk fibroin scaffolds offer a promising alternative with similar cell attachment properties and a significantly better load bearing ability. A recent study has shown that silk fibroin scaffolds generated by creating a porous structure using sucrose and hexafluoro isopropanol leaching generates a template that is better suited for cartilage tissue engineering as compared to the traditional salt water leaching technique [37]. In another study, good chondrogenic differentiation of mesenchymal stem cells was observed when cultured on silk fibroin scaffolds when compared to type I collagen or crosslinked collagen scaffolds. This template also promoted uniform deposition of cartilaginous matrix with an increased upregulation of type II collagen and aggrecan when compared to peripheral deposition of cartilaginous matrix in the collagen scaffolds [38]. In the presence of chondrocytes and a signaling molecule like TGFβ, silk fibroin scaffolds facilitated the formation of cartilage - like tissue in vitro [39]. Chondrogenic differentiation was assessed by upregulation of cartilage specific genes and the presence of cartilage specific ECM proteins. These studies indicated that silk fibroin can serve as excellent chondrogenic and osteogenic scaffold.
Spider silk as a scaffold for tissue engineering
Spider silk requires special mention as it is different from the widely used silk worm silk. Spider silk is one of nature’s engineering triumphs, stronger on a peer-weight basis than steel. Spider silk offers higher mechanical properties than silk worm silk, but cannot be obtained in abundant amounts as the latter. The increased mechanical strength of spider silk stems from the high tensile strength of the polymer strands. In a recent study Lee et al. have shown that spider silk can be strengthened by infiltrating it with metal ions using a technique called “atomic layer deposition” [40]. Using this technique, there is good control of deposition thickness in the range of 1 picometer thereby altering mechanical property of the material at the atomic level. This proves that manipulation of the nanoscale properties can control the macro scale effectiveness of the material.
Spider silk has a great potential as a scaffold for bone tissue engineering, however, the predatory nature of the insect seriously hinders the amount of silk that can be obtained. Development of recombinant silk proteins has to an extent rekindled interest in spider silk. Amino acid sequence show the presence of multiple repeats in silk protein sequence that can be exploited to create recombinant peptides and proteins that can be made to self assemble [41]. These peptides also polymerize into β sheets like silk worm silk. Additionally, the composition of the peptides can also be altered to suit biological needs. Recently, a novel protein that combines the unique self-assembly and mechanical features of spider silks with the hydroxyapatite nucleating ability of the C-terminal domain of DMP1 (dentin matrix protein 1) [43]. The consensus sequence from spider dragline silk and the C-terminal domain of DMP1 were combined at the genetic level by synthetic gene construction. The clone was expressed and purified and the in-vitro nucleating ability of the recombinant protein was studied along with a control system without the DMP1 domain. The materials formed were characterized to confirm both silk structure and the formation of hydroxyapatite, suggesting a new approach to the formation of novel molecular – level organic-inorganic biomaterial composite systems. This hybrid system would find potential applications in bone tissue engineering [42].
Fibrin gels as Biomaterials
Fibrin is the major protein constituent of blood clots and is a significant component of the provisional extracellular matrix involved in wound healing. Fibrin and its precursor, fibrinogen are dimers of trimers, with each trimer being composed of an α, β and γ chain. Fibrin gels are generated by gelling fibrinogen in the presence of thrombin. The mechanical properties of the gel are governed by the initial concentration of the fibrinogen and also the concentration of thrombin. Additionally, the gelling conditions also vary depending on the enzyme used. Fibrin gels support the attachment of fibroblasts and also aid in nerve cell regeneration, however, they have been rarely used in hard tissue engineering primarily due to the lack of mechanical stability of these gels. However, mesenchymal cells embedded in fibrin glue and implanted subcutaneously in mice have been shown to differentiate into bone forming cells [43]. Additionally, fibrin gels embedded with mesenchymal cells and either β tricalcium phosphate or bone cement has been used successfully in bone tissue engineering [44]. Recently, fibrin gels strengthened by incorporation of calcium phosphate nanocrystals from aqueous solutions or by direct deposition of hydroxyapatite on the fibrin fibers has been shown to function as a template with microporous structures that facilitated mesenchymal cell attachment and differentiation as characterized by increased alkaline phosphatase activity and expression of bone specific marker genes. Additionally, these scaffolds promoted bone formation in a mouse calvarial defect model [45]. This again demonstrates that functionality of these scaffolds can be enhanced by nanoscale modifications.
Fibrin gels in cartilage tissue engineering
Fibrin gels have been extensively used as a base material in conjunction with various other naturally occurring and synthetic biomaterials for cartilage tissue engineering. The soft and yielding nature of the hydrogel offers the perfect mechanical properties required for cartilage tissue regeneration. One clinical study used autologous chondrocyte implantation by means of embedding the chondrocytes in fibrin scaffold. The results from this study showed that the scaffold did not elicit any graft related complications. Furthermore, the functionality was improved and a nearly normal cartilage tissue formed within 12 months post implantation [46]. Recently, PGA scaffolds containing fibrin glue was tested for its ability to form cartilage-like tissue from human septal chondrocytes. The study found that scaffolds containing fibrin glue showed increased cellular proliferation. However, GAG production was not significantly affected by the inclusion of fibrin suggesting that the fibrin component of the scaffold facilitated cell attachment and proliferation [47]. Polyurethane foams along with polymerized fibrin glue have also been shown to induce chondrocytic differentiation of intervertebral disc cells and has been suggested as a possible scaffold for treatment of invertebrate disc failure. Results demonstrated that the scaffold promoted the expression of GAG and chondrocytic ECM proteins as well as upregulation of chondrocytic markers [48].
Extracellular matrix as Biomaterials
Tissues in our body are built of different types of cells embedded within dynamic ECM gels, which are composed of many different types of proteins and sugars secreted by cells [49, 50]. The ECM comes in many different ‘flavors” that are specific to a particular cell and tissue type. To expect in vivo like cellular response it is mandatory to recreate extracellular matrix (ECM) mimicking microenvironment (Fig 4A&B). It has been realized that in addition to spatial framework, tissue specific ECM cues are essential to regenerate an organ or tissue. It is well established that ECM is dynamic and plays an instructive role in building a tissue and in regenerating it after damage, unlike the earlier notion that ECM functions as a “space filler” or a mechanical scaffold that keeps cells in the right place [51]. Three-dimensional natural extracellular matrix materials provide physiologically relevant cellular environments as they are a rich source of bioactive molecules and thus have been used extensively to engineer tissues.
Figure 4. The natural ECM matrix.
Fig 4A: SEM image of the ECM matrix synthesized by osteoblast cells. Note the cell process on the matrix.
Fig 4B: SEM image of the matrix at higher magnification: Note the thick fibrous and porous architecture of the natural ECM.
The ECM is mainly composed predominantly of hierarchically arranged collagen, laminin, fibronectin and proteoglycans in a complex topography in the nanometer range. Fibers obtained from ECM proteins are characterized by ultra fine continuous fibers, high surface- to- volume ratio, high porosity and variable pore size (Fig 4A&B). An ideal scaffold for tissue engineering, need to have interconnected highly porous structures to facilitate cellular migration and transport of nutrients and metabolic wastes to allow the formation of new tissue. Thus, ECM derived from cell lines has several advantages for tissue regeneration. However, there is currently no convenient methodology of presenting cell line derived ECM on biological scaffolds to provide it with structure and form. In a recent study Narayanan et al. have shown that the reconstituted ECM scaffolds retained the bioactivity of the intact ECM by demonstrating that the hMSCs seeded on this scaffold had differentiated into bone cells with the ability to “remodel” the cell-scaffold construct into a bone phenotype [52]. Evans et al. has recently demonstrated that ECM derived from an osteogenic cell line significantly promotes osteogenesis in embryonic stem cells when compared to ECM derived from non-bone like cell lines or type I collagen alone [53].
Basement membranes are unique extra cellular matrices that support cell adhesion and provide an environment for the cells to interact with the surroundings. The ECM is composed of specific proteins, several functional groups and growth factor reservoirs Along with many tropic agents that are responsible for cellular functions. Several cellular activities such as adhesion, proliferation, migration, differentiation and cell shape are influenced by the ECM in which the cells reside [54]. The nanotopographic features of the natural basement membrane provide the physical and chemical cues required by cells. These observations reinforce the advantage of using ‘whole’ ECM, rather than isolated ECM components in a scaffold. Thus, ECM based scaffolds provide not only physical support but also demonstrate controlled presentation of the appropriate biological cues at the nanoscale. However, the exact interplay of the different components and factors in the natural environment being as yet, unknown.
Designer Biomaterials based on the principles of biomimicry
Thus far, we have reviewed naturally occurring biomaterials having a functional role in tissue engineering. ECM proteins have the potential to function as a scaffold for hard tissue regeneration as they possess the required properties by themselves or in conjunction with other components to guide tissue regeneration. However, the major concern with these materials is that they do not possess all the required properties for hard tissue engineering. Bone tissue engineering requires scaffolds to be designed with optimal properties including strength, toughness, porosity, controlled rate of degradation, nontoxic degradation products, minimal inflammatory response, moldability, osteoconductivity and osteoinductivity. Biomimicry is an emerging discipline that studies nature’s best ideas and then imitates these designs and principles. This principle is particularly useful for the development of nanoscale materials for tissue engineering. Using the principles of biomimicry, several published reports have demonstrated the synthesis of peptide based “designer scaffolds” with biologically active peptide sequences in order to create instructive synthetic extracellular matrices.
Biomaterials with cell-instructive properties
A new class of biomaterials is currently being designed to mimic cellular and extracellular protein domains, with emphasis on presentation of signals in a controlled spatio-temporal manner. He et al. [1] have investigated the effect of grafting RGD peptide which would provide sites for cell attachment to the substrate and the BMP peptide which would interact with the type I and type II transmembrane receptors when grafted to the inert PLEOF hydrogel (Poly- lactide-co-ethylene oxide-co-fumarate) substrate. The density of RGD and BMP peptides on the surface did influence the differentiation and mineralization of bone marrow stromal cells. They demonstrate that the ligands act synergistically to enhance osteogenic differentiation when the RGD and BMP peptide density on the hydrogel surface was 1.62 and 5.2 pmol/cm2 respectively. Thus, surface nanoscale features were found to steer osteoblast adhesion and influence cell signaling.
Biomaterials with nanotopographical features
It is clear from the literature that nanotopographical features significantly alter cell behavior. Nanotopographical features namely pores, ridges, groves, fibers, nodes and combinations of these features influence cellular adhesion, morphology, proliferation, endocytic activity, motility and gene expression of various cell types [54]. Various nanotopographical features have been created and used as new generation tissue engineering scaffolds. Studies have demonstrated that nanostructured materials with cell favorable surface properties may promote greater amounts of specific protein interactions to efficiently stimulate new bone growth compared to conventional materials [2, 55, 56]. Material surface properties mediate adsorption of specific proteins such as fibronectin, vitronectin and laminin adsorption necessary for cell adherence and thus regulate cell behavior and regulate tissue regeneration [57]. Nanophase ceramics especially nano-hydroxyapatite have been extensively used as a biomaterial to promote mineralization. Due to their grain sizes and high surface fraction of grain boundaries, osteoblast function is increased. Studies have shown that nanophase HA about 67 nm grain size significantly enhanced osteoblast adhesion and inhibited fibroblast adhesion when compared with conventional HA particles of 179 nm grain size [58]. The advantage of using nano-hydroxyapatite particles as a biomaterial is because they have similar mechanical properties to bone and also functioned as an osteo-conductive coating.
Although biomaterials are typically used to guide the behavior of cells transplanted with the material or cells in the tissue into which the material is implanted, it has also become apparent that biomaterials can be designed to manipulate specific cell populations that reside in the host at a significant distance from the implant site [59]. Thus, substrate micro-topography has been shown to influence cell adhesion and contact guidance as these dimensions are comparable with cellular dimensions of 10–30 μm [60], while surface nanotopography mainly influences the orientation of cells on surfaces [61].
Biomaterials based on self-assembled chemistries
The natural environment of cells extracellular matrix is extremely complex and therefore hard to recreate [50, 62]. In nature, sophisticated functional materials are created through hierarchical self-assembly of simple nanoscale motifs [63]. Self-assembled biomolecular structures are particularly useful in tissue engineering due to their versatile chemistry, molecular recognition properties and biocompatibility [64, 65]. Natural peptide and protein-based self-assembling systems are found extensively throughout biology [66, 67]. These natural systems offer inspiration in the effort to engineer novel assemblies through de -novo design. Molecular self-assembly is emerging as a viable “bottom-up” approach for fabricating nanostructures for tissue engineering. Self assembling peptides can be designed so that their molecular and nanoscale structures are compatible for cell culture and enhance the differentiation of precursor cells.
With a growing interest and demand for materials programmed at the nano-to-micrometer scales, biomolecular self-assembly is attracting considerable attention as an efficient tool for building new supramolecular architectures and composites [68, 69]. Self-assembly consists of the spontaneous organization of molecules under thermodynamic equilibrium conditions into structurally stable arrangements by the driving force of non-covalent interactions, including hydrogen bonds, ionic bonds, electrostatic interaction, van der Waals interaction.
Self-assembling natural materials such as DNA have inspired the development of new types of nanomaterial with precisely defined structures [2, 76] based on the highly regulated base-pairing property of DNA. Using self-assembly process, it is possible to create a biologically inspired 3-D scaffold with self-assembly based biomimetic features suitable for reconstructing 3-D bone and cartilage. Recently, self-assembly of DNA base pairs (Guanine–Cytosine) in aqueous solution have been reported for bone tissue engineering. Tailorable amino acid and peptide side chains (such as lysine, RGD and peptide Lys-Arginine-Serine-Arginine) were incorporated to promote osteoblast adhesion and are excellent mineralization templates were incorporated [2, 76].
The power of self-assembling protein and peptide scaffolds in tissue engineering is highlighted below. Nanostructured self-assembled chemistries have been used for cartilage tissue engineering. Kisiday et al. designed a self-assembling peptide KLD-12 (Lys-Leu-Asp) hydrogel for cartilage repair [77]. The repeating sequence arranged as a β-sheet promoting self-assembly. The alternating hydrophilic and hydrophobic domains facilitated the β-sheet formation. This 3-D scaffold when used for chondrocyte encapsulation, promoted chondrocyte differentiation and synthesis of a cartilage-like ECM matrix.
RAD16-I is another self-assembling peptide that could form nanofibres. Due to their nanopore size and biomechanical properties RAD16-I has been frequently used to culture cells in the laboratory [64, 78]. Osteo-induced mouse embryonic fibroblast 3D cultures were able to develop mineralized matrix when grown on this scaffold. Further, the cells synthesized collagen type I and upregulated the expression of the transcription factor Runx2, suggesting that the cells acquired an osteoblast-like phenotype [79]. Further, RAD16-I applied in small bone defects (3 mm) in mice calvaria promoted bone regeneration [80].
Nature has provided a diversity of peptide motifs that can serve as building blocks capable of self-assembling into macroscale materials with pre-defined structure and properties [81, 82]. Templates for tissue engineering have been built with three major protein-folding motifs: α-helices [70], β-pleated structures [52, 71–73] and collagen triple helices [74]. Exploiting the properties of these motifs has been demonstrated as a new route for novel material synthesis[83, 84]. One of the most studied motifs is leucine zipper (coiled-coil motif). Petka et al. [85] designed a tri-block artificial protein with an alanylglycine-rich coil between two terminal leucine zipper motifs. This protein forms a stable and reversible hydrogel under physiological condition. Such strategy is particularly interesting because multiple bioactive motifs can be potentially incorporated into the construct and the leucine zipper self-assembling properties will be unaffected if it is designed appropriately. Coiled coils are characterized by heptad repeated units designated as abcdefg, where the a and d positions are occupied by hydrophobic residues such as leucine and the e and g positions are occupied by charged residues [86, 87]. These domains fold into α-helices and hydrophobic interactions are responsible for formation of oligomeric clusters. These hydrogels are reversible in response to high pH or high temperature, where the leucine zipper domains are denatured. Using recombinant DNA technology several domains necessary for the formation of a functional tissue can be engineered with leucine zipper motives[88–93]. Thus, novel materials can be synthesized by emulating the self-assembly strategy of nature. Self-assembly would allow controlled organization of the organic/inorganic interface based on molecular recognition elements, resulting in hierarchical organization with desirable properties at multiple length scales. Development of such templates is necessary for the development of hierarchically structured materials like bone and dentin.
Using the principle of biomimicry, we demonstrated a novel strategy for synthesizing de novo cell interactive functional materials, which can also promote template-driven mineralization of crystalline apatites for hard tissue regeneration. We show that self-assembling leucine zipper polypeptide (LZ) can be designed to accommodate motifs from the hydroxyapatite nucleating domain and cell adhesive motifs of dentin matrix protein 1 (DMP1)[94] (Fig. 5A). Although, DMP1 was initially isolated from the dentin matrix and was thought to be unique to dentin and named accordingly, it has been found to be present in all mineralized tissues of the vertebrate system. The C-terminal polypeptide of DMP1 contains the HAP nucleating domain as well as an RGD motif for cell-adhesion, which makes it a highly desirable polypeptide for hard tissue regeneration. Cells seeded on LZ-DMP1 coated substrate promoted integrin clustering, which in turn recruited actin filaments and directed focal adhesion complex formation (Fig B, C&D). Furthermore, this self-assembled biomaterial can induce and assemble mineral formation at the nanoscale level. Thus, integration of biological self-organization and inorganic assembly components is important to synthesize complex materials that exhibit order from the molecular to the macroscopic scale.
Figure 5. Leucine zipper-DMP1 hydrogel promotes cell-matrix dialogue.

Fig 5A is a schematic representation of the leucine zipper-DMP1 chimeric protein.
Fig 5B is a light microscopy image showing cell attachment on a LZ-DMP1 substrate and less on the glass plate (control).
Fig 5C & D shows actin stress fibers and cell-cell communication when coated on LZ-DMP1 substrate.
Fig 5E is a scanning electron micrograph of a leucine zipper hydrogel imaged using a field emission scanning electron microscope. The image shows the porous nature of the hydrogel.
Inspired by nature, researchers have sought to develop hydrogels that can mimic some of the key structural and biochemical functions of the natural extracellular milieu (Fig 5E). Hydrogels are 3-dimensional hydrophilic polymer networks that store large amounts of water. One common feature of different hydrogel systems is the presence of cross-links, either chemical or physical. Based on their composition, hydrogels can be categorized as synthetic polymer based hydrogels, bio-polymer based hydrogels or hybrid hydrogel systems, which contain both synthetic and biomolecular moieties [75]. “Smart hydrogels” are currently being developed that try to mimic the temporal and spatial variability of the cells natural environment and interact with surrounding tissues by biomolecular recognition.
Biomimetic gels containing building blocks that are sensitive to cell-secreted enzymes [95] and can break down proteins with the aim of mimicking the dynamic character of the ECM during remodeling would be a good design to be used for bone tissue engineering. Polymer gels, PEG based hydrogels have been rendered chemically degradable through hydrolytic breakdown of ester bonds and have been developed with cleavage sites for cell-secreted matrix metalloproteinase’s or plasmin. This cell-regulatable breakdown of the matrix allows cell migration and proliferation in a manner determined by the cells [96].
Using elegant approaches based on supramolecular chemistry, peptide amphiphiles (PA) have been synthesized and used for bone and cartilage regeneration [97]. Peptide amphiphiles can self-assemble from aqueous media into supramolecular nanofibres of high aspect ratio [98]. These molecules consist of a peptide segment covalently bonded to a more hydrophobic segment such as an alkyl tail [98]. Peptide amphiphiles are charged molecules that self-assemble in the presence of ions into cylindrical nanofibres, which are formed by hydrogen bonding among peptide segments into β-sheets and hydrophobic collapse of their alkyl segments. An added advantage of PAs is that they can have a terminal bio-signaling peptide, which upon self-assembly becomes exposed at very high densities on the surface of the nanofibers. In a recent study Shah et al. have designed and evaluated a unique self-assembling PA molecule which includes a TGF-β binding domain for use in articular cartilage regeneration [75]. Studies have shown that these nanofibers can also direct mineralization of hydroxyapatite in which the crystallographic c-axes of hydroxyapatite are aligned with the long axes of the fibers.
Diblock copolypeptide amphiphiles containing charged and hydrophobic segments were synthesized by Nowak et al.[99] and these protein gels showed high thermal stability and retain their mechanical strength up to temperatures of about 90ºC. They determined that gelation depends not only on the amphiphilic nature of the polypeptides, but also on chain conformations-α-helix, β-strand or random coil. Thus, self-assembly process is a promising technique that could create a biologically inspired 3-D scaffold suitable for bone and cartilage repair or regeneration [100].
Dinca et al [65] have demonstrated a new method for the controlled self-assembly of peptides in three dimensions. The method is based on biotin-avidin and thiol chemistry and ORMOCER (a photostructurable organic-inorganic hybrid) is used as a substrate. The advantage is thiol functionalized biomolecules can be easily produced in the laboratory scale and can even be commercially available, so this method should be applicable not only to peptides, but to other self-assembling biomolecules as well. Dinca et al [65] further suggested that a combination of larger scaffolds with well-defined biodegradable peptide supports in a “scaffold on scaffold” format could possibly be used as a support to allow the directed growth of several types into ordered arrays of functional biological units. This methodology would be particularly applicable in tissue engineering. Peptide networks could form excellent cell supports in the form of injectable hydrogels [101].
To achieve in vivo like cellular response it is mandatory to recreate extracellular matrix mimicking microenvironment. In human tissues, cells often grow in a 3-D network. This architecture permits cells to interact with both their surroundings and each other in a way fitting to their environment. 3-D cell culture scaffolds are a better representation of the natural environment experienced by the cells in the living organism (Fig 3A & B). Therefore, mimicking natural conditions allows for intercellular interactions with more realistic biochemical and physiological responses. Currently, in most of the published studies cells are grown on flat surfaces, this creates an artificial and unnatural environment as it does not resemble the in-vivo architecture where cells flourish at their best. Polypeptides can be made to self-assemble thereby producing a 3D system with all the required characteristics for tissue engineering. Self-assembling diphenylalanine derivative hydrogel showed many benefits for 3D cell culture. Living cells suspended within the dispersion medium were easily immobilized with addition of the gelling agent. Further, immobilization of suspended cells allowed for a 3D dispersion of cells throughout the gel, permitting the cells to adopt a 3D structure rather then the elongated conformation seen in surface cultures [102]. Fmoc peptide hydrogel was also used to culture chondrocytes and measurable growth was observed after 7 days [103]. Thus the benefit of using hydrogels for 3D cell culture has important relevance to bone and tissue engineering.
The most challenging, but perhaps the ultimate design of a biomaterial for hard tissue engineering is to create multi-component and multifunctional material that affects tissue regeneration on multiple levels. Such a biomaterial should enhance cell viability, promote mobilization of endogenous cells involved in repair, deliver diffusible cytokines and display regulatory proteins to stimulate tissue-specific differentiation. Additionally, the material should posses the ability to be modified at the nanoscale to bring out the desired effect. The overall goal is to achieve mechanical integrity and robust hard tissue repair with biomaterials that are able to respond appropriately to surrounding cues and physical forces similar to those encountered in healthy mineralized tissues. Protein-based nanostructured templates possess self-assembling characteristics and chemical versatility for tissue engineering of bones and teeth and may provide what we have so long sought: “a Prometheus for modern times”.
Acknowledgments
We gratefully acknowledge support from National Institutes of Health Grant DE 11657 and DE 16533. The authors would like to thank Mr. Ari Salinger for compiling the published papers in this area.
Biographies

Anne George received her Ph.D. in Physical Chemistry from Madras University, India in 1983. She then did her Post-Doctoral work with Dr. Arthur Veis at Northwestern University on determining the structure of type I collagen in-solution using Fourier Transform Infrared Spectroscopy. She joined as an Assistant Professor in 1993 at Northwestern University where she started work on the cloning of the dentin matrix proteins. She was instrumental in identifying the family of dentin matrix proteins from the rat odontoblasts. She then moved to the University of Illinois at Chicago in 1998 as an Associate Professor and became a full Professor in 2003 and continued her work on noncollagenous proteins and their role in biomineralization. She is now an Allan G. Brodie endowed professor at the University of Illinois at Chicago and head of the Brodie Tooth Development and Regenerative Medicine Research laboratory. Her current interest is in hard tissue engineering with designer peptide scaffold.

Sriram Ravindran received his B. Tech degree in chemical engineering in 2001 from India and then a Ph.D in Bioengineering in 2005 from the University of Illinois at Chicago. In 2006 he joined the Brodie Tooth Development Genetics and Regenerative Medicine Research Laboratory as a Research Associate. His research interests include identification and functional analysis of the proteins involved in bone and teeth mineralization and in designing bioploymeric scaffolds for hard tissue engineering. He has published papers in the area of understanding the biology of mineralized matrix formation and synthesis of scaffolds for tissue engineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He X, Ma J, Jabbari E. Langmuir. 2008;24:12508–12516. doi: 10.1021/la802447v. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Rodriguez J, Raez J, Myles AJ, Fenniri H, Webster TJ. Nanotechnology. 2009;20:175101. doi: 10.1088/0957-4484/20/17/175101. [DOI] [PubMed] [Google Scholar]
- 3.He X, Jabbari E. Biomacromolecules. 2007;8:780–792. doi: 10.1021/bm060671a. [DOI] [PubMed] [Google Scholar]
- 4.Ortiz C, Boyce MC. Science. 2008;319:1053–1054. doi: 10.1126/science.1154295. [DOI] [PubMed] [Google Scholar]
- 5.Fratzl P, Barth FG. Nature. 2009;462:442–448. doi: 10.1038/nature08603. [DOI] [PubMed] [Google Scholar]
- 6.Fratzl P. J R Soc Interface. 2007;4:637–642. doi: 10.1098/rsif.2007.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aizenberg J, Weaver JC, Thanawala MS, Sundar VC, Morse DE, Fratzl P. Science. 2005;309:275–278. doi: 10.1126/science.1112255. [DOI] [PubMed] [Google Scholar]
- 8.Veis A. Science. 2005;307:1419–1420. doi: 10.1126/science.1109440. [DOI] [PubMed] [Google Scholar]
- 9.George A, Veis A. Chem Rev. 2008;108:4670–4693. doi: 10.1021/cr0782729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George A, Sabsay B, Simonian PA, Veis A. J Biol Chem. 1993;268:12624–12630. [PubMed] [Google Scholar]
- 11.Osteoporos Int. 2003;14(Suppl 5):S16–20. doi: 10.1007/s00198-003-1468-2. discussion S20–11. [DOI] [PubMed] [Google Scholar]
- 12.Boskey AL, Young MF, Kilts T, Verdelis K. Cells Tissues Organs. 2005;181:144–153. doi: 10.1159/000091376. [DOI] [PubMed] [Google Scholar]
- 13.Groeneveld EH, van den Bergh JP, Holzmann P, ten Bruggenkate CM, Tuinzing DB, Burger EH. J Biomed Mater Res. 1999;48:393–402. doi: 10.1002/(sici)1097-4636(1999)48:4<393::aid-jbm1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien FJ, Harley BA, Yannas IV, Gibson LJ. Biomaterials. 2005;26:433–441. doi: 10.1016/j.biomaterials.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 15.Dickson G, Buchanan F, Marsh D, Harkin-Jones E, Little U, McCaigue M. Technol Health Care. 2007;15:57–67. [PubMed] [Google Scholar]
- 16.Gonen-Wadmany M, Oss-Ronen L, Seliktar D. Biomaterials. 2007;28:3876–3886. doi: 10.1016/j.biomaterials.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno S, Glowacki J. Biomaterials. 1996;17:1819–1825. doi: 10.1016/0142-9612(96)00041-5. [DOI] [PubMed] [Google Scholar]
- 18.Allemann F, Mizuno S, Eid K, Yates KE, Zaleske D, Glowacki J. J Biomed Mater Res. 2001;55:13–19. doi: 10.1002/1097-4636(200104)55:1<13::aid-jbm20>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 19.Stone KR, Steadman JR, Rodkey WG, Li ST. J Bone Joint Surg Am. 1997;79:1770–1777. doi: 10.2106/00004623-199712000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Pabbruwe MB, Kafienah W, Tarlton JF, Mistry S, Fox DJ, Hollander AP. Biomaterials. 31:2583–2591. doi: 10.1016/j.biomaterials.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Amos JR, Li S, Yost M, Phloen H, Potts JD. Biorheology. 2009;46:439–450. doi: 10.3233/BIR-2009-0553. [DOI] [PubMed] [Google Scholar]
- 22.Kawazoe N, Inoue C, Tateishi T, Chen G. Biotechnol Prog. 2009 doi: 10.1002/btpr.375. [DOI] [PubMed] [Google Scholar]
- 23.Pek YS, Gao S, Arshad MS, Leck KJ, Ying JY. Biomaterials. 2008;29:4300–4305. doi: 10.1016/j.biomaterials.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 24.Geiger M, Li RH, Friess W. Adv Drug Deliv Rev. 2003;55:1613–1629. doi: 10.1016/j.addr.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Ravindran S, Song Y, George A. Tissue Eng Part A. 16:327–342. doi: 10.1089/ten.tea.2009.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KW, Thomas RL, Lee C, Park HJ. J Food Prot. 2003;66:1495–1498. doi: 10.4315/0362-028x-66.8.1495. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Jayarama Reddy V, Wong SY, Li X, Su B, Ramakrishna S, Lim CT. Tissue Eng Part A. doi: 10.1089/ten.TEA.2009.0221. [DOI] [PubMed] [Google Scholar]
- 28.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 29.Gellynck K, Verdonk PC, Van Nimmen E, Almqvist KF, Gheysens T, Schoukens G, Van Langenhove L, Kiekens P, Mertens J, Verbruggen G. J Mater Sci Mater Med. 2008;19:3399–3409. doi: 10.1007/s10856-008-3474-6. [DOI] [PubMed] [Google Scholar]
- 30.Min BM, Lee G, Kim SH, Nam YS, Lee TS, Park WH. Biomaterials. 2004;25:1289–1297. doi: 10.1016/j.biomaterials.2003.08.045. [DOI] [PubMed] [Google Scholar]
- 31.Fini M, Motta A, Torricelli P, Giavaresi G, Nicoli Aldini N, Tschon M, Giardino R, Migliaresi C. Biomaterials. 2005;26:3527–3536. doi: 10.1016/j.biomaterials.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 32.Meinel AJ, Kubow KE, Klotzsch E, Garcia-Fuentes M, Smith ML, Vogel V, Merkle HP, Meinel L. Biomaterials. 2009;30:3058–3067. doi: 10.1016/j.biomaterials.2009.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meinel L, Betz O, Fajardo R, Hofmann S, Nazarian A, Cory E, Hilbe M, McCool J, Langer R, Vunjak-Novakovic G, Merkle HP, Rechenberg B, Kaplan DL, Kirker-Head C. Bone. 2006;39:922–931. doi: 10.1016/j.bone.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Kim HJ, Kim UJ, Kim HS, Li C, Wada M, Leisk GG, Kaplan DL. Bone. 2008;42:1226–1234. doi: 10.1016/j.bone.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Vepari C, Jin HJ, Kim HJ, Kaplan DL. Biomaterials. 2006;27:3115–3124. doi: 10.1016/j.biomaterials.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Kim JY, Choi JY, Jeong JH, Jang ES, Kim AS, Kim SG, Kweon HY, Jo YY, Yeo JH. BMB Rep. 43:52–56. doi: 10.5483/bmbrep.2010.43.1.052. [DOI] [PubMed] [Google Scholar]
- 37.Makaya K, Terada S, Ohgo K, Asakura T. J Biosci Bioeng. 2009;108:68–75. doi: 10.1016/j.jbiosc.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Hofmann S, Knecht S, Langer R, Kaplan DL, Vunjak-Novakovic G, Merkle HP, Meinel L. Tissue Eng. 2006;12:2729–2738. doi: 10.1089/ten.2006.12.2729. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Blasioli DJ, Kim HJ, Kim HS, Kaplan DL. Biomaterials. 2006;27:4434–4442. doi: 10.1016/j.biomaterials.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 40.Lee SM, Pippel E, Gosele U, Dresbach C, Qin Y, Chandran CV, Brauniger T, Hause G, Knez M. Science. 2009;324:488–492. doi: 10.1126/science.1168162. [DOI] [PubMed] [Google Scholar]
- 41.Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Wong C, George A, Kaplan DL. Biomaterials. 2007;28:2358–2367. doi: 10.1016/j.biomaterials.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Isogai N, Landis WJ, Mori R, Gotoh Y, Gerstenfeld LC, Upton J, Vacanti JP. Plast Reconstr Surg. 2000;105:953–963. doi: 10.1097/00006534-200003000-00019. [DOI] [PubMed] [Google Scholar]
- 44.Yamada Y, Boo JS, Ozawa R, Nagasaka T, Okazaki Y, Hata K, Ueda M. J Craniomaxillofac Surg. 2003;31:27–33. doi: 10.1016/s1010-5182(02)00143-9. [DOI] [PubMed] [Google Scholar]
- 45.Osathanon T, Linnes ML, Rajachar RM, Ratner BD, Somerman MJ, Giachelli CM. Biomaterials. 2008;29:4091–4099. doi: 10.1016/j.biomaterials.2008.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim MK, Choi SW, Kim SR, Oh IS, Won MH. Knee Surg Sports Traumatol. Arthrosc. 2009 doi: 10.1007/s00167-009-0905-y. [DOI] [PubMed] [Google Scholar]
- 47.Watson D, Sage A, Chang AA, Schumacher BL, Sah RL. Am J Rhinol Allergy. 24:e19–22. doi: 10.2500/ajra.2010.24.3433. [DOI] [PubMed] [Google Scholar]
- 48.Mauth C, Bono E, Haas S, Paesold G, Wiese H, Maier G, Boos N, Graf-Hausner U. Eur Cell Mater. 2009;18:27–38. doi: 10.22203/ecm.v018a03. discussion 38–29. [DOI] [PubMed] [Google Scholar]
- 49.Lutolf MP. Integr Biol (Camb) 2009;1:235–241. doi: 10.1039/b902243k. [DOI] [PubMed] [Google Scholar]
- 50.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scadden DT. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 52.Narayanan K, Leck KJ, Gao S, Wan AC. Biomaterials. 2009;30:4309–4317. doi: 10.1016/j.biomaterials.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 53.Evans ND, Gentleman E, Chen X, Roberts CJ, Polak JM, Stevens MM. Biomaterials. 31:3244–3252. doi: 10.1016/j.biomaterials.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 54.Kumbar SG, James R, Nukavarapu SP, Laurencin CT. Biomed Mater. 2008;3:034002. doi: 10.1088/1748-6041/3/3/034002. [DOI] [PubMed] [Google Scholar]
- 55.Webster TJ, Ahn ES. Adv Biochem Eng Biotechnol. 2007;103:275–308. doi: 10.1007/10_021. [DOI] [PubMed] [Google Scholar]
- 56.Webster TJ, Ergun C, Doremus RH, Lanford WA. J Biomed Mater Res A. 2003;67:975–980. doi: 10.1002/jbm.a.10160. [DOI] [PubMed] [Google Scholar]
- 57.Tran N, Webster TJ. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:336–351. doi: 10.1002/wnan.23. [DOI] [PubMed] [Google Scholar]
- 58.Webster TJ, Ergun C, Doremus RH, Siegel RW, Bizios R. J Biomed Mater Res. 2000;51:475–483. doi: 10.1002/1097-4636(20000905)51:3<475::aid-jbm23>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 59.Huebsch N, Mooney DJ. Nature. 2009;462:426–432. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Curtis A, Wilkinson C. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 61.Justesen J, Lorentzen M, Andersen LK, Hansen O, Chevallier J, Modin C, Fuchtbauer A, Foss M, Besenbacher F, Duch M, Pedersen FS. J Biomed Mater Res A. 2009;89:885–894. doi: 10.1002/jbm.a.32032. [DOI] [PubMed] [Google Scholar]
- 62.Khetan S, Burdick J. J Vis Exp. 2009 doi: 10.3791/1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitesides GM, Grzybowski B. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S, Gelain F, Zhao X. Semin Cancer Biol. 2005;15:413–420. doi: 10.1016/j.semcancer.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Dinca V, Kasotakis E, Catherine J, Mourka A, Ranella A, Ovsianikov A, Chichkov BN, Farsari M, Mitraki A, Fotakis C. Nano Lett. 2008;8:538–543. doi: 10.1021/nl072798r. [DOI] [PubMed] [Google Scholar]
- 66.Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Proc Natl Acad Sci U S A. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stephen M. Chem Commun (Camb) 2004:1–4. doi: 10.1039/b310010n. [DOI] [PubMed] [Google Scholar]
- 68.Ryadnov MG, Bella A, Timson S, Woolfson DN. J Am Chem Soc. 2009;131:13240–13241. doi: 10.1021/ja905539h. [DOI] [PubMed] [Google Scholar]
- 69.Gribbon C, Channon KJ, Zhang W, Banwell EF, Bromley EH, Chaudhuri JB, Oreffo RO, Woolfson DN. Biochemistry. 2008;47:10365–10371. doi: 10.1021/bi801072s. [DOI] [PubMed] [Google Scholar]
- 70.Woolfson DN, Ryadnov MG. Curr Opin Chem Biol. 2006;10:559–567. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 71.Branco MC, Schneider JP. Acta Biomater. 2009;5:817–831. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Biomaterials. 2009;30:1339–1347. doi: 10.1016/j.biomaterials.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rudra JS, Tian YF, Jung JP, Collier JH. Proc Natl Acad Sci U S A. 107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gauba V, Hartgerink JD. J Am Chem Soc. 2008;130:7509–7515. doi: 10.1021/ja801670v. [DOI] [PubMed] [Google Scholar]
- 75.Xu C, Kopecek J. Pharm Res. 2008;25:674–682. doi: 10.1007/s11095-007-9343-z. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Chen Y, Rodriguez J, Fenniri H, Webster TJ. Int J Nanomedicine. 2008;3:323–333. doi: 10.2147/ijn.s2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kisiday JD, Lee JH, Siparsky PN, Frisbie DD, Flannery CR, Sandy JD, Grodzinsky AJ. Ann Biomed Eng. 2009;37:1368–1375. doi: 10.1007/s10439-009-9699-9. [DOI] [PubMed] [Google Scholar]
- 78.Zhang S. Adv Cancer Res. 2008;99:335–362. doi: 10.1016/S0065-230X(07)99005-3. [DOI] [PubMed] [Google Scholar]
- 79.Garreta E, Genove E, Borros S, Semino CE. Tissue Eng. 2006;12:2215–2227. doi: 10.1089/ten.2006.12.2215. [DOI] [PubMed] [Google Scholar]
- 80.Misawa H, Kobayashi N, Soto-Gutierrez A, Chen Y, Yoshida A, Rivas-Carrillo JD, Navarro-Alvarez N, Tanaka K, Miki A, Takei J, Ueda T, Tanaka M, Endo H, Tanaka N, Ozaki T. Cell Transplant. 2006;15:903–910. doi: 10.3727/000000006783981369. [DOI] [PubMed] [Google Scholar]
- 81.Rexeisen EL, Fan W, Pangburn TO, Taribagil RR, Bates FS, Lodge TP, Tsapatsis M, Kokkoli E. Langmuir. 2009 [Google Scholar]
- 82.Sun J, Zheng Q. J Huazhong Univ Sci Technolog Med Sci. 2009;29:512–516. doi: 10.1007/s11596-009-0424-6. [DOI] [PubMed] [Google Scholar]
- 83.Hirschmann RF, Nicolaou KC, Angeles AR, Chen JS, Smith AB., 3rd Acc Chem Res. 2009;42:1511–1520. doi: 10.1021/ar900020x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kokubun K, Kashiwagi K, Yoshinari M, Inoue T, Shiba K. Biomacromolecules. 2008;9:3098–3105. doi: 10.1021/bm800638z. [DOI] [PubMed] [Google Scholar]
- 85.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 86.Stevens MM, Allen S, Davies MC, Roberts CJ, Sakata JK, Tendler SJ, Tirrell DA, Williams PM. Biomacromolecules. 2005;6:1266–1271. doi: 10.1021/bm049369x. [DOI] [PubMed] [Google Scholar]
- 87.Stevens MM, Allen S, Sakata JK, Davies MC, Roberts CJ, Tendler SJ, Tirrell DA, Williams PM. Langmuir. 2004;20:7747–7752. doi: 10.1021/la030440e. [DOI] [PubMed] [Google Scholar]
- 88.Liu B, Lewis AK, Shen W. Biomacromolecules. 2009 doi: 10.1021/bm900908g. [DOI] [PubMed] [Google Scholar]
- 89.Fischer SE, Mi L, Mao HQ, Harden JL. Biomacromolecules. 2009;10:2408–2417. doi: 10.1021/bm900202z. [DOI] [PubMed] [Google Scholar]
- 90.Wheeldon IR, Campbell E, Banta S. J Mol Biol. 2009;392:129–142. doi: 10.1016/j.jmb.2009.06.075. [DOI] [PubMed] [Google Scholar]
- 91.Wheeldon IR, Gallaway JW, Barton SC, Banta S. Proc Natl Acad Sci U S A. 2008;105:15275–15280. doi: 10.1073/pnas.0805249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fischer SE, Liu X, Mao HQ, Harden JL. Biomaterials. 2007;28:3325–3337. doi: 10.1016/j.biomaterials.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 93.Ryadnov MG, Papapostolou D, Woolfson DN. Methods Mol Biol. 2008;474:35–51. doi: 10.1007/978-1-59745-480-3_3. [DOI] [PubMed] [Google Scholar]
- 94.Gajjeraman S, He G, Narayanan K, George A. Adv Funct Mater. 2008;18:3972–3980. doi: 10.1002/adfm.200801215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 96.Lin CC, Anseth KS. Pharm Res. 2009;26:631–643. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui H, Webber MJ, Stupp SI. Biopolymers. 94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nowak AP, Breedveld V, Pakstis L, Ozbas B, Pine DJ, Pochan D, Deming TJ. Nature. 2002;417:424–428. doi: 10.1038/417424a. [DOI] [PubMed] [Google Scholar]
- 100.Semino CE. J Dent Res. 2008;87:606–616. doi: 10.1177/154405910808700710. [DOI] [PubMed] [Google Scholar]
- 101.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc Natl Acad Sci U S A. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liebmann T, Rydholm S, Akpe V, Brismar H. BMC Biotechnol. 2007;7:88. doi: 10.1186/1472-6750-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, Ulijn RV. Acta Biomater. 2009;5:934–943. doi: 10.1016/j.actbio.2009.01.006. [DOI] [PubMed] [Google Scholar]